Abstract
Age-related hearing loss (ARHL) is the most prevalent sensory impairment in the elderly. However, the pathogenesis of ARHL remains unclear. This study was aimed to explore the potential inflammation-related genes of ARHL and suggest novel therapeutic targets for this condition. Initially, a total of 105 Inflammatory related differentially expressed genes (IRDEGs) were obtained by overlapping the differentially expressed genes from the GSE49522 and GSE49543 datasets with Inflammatory related genes. The IRDEGs were mainly enriched in MAPK, PI3K-Akt, Hippo and JAK-STAT pathways by analysis of Gene Ontology and Kyoto Encyclopedia of Genes and Genomes. We then identified 10 key IRDEGs including Alox5ap, Chil1, Clec7a, Dysf, Fcgr3, etc. using Least absolute shrinkage and selection operator regression analysis and converted them into human genes. The ROC curve indicated that Alox5ap expression presented a high accuracy in distinguishing between different groups. By CIBERSORT algorithm, 8 humanized key IRDEGs were correlated with the infiltration abundance of 3 immune cells. Finally, it showed that the Alox5ap expression was significantly more effective compared to other variables in the diagnostic model of ARHL. This study suggests that inflammation might play a role in the development of ARHL, providing a deeper understanding of the underlying causes of this disease.
Similar content being viewed by others
Introduction
Age-related hearing loss (ARHL), also referred to as presbycusis, is the most common chronic sensory deficit among older individuals. It is described as a gradual, bilateral, symmetrical sensorineural hearing loss that occurs with age, particularly affecting higher frequencies1. Emerging evidence suggests that individuals with ARHL have more chance to experience social isolation, depression, and cognitive decline2,3. Besides the health and social burden, ARHL also causes a remarkable economic cost. Due to the projected rise in life expectancy, the yearly expenses linked to ARHL are expected to reach approximately $60 billion by the year 20304. Given the enormous economic and social burden resulting from ARHL, there is a pressing requirement for successful interventions to alleviate auditory decline associated with aging.
By 2017, it was estimated that age-related and other acquired hearing loss affected approximately 1.3 billion humans worldwide, while the population with hearing loss due to other congenital anomalies reached 25 million5. However, compared with hereditary hearing loss, there is no clear explanation for the etiology and pathogenesis of ARHL. Knowledge of the potential molecular mechanisms of this condition could help explore promising therapeutic interventions. Genetic and environmental factors usually jointly participate in the occurrence and development of this disease. Many studies of the pathophysiological mechanisms underlying ARHL focused on mitochondrial dysfunction6, oxidative stress7, and apoptosis8,9,10, while there has been little attention paid to inflammation and immune responses in aging inner ear. Consequently, our research primarily aimed to thoroughly identify inflammation-related genes associated with ARHL and comprehensively understand the relevant biological processes and pathways.
For an extended period of time, the cochlea was considered an “immune-privileged” organ because of the presence of the blood-labyrinth barrier (BLB)11. Recent studies have uncovered the existence of resident macrophages in various parts of the cochlea, such as the lateral wall, basilar membrane, and spiral limbus12. These macrophages can respond to various harmful stimuli, such as mitochondrial damage, ischemia, noise exposure, and surgical stress13,14,15. Evidence suggests that chronic, low-level inflammation may be associated with age-related hearing issues16. Throughout the aging process, there is a persistent, non-infectious, low-grade inflammation that contributes to the development of age-related illnesses17. ARHL has been linked to plenty comorbid conditions, including frailty, dementia, Alzheimer’s disease, and type II diabetes1,18,19,20,21. A common characteristic of these conditions is the presence of chronic inflammation in the affected organs22. Several recent reviews evaluate the possible link between inflammation and ARHL systematically, suggesting that the immune response and inflammation are significant factors in the onset and development of ARHL23,24. Cheng et al. performed comprehensive bioinformatic and machine learning methods to identify the critical genes of ARHL and found that these genes were mainly associated with inflammation, immune response, and infection25. Increasing evidence indicates that inflammaging serves as a crucial pathophysiological mechanism underlying ARHL26. Recent studies reveal the effectiveness of anti-inflammatory treatment in mouse model of presbycusis27,28,29. However, Inflammatory related genes (IRGs) of ARHL remain largely unknown and require additional investigation, which would identify potential biomarkers for ARHL treatment.
In this study, we acquired data from the Gene Expression Omnibus (GEO) database, specifically GSE49522, GSE49543, and GSE6045. Subsequently, we identified the Inflammatory related differentially expressed genes (IRDEGs) linked to ARHL and performed enrichment analysis on IRDEGs using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG). Furthermore, we utilized Least absolute shrinkage and selection operator (LASSO) regression analysis to pinpoint key IRDEGs and converted them into human genes. We then used CIBERSORT to assess changes in immune infiltration between ARHL group and controls. The MCPcounter algorithm was applied to evaluate the relationship between our identified humanized key genes and immune infiltration levels. Additionally, we conducted Gene Set Enrichment Analysis (GSEA) and Gene Set Variation Analysis (GSVA) on both high and low-risk groups and established interaction networks for mRNA-miRNA, mRNA-transcription factor (TF), and mRNA-RNA-binding protein (RBP). Finally, the clinical diagnostic significance of IRDEGs in ARHL was studied using Logistic regression analysis. Therefore, this comprehensive bioinformatics investigation sheds light on potential IRGs, offering insights into the underlying mechanisms of ARHL and proposing new therapeutic targets for this condition.
Methods
Data download
We obtained datasets related to ARHL from GSE49522, GSE4954330 and GSE604531 in the GEO database using the GEOquery32 package. These datasets, characterized by their large sample sizes and reliable experimental designs, effectively reflect the expression of genes associated with ARHL, thereby providing a solid foundation for our research.
GSE49522 is derived from Mus musculus, with the data platform file being GPL339 [MOE430A] Affymetrix Mouse Expression 430A Array. We used the expression profile data of 5 cases of severe presbycusis samples (group: ARHL) and 8 cases of normal samples (group: Normal) in this study.
The dataset GSE49543 comes from Mus musculus, and the data platform file is GPL339 [MOE430A] Affymetrix Mouse Expression 430A Array. We included the expression profile data of 6 cases of severe presbycusis samples (group: ARHL) and 9 cases of normal samples (group: Normal) in this study.
The dataset GSE6045 comes from Mus musculus, and the data platform file is GPL339 [MOE430A] Affymetrix Mouse Expression 430A Array. This study included the expression profile data of 3 samples categorized as having moderate presbycusis (grouping: Moderate presbycusis) and 3 samples categorized as having severe presbycusis (grouping: Severe presbycusis).
IRGs were obtained from the GeneCards database33, a resource that offers thorough details on human genes. A search utilizing the keyword “Inflammatory” yielded 732 IRGs that met the criteria of being “Protein Coding” with a Relevance score > 5. In addition, we used “Inflammatory” as the search keyword to collect 200 IRGs in the published literature on PubMed website34,35. After merging and deduplication, we finally obtained 768 IRGs. See Supplementary Table S1 for specific information.
Differentially expressed genes related to presbycusis
When identifying differentially expressed genes, standard differential expression analysis methods were used to accurately find IRDEGs and lay the foundation for subsequent enrichment analysis. To uncover the intrinsic mechanisms of distinct genes in ARHL and the associated biological features and pathways, we combined the ARHL data from GSE49522 and GSE49543, followed by batch effect removal using the R package (sva package). The limma package standardized the merged dataset to create the presbycusis dataset.
After merging the dataset, differential analysis was conducted using the limma package36 to identify differentially expressed genes (DEGs) between the Normal and ARHL groups, with | logFC | > 0 and P < 0.05 as the criteria. Our study primarily aimed to identify inflammation-related genes associated with ARHL, so the choice to use a threshold of | logFC | > 0 in the initial step was to include as many relevant genes as possible to suit the specific objectives and context of our study. Moreover, | logFC | > 0 was also employed as the screening criterion in relevant literature37 to identify differentially expressed genes with biological significance. First, this threshold allows us to capture genes that may be important in presbycusis research but whose expression does not change much. Secondly, considering the diversity of samples, we believe that the inclusion of these genes will help to comprehensively understand the molecular mechanism of ARHL. The genes identified through this method were designated as DEGs for subsequent investigation. Specifically, genes with logFC > 0 and P < 0.05 were classified as up-regulated DEGs (up regulated genes), while those with logFC < 0 and P < 0.05 were categorized as down-regulated DEGs (down regulated genes).
To acquire the IRDEGs linked to ARHL, we calculated the intersection of the merged dataset’s DEGs and IRGs and visualized the results using a Venn diagram. The outcomes from the differential analysis were generated using the R package ggplot2, showcasing a volcano plot. A heat map depicting IRDEGs was crafted utilizing the R package pheatmap.
Functional enrichment analysis (GO) and pathway enrichment analysis (KEGG) of differential expressed genes
GO and KEGG enrichment analysis help us understand the biological functions of IRDEGs and their role in specific biological processes. Analyzing GO38 is a commonly used technique for extensive investigation into functional enrichment, encompassing biological process (BP), molecular function (MF) and cellular components (CC). KEGG39 stands out as a popular repository housing data on genomes, biological pathways, ailments, pharmaceuticals, among others. Utilizing the R package clusterProfiler40, we conducted GO annotation analysis on IRDEGs. Criteria employed for entry screening included P.value < 0.05 and FDR value (q.value) < 0.05, with Benjamini-Hochberg (BH) serving as the correction method for P values.
GSEA
GSEA41 aims to assess the pattern of gene distribution within a predefined gene set listed in the gene table arranged by their correlation with the phenotype, in order to determine their impact on the phenotype. In this investigation, the logFC metric serves as the marker for molecules ranking to assess potential significant enrichment within the predefined gene set. Subsequently, the clusterProfiler toolkit is utilized to conduct enrichment analysis on all genes associated with the phenotype. The parameters utilized in the GSEA enrichment analysis encompass: the initial seed set at 2020, conducting 1000 calculations, a gene set size ranging from a minimum of 10 to a maximum of 500 genes, and employing BH as the method for P value correction. The gene set “c2.cp.v7.2.symbols.gmt” from the Molecular Signatures Database (MSigDB) was acquired for GSEA analysis on the expression of genes within the combined dataset. The standard for significant enrichment classification was defined as P.value < 0.05 and FDR value (q.value) < 0.25.
GSVA
GSVA42 is a non-parametric and unsupervised method that is used to analyze gene expression differences between samples by converting the expression matrix of genes into the expression matrix of gene sets. Quantitative matrix was taken to evaluate the enrichment of gene sets in the microarray nuclear transcriptome. In order to assess the enrichment of various pathways across samples, we utilized the gene set “m2.all.v2022.1.Mm.symbols.gmt” from the MSigDB database. By conducting GSVA on the combined dataset, we were able to calculate differences in functional enrichment between groups.
The use of GSEA and GSVA allows us to evaluate the differences in biological pathways between high- and low-risk groups.
LASSO regression analysis to screen IRDEGs
LASSO regression analysis can be used to perform variable selection and regularization at the same time to identify the most critical IRDEGs, reduce model complexity, and avoid overfitting. To identify IRDEGs, we initially utilized the glmnet package for conducting LASSO regression43 on IRDEGs by using the “binomial” as the parameter family and a ten-fold cross-validation method ADDIN EN. CITE. We performed 1000 iterations to avoid overfitting. LASSO regression is a popular machine learning algorithm for model development by incorporating a penalty term (lambda × absolute slope) and using regularization within linear regression to address overfitting and enhance the model generalization.
We then determined the penalty coefficient (lambda) for the identified IRDEGs in the LASSO regression diagnostic model and computed the risk score of the diagnostic model based on these IRDEGs. Furthermore, we utilized the R package RCircos to map the identified IRDEGs to human genes, with annotations of the positions of human chromosomes. Subsequently, we focused on analyzing the IRDEGs that were identified through the LASSO regression analysis.
Immune infiltration assay (CIBERSORT)
CIBERSORT is a tool available on the website (https://cibersortx.stanford.edu/)(44) that utilizes linear support vector regression to analyze the expression matrix of human immune cell subtypes. By assessing gene expression signatures of 22 known immune cell subtypes, this tool can determine the infiltration status of immune cells in sequencing samples. In our research, we utilized the CIBERSORT algorithm to assess immune cell infiltration in a combined dataset. Initially, we compared the infiltration patterns of 22 immune cell types in both the ARHL and Normal groups, creating a visual representation of the differences. Subsequently, we selected the immune cells with significant infiltration differences to draw cell maps which presented correlations among immune cells.
Immune infiltration analysis (MCPcounter)
The presence of immune and other stromal cells within the cochlea was assessed using MCPcounter45. This tool provides estimates of the quantities of eight immune cell types, specifically CD4+ T cells, CD8+ T cells, natural killer (NK) cells, B lymphocytes, monocytes, dendritic cells, neutrophils, and Toxic lymphocytes (comprising CD8+ T cells and cytotoxic innate lymphoid NK cells). Additionally, MCPcounter also calculated the quantities of two types of non-immune stromal cells-fibroblasts and endothelial cells. A Spearman correlation analysis was conducted according to MCPcounter abundance estimation and the expression of antigen genes, with statistical significance defined as p < 0.05.
The application of CIBERSORT and MCPcounter algorithms allows us to quantitatively analyze the infiltration of different immune cells in the ARHL group and control group, thereby exploring the relationship between immune response and ARHL, which is crucial for understanding the underlying mechanism of ARHL.
Clinical evaluation of IRDEGs
In order to investigate the diagnostic significance of IRDEGs for ARHL, a Logistic regression analysis was conducted on the IRDEGs expression in the merged dataset to develop a Logistic regression model. Subsequently, the R package rms was utilized to create a nomogram based on the outcomes of the Logistic regression analysis. The nomogram consists of a series of disconnected line segments plotted on a Cartesian coordinate system to represent the functions between multiple independent variables. Through the multi-factor regression analysis, each variable in the model was assigned a score and described using a specific scale. By summing the scores, the probability of event occurrence could be estimated.
The calibration plot is used to assess how well the model’s predictions align with the actual results by displaying the relationship between the predicted and observed probabilities in various scenarios. This graph is primarily utilized to evaluate models created using Logistic regression and to analyze how well they fit the observed data.
DCA, or decision curve analysis, is a straightforward approach to assess the performance of clinical prediction models, diagnostic tests, and molecular markers. Finally, the R package ggDCA was utilized for creating DCA plots to evaluate the accuracy and resolution of the Logistic regression model.
Construction of mRNA-miRNA, mRNA-TF, mRNA-RBP interaction network
The construction of mRNA-miRNA, mRNA-TF, and mRNA-RBP interaction networks further reveals the regulatory relationships between key genes, thereby providing important clues for the biological mechanisms and potential therapeutic targets of ARHL.
The ENCORI46 database (https://starbase.sysu.edu.cn/) represents the upgraded version 3.0 of the starBase database, which showcases various interactions including miRNA-ncRNA, miRNA-mRNA, ncRNA-RNA, RNA-RNA, RBP-ncRNA, and RBP-mRNA. These interactions are derived from data mining analyses of CLIP-seq and degradome sequencing (specifically for plants), which provides diverse visual interfaces for exploring microRNA targets. For predicting miRNA target genes and facilitating functional annotation, researchers rely on the miRDB47 database. By utilizing the ENCORI and miRDB databases, we were able to forecast miRNAs that interact with IRDEGs (mRNA). Subsequently, we compared the results from both databases and constructed the mRNA-miRNA interaction network based on the intersecting outcomes.
We also used the ENCORI database to predict RBPs interacting with IRDEGs (mRNA), and then used the results to draw the mRNA-RBP interaction network.
Utilizing the ENCORI database, we conducted predictions for RBPs that interact with IRDEGs (mRNA) and constructed the mRNA-RBP interaction network.
Within the CHIPBase database (version 3.0) (https://rna.sysu.edu.cn/chipbase/)(48), we identified numerous binding motif matrices and their corresponding binding sites based on ChIP-seq data of DNA-binding proteins. This database also contains information on the transcriptional regulatory relationships between millions of TFs and genes. By exploring the CHIPBase database (version 3.0), we were able to identify TFs that are bound to IRDEGs (mRNA).
Functional similarity analysis
Comparing GO annotations semantically provides a quantitative approach to calculate the similarity between genes and genomes, serving as a crucial foundation for numerous bioinformatics analysis techniques. The GOSemSim package is utilized to calculate the GO semantic similarity of IRDEGs, followed by assessing their geometric average at the BP, CC and MF levels to derive the final score. Subsequently, the results of the functional similarity analysis are visually represented using the ggplot package.
Spatial structures of protein
AlphaFold Protein Structure Database (https://www.alphafold.ebi.ac.uk/) is based on artificial intelligence computing models, combines physical and biological data of proteins, and uses multiple sequence alignments to design deep learning algorithms. The spatial model of the protein can be regularly predicted based on the amino acid sequence of a protein in this database. We used the AlphaFold Protein Structure Database to predict and display the protein spatial structure of IRDEGs.
Receiver operating characteristic (ROC) curve
The ROC curve serves as a valuable tool in analyzing data, aiding in the selection of the most optimal model, discarding alternatives, or identifying the ideal threshold within a given model. This graphical representation offers a comprehensive evaluation of continuous variables’ sensitivity and specificity while showcasing the nuanced relationship between the two. The area under the ROC curve typically ranges from 0.5 to 1, with values closer to 1 indicating superior diagnostic efficacy. AUC values falling between 0.5 and 0.7 signify lower accuracy, while those ranging from 0.7 to 0.9 indicate a certain level of accuracy. AUC values exceeding 0.9 reflect a relatively high level of accuracy. To assess the diagnostic potential of IRDEGs expression in disease classification, we utilized the pROC package to construct ROC curves for distinct groups (Normal/ ARHL) within the merged dataset and calculated corresponding AUC values.
Statistical analysis
To compare continuous variables between two groups, the independent Student t test is employed for normally distributed data, while the Mann-Whitney U test (i.e. Wilcoxon rank sum test) is utilized for non-normally distributed data. For comparing and analyzing categorical variables between two groups, either the Chi-square test or Fisher’s exact test was conducted. The Spearman correlation analysis was utilized to compute correlation coefficients among various molecules. Unless stated otherwise, all P values are considered statistically significant if P < 0.05, and all tests are two-sided.
Result
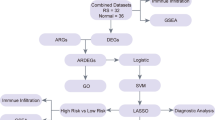
Technology roadmap
It was shown in Fig. 1.
ARHL: age-related hearing loss; GSEA: Gene Set Enrichment Analysis; GSVA: Gene Set Variation Analysis; GO: Gene Ontology; KEGG: Kyoto Encyclopedia of Genes and Genomes; ROC: receiver operating characteristic curve; PCA: Principal Component Analysis; IRDEGs: Inflammatory related differentially expressed genes; DEGs: differentially expressed genes; IRGs: Inflammatory related genes; TF: transcription factor; RBP: RNA-binding protein.
Data set merge correction
The batch effect was eliminated from the two ARHL datasets (GSE49522 and GSE49543) using the Sva package, followed by standardization of the merged dataset using the limma package (Fig. 2A-B). The results indicate that the batch effect of samples from various sources is basically eliminated after the process of removing batch. The merged dataset includes 11 samples from mice with severe presbycusis (grouping: ARHL) and 17 samples of normal controls (grouping: Normal). The principal component analysis (PCA) on the expression matrix of the merged dataset also indicates the removal of the batch effect originating from various sample sources (Fig. 2C-D).
Afterwards, the data set GSE6045 (Fig. 2E-F) was standardized. Data cleaning operations, including probe annotations, were carried out, resulting in the creation of a boxplot to visualize the distribution of the data pre and post normalization. Upon standardizing the data, the patterns of expression among various samples within the dataset appeared to align.
A–B. The boxplot graph before (A) and after (B) normalization of the merged dataset. C–D. The PCA graph of merged dataset before (C) and after (D) the process of removing the batch effect. (E) Distribution box plot of gene expression among samples in the GSE6045 data set before correction. (F) Box plot of the distribution of gene expression among samples in the corrected dataset GSE6045. PCA: Principal Component Analysis.
Analysis of differentially expressed genes related to presbycusis
Differential analysis was conducted on the combined dataset utilizing the limma package to identify genes that were expressed differently between the ARHL group and the Normal group. We found a total of 1067 genes that met the criteria of |logFC| > 0 and P.value < 0.05. Out of these genes, 525 showed high expression in the ARHL group (indicating low expression in the normal group, with a logFC greater than 0), while 542 genes exhibited low expression in the ARHL group (indicating high expression in the normal group, with a logFC less than 0). A volcano map was generated to visualize the results of the differential analysis (Fig. 3A).
By intersecting the total differentially expressed genes with IRGs, we identified 105 IRDEGs. A Venn diagram was constructed to represent the overlap of these genes (Fig. 3B).
Subsequently, we analyzed the expression differences of the 105 IRDEGs across different groups within the merged dataset (Fig. 3C) and utilized the R package pheatmap to create a heat map illustrating the results. The findings from the heat map indicate a clear clustering pattern of the 105 IRDEGs across different groups in the merged dataset.
(A) Volcano map of the differentially expressed genes in merged dataset. (B) Venn diagram of DEGs and IRGs in the merged dataset. (C) Heat map of 105 IRDEGs in different groups. DEGs: differentially expressed genes. IRGs: Inflammatory related genes. IRDEGs: Inflammatory related differentially expressed genes.
Functional enrichment analysis (GO) and pathway enrichment (KEGG) analysis of IRDEGs
To investigate the molecular functions, cellular components, biological processes, biological pathways, and their connection to presbycusis in 105 IRDEGs, we initially conducted GO (Supplementary Table S2) and KEGG (Supplementary Table S3) enrichment analyses. The significance criteria for the enrichment items were defined as a P.value < 0.05 and an FDR value (q.value) < 0.05, making them statistically significant. The results of the analyses were visually presented through bubble diagrams (Fig. 4A-B) and ring network diagrams (Fig. 4C-D), followed by a bar graph displaying the GO functional enrichment analysis of IRDEGs alongside logFC (Fig. 4E-F).
It was shown that 105 IRDEGs were mainly enriched in biological processes (biological process, BP) including I-kappaB kinase/NF-kappaB signaling (GO: 0007249), receptor signaling pathway via JAK-STAT (GO: 0007259), negative regulation of MAPK cascade (GO: 0043409), and autophagy (GO: 0006914), in cell components (cellular component, CC) including extracellular exosome (GO: 0070062), membrane microdomain (GO: 0098857), membrane raft (GO: 0045121), and receptor complex (GO: 0043235), and in molecular functions (molecular function, MF) including immune receptor activity (GO: 0140375), cytokine receptor binding (GO: 0005126), cytokine binding (GO: 0019955), cytokine receptor activity (GO: 0004896).
The KEGG enrichment analysis predominantly highlighted pathways such as NF-kappa B signaling pathway (mmu04064), PI3K-Akt signaling pathway (mmu04151), JAK-STAT signaling pathway (mmu04630), MAPK signaling pathway (mmu04010), Hippo signaling pathway (mmu04390), Necroptosis (mmu04217), and Apoptosis-multiple species (mmu04215).
The pathway map generated using the R package Pathview illustrated the outcomes of the KEGG enrichment analysis, specifically focusing on pathways like the MAPK signaling pathway, PI3K-Akt signaling pathway, Hippo signaling pathway, and JAK-STAT signaling pathway (Supplementary Figure S1).
A–B. Results of GO functional enrichment analysis (A) and KEGG pathway enrichment analysis (B) of IRDEGs are displayed in bubble diagrams. C–D. Results of GO function enrichment analysis (C) and KEGG pathway enrichment analysis (D) of IRDEGs are displayed in a circular network diagram. E–F. Results of GO functional enrichment analysis (E) and KEGG pathway enrichment analysis (F) of IRDEGs are shown in histograms. GO: Gene Ontology. KEGG: Kyoto Encyclopedia of Genes and Genomes. IRDEGs: Inflammatory related differentially expressed genes. BP: biological process. CC: cellular component. MF: molecular function. The screening criteria for GO and KEGG enrichment items were P.value < 0.05 and FDR value (q.value) < 0.05.
GSEA and GSVA enrichment analysis of merged dataset
To explore the impact of gene expression levels on presbycusis, we conducted a thorough analysis of the correlation between gene expression and relevant biological processes, affected cell components and molecular functions between different groups (Normal/ARHL) through GSEA enrichment analysis. Our findings (Supplementary Figure S2A) revealed that all genes in the combined dataset exhibited notable enrichment in Lair Pathway (Supplementary Figure S2B), Mhc Pathway (Supplementary Figure S2C), Local Acute Inflammatory Response Pathway involving Cells and Molecules (Supplementary Figure S2D), Il23 Pathway (Supplementary Figure S2E), and Inflammatory Bowel Disease Signaling Pathway (Supplementary Figure S2F) (Supplementary Table S4).
Furthermore, the results from the GSVA analysis indicated significant disparities in the PTC1 pathway and other gene sets between the ARHL and normal groups within the combined dataset (Supplementary Figure S2G, Supplementary Table S5).
LASSO regression analysis to screen IRDEGs
LASSO regression analysis was used to obtain key IRDEGs from 105 IRDEGs in the merged dataset (Supplementary Figure S3A). Subsequently, we illustrated the expression patterns of these key IRDEGs in different groups (ARHL/Normal) using a forest plot (Supplementary Figure S3B). The screened 10 key genes are Alox5ap, Chil1, Clec7a, Dysf, Fcgr3, Fgf1, H2-K1, Il10ra, Rgs1, and Tnnt2 as shown in Supplementary Figure S3B. The outcomes of the LASSO regression model were visually represented through a locus plot of the LASSO variables (Supplementary Figure S3C).
Expression analysis of key genes in merged dataset
In the merged dataset, we illustrated the differential expression of 10 key genes (Alox5ap, Chil1, Clec7a, Dysf, Fcgr3, Fgf1, H2-K1, Il10ra, Rgs1, and Tnnt2) within distinct groups (Normal/ARHL) through grouped violin plot (Supplementary Figure S4A). Our statistical analysis indicated that 9 out of the 10 key genes showed significant differences in expression levels between the two groups (p value < 0.05). Among them, the expression levels of Alox5ap, Chil1, Clec7a, Fcgr3, H2 − K1, and Tnnt2 were extremely significantly different (p value < 0.001) between different groups (Normal/ARHL), and the expression levels of Il10ra and Rgs1 showed highly significant discrepancies (p value < 0.01). The expression level of Dysf was statistically significantly different between the Normal and ARHL groups (p value < 0.05).
Correlation analysis was conducted according to the full expression matrix of the 10 key genes within the combined dataset, yielding a correlation heat map (Supplementary Figure S4B). The findings indicated significantly positive correlations between key genes Clec7a and Fcgr3, Clec7a and H2 − K1, H2 − K1 and Fcgr3, etc. The key genes Fgf1 and Alox5ap, H2 − K1 and Dysf, Rgs1 and Fgf1 have significantly negative correlations.
We then converted 10 key genes into human genes and annotated their positions on the human chromosome in a circle diagram (Supplementary Figure S4C). The illustration reveals that the key genes―FCGR2A, RGS1, TNNT2, and CHI3L1―are located on chromosome 1, while DYSF and FGF1 are located on chromosome 2 and 5, respectively.
Functional similarity analysis was carried out on the 10 key genes. By computing the semantic similarity among GO terms, sets of GO terms, gene products, and gene clusters using the GOSemSim package, we visualized the results of the analysis through a boxplot (Supplementary Figure S4D). The data revealed that Fgf1 exhibited the highest functional similarity values with the other critical genes.
Subsequently, ROC curves were plotted for the 10 key genes in the merged dataset, and the findings were depicted in Supplementary Figure S5. The ROC curve indicated that the expression level of Alox5ap in the merged dataset presented a high accuracy between different groups (AUC > 0.9). The expression levels of key genes―Chil1, Tnnt2, Rgs1, Il10ra, Dysf, and Fgf1―in the integrated merged dataset displayed a certain accuracy (0.7 < AUC < 0.9) between different groups (Normal/ARHL).
Expression analysis of key genes in dataset GSE6045
The expression levels of the 10 key genes were analyzed between different groups (Moderate presbycusis/Severe Presbycusis) in GSE6045, as shown in Supplementary Figure S6.
Utilizing the comprehensive expression matrix of these key genes within the dataset, a correlation analysis was conducted, resulting in the creation of a correlation heat map (Supplementary Figure S7A). The findings indicated a significantly positive correlation between Chil1 and Clec7a, as well as Chil1 and Fcgr3. Conversely, there was a notably negative correlation between Chil1 and Tnnt2.
Next, the ROC curves were generated for the key genes in the dataset GSE6045, and the results were displayed in Supplementary Figure S7B-D. The ROC curve analysis demonstrated that the expression levels of the Dysf, Rgs1, and Fgf1 genes possess a certain accuracy (0.7 < AUC < 0.9) in distinguishing between Moderate presbycusis and Severe Presbycusis groups.
Immune infiltration analysis (CIBERSORT and MCPcounter)
Based on the findings from the immune infiltration analysis by CIBERSORT algorithm, a histogram (Supplementary Figure S8A) was created to visualize the infiltration patterns of 22 immune cells present in each sample within the combined dataset.
A comparative analysis of the abundance of immune cell infiltration between different groups (Normal/ARHL) within the merged dataset was performed, revealing statistically significant differences (P < 0.05) in the infiltration levels of 3 immune cell types (Dendritic cells resting, Macrophages M1, and T cells follicular helper) (Supplementary Figure S8B).
To further explore the relationship between the 10 humanized key genes and the infiltration abundance of the 3 identified immune cell types, a correlation heat map was generated (Supplementary Figure S8C). This analysis highlighted correlations between 8 key genes (ALOX5AP, CHI3L1, CLEC7A, DYSF, FCGR2A, HLA − A, IL10RA, and RGS1) and the infiltration levels of the 3 aforementioned immune cell types. The results showed that there was a positive correlation between Dendritic cells resting and CLEC7A, Macrophages M1 and RGS1, T cells follicular help and FCGR2A, etc.
Using the MCPcounter algorithm, the correlation between the 10 humanized key genes and the immune cell infiltration levels in the combined dataset was calculated (Supplementary Figure S8D). This analysis revealed varied correlations between the key genes and different immune cell types, including positive correlations between CD8 T cells and IL10RA, as well as Neutrophils and CLEC7A. Conversely, negative correlations were observed between Fibroblasts and CHI3L1, Fibroblasts and FCGR2A, Fibroblasts and HLA − A, etc.
GSEA and GSVA enrichment analysis of high and low risk groups
To evaluate the impact of gene expression on presbycusis, we initially categorized the disease group from the combined dataset into high and low risk groups based on the median riskScore value derived from the LASSO regression model. Subsequently, we conducted an analysis of the correlation between gene expression and relevant biological processes, cellular components, and molecular functions in the high and low risk groups using GSEA enrichment analysis. The results indicated that the genes in both the high and low risk groups were significantly enriched in Classic Pathway (Supplementary Figure S9B), Neutrophil Degranulation (Supplementary Figure S9C), Mitochondrial Translation (Supplementary Figure S9D), Ribosome (Supplementary Figure S9E), Translation (Supplementary Figure S9F) and other pathways.
The outcomes of the GSVA enrichment analysis performed on the combined dataset (Supplementary Figure S9G) revealed distinctions in the AKAP13 pathway and other gene sets between the high and low risk groups.
Construction of mRNA-miRNA, mRNA-TF, and mRNA-RBP interaction network
Using the mRNA-miRNA data in the ENCORI database and the miRDB database, the prediction was made for the miRNAs that interacted with the 10 key genes (Alox5ap, Chil1, Clec7a, Dysf, Fcgr3, Fgf1, H2-K1, Il10ra, Rgs1, and Tnnt2). To visualize the overlapping outcomes from both databases, the mRNA-miRNA interaction network was illustrated using Cytoscape software (Supplementary Figure S10A). The network comprised 7 key genes (mRNA) (Alox5ap, Chil1, Fcgr3, Fgf1, H2-K1, Il10ra, and Rgs1), along with 67 miRNA molecules, forming a total of 74 mRNA-miRNA pairs (Supplementary Table S6).
TFs were identified by querying the CHIPBase database (version 3.0) for binding to key genes. The Cytoscape software was utilized to generate a visual representation of the mRNA-TF interaction network (Supplementary Figure S10B). We finally obtained 6 key genes (mRNA) (Tnnt2, Alox5ap, Dysf, Fcgr3, H2-K1, and Rgs1) and 94 TFs, resulting in a total of 111 unique pairs of mRNA-TF interactions (Supplementary Table S7).
Predicted interactions between RBPs and 10 key genes (Alox5ap, Chil1, Clec7a, Dysf, Fcgr3, Fgf1, H2-K1, Il10ra, Rgs1, and Tnnt2) were identified using mRNA-RBP data within the ENCORI database. The resulting mRNA-RBP network was visualized using Cytoscape software (Supplementary Figure S10C). Our network includes 7 key genes (mRNA) (Alox5ap, Dysf, Fcgr3, Fgf1, H2-K1, Il10ra, and Tnnt2) connected to 28 different RBP molecules, resulting in a total of 45 unique mRNA-RBP pairs (Supplementary Table S8).
Protein spatial structure
We used the AlphaFold Fold Protein Structure Database to forecast the spatial structure of proteins for 10 key genes (Alox5ap, Chil1, Clec7a, Dysf, Fcgr3, Fgf1, H2-K1, Il10ra, Rgs1, and Tnnt2) and finally obtained the spatial structure diagram of proteins.
Clinical evaluation of key genes
The Logistic regression analysis was performed on the expression of 10 key genes (Alox5ap, Chil1, Clec7a, Dysf, Fcgr3, Fgf1, H2-K1, Il10ra, Rgs1, and Tnnt2) in order to build a Logistic regression model. The genes that demonstrated statistical significance with p-values less than 0.05 are illustrated in a forest plot, as shown in Supplementary Figure S12A.
Subsequently, a nomogram was developed to assess the diagnostic potential of the model through nomogram analysis (Supplementary Figure S12B). The findings indicated that the expression of the model gene Alox5ap was significantly more effective than that of other variables in the diagnostic model of ARHL. The gene Fgf1 exhibited markedly poorer performance in the diagnostic model for ARHL compared to other variables.
We presented the ROC curve analyzing the linear predictors from the Logistic regression model across diverse groups (Normal/ARHL) within the combined dataset (Supplementary Figure S12C). The visualization highlights the strong diagnostic performance of the Logistic regression model in the merged dataset.
It was displayed in Supplementary Figure S12D that the ROC curve of the linear predictors between different groups (Moderate presbycusis/Severe Presbycusis) in the dataset GSE6045. The graphic demonstrates the effective diagnostic capabilities of the Logistic regression model within GSE6045.
To assess the precision and effectiveness of the Logistic regression model, we constructed a Calibration Curve. The predictive accuracy of the model is assessed by comparing the actual probabilities in the fitting situation with the model’s predicted probabilities in various situations (Supplementary Figure S13).
The clinical utility of the Logistic regression model was evaluated through DCA based on the model genes in the integrated merged dataset, as depicted in Supplementary Figure S14.
Discussion
ARHL is the predominant type of sensorineural hearing impairment in adults and significantly affects quality of life, economic participation and overall well-being in older adults, yet its pathophysiological mechanisms remain poorly understood and require further investigation49. Inflammation plays a crucial role in various age-related illnesses. Few studies have investigated its possible effect in the pathogenesis of ARHL. Therefore, exploring the specific role of the key inflammatory molecules and immune cells in the pathogenesis of ARHL will help to provide potential therapeutic options for this condition. For the first time, we adopted comprehensive, value-free, and effective bioinformatics methods to prospectively explore the connections between inflammation and ARHL.
Inflammation is considered as a key indicator of aging and is strongly associated with various age-related diseases, such as cardiovascular disease, diabetes, arthritis, chronic kidney disease, Parkinson’s disease, and Alzheimer’s disease50,51,52. Many pro-inflammatory biomarkers play important roles in this process, such as IL-1, IL-6, IL-8, IL-17, 5-LOX, etc. To our best knowledge, inflammation has been shown to be a major cause of noise-induced hearing loss53. TNF-α is significantly elevated in patients with sudden sensorineural hearing loss54. As for the association to ARHL, TNF-α and IL-6 were detected an increase in the cochlea of aging mouse55. Elevated levels of IL-6 have also been seen in elderly individuals56. These studies indicate that inflammation might be one of the pathogeneses of ARHL.
We intersected the DEGs of the merged dataset and IRGs to obtain the IRDEGs associated with ARHL. A total of 105 genes were identified as IRDEGs and enriched for multiple processes related to inflammation, including NF-kappa B signaling, PI3K-Akt signaling, JAK-STAT signaling, MAPK signaling, Hippo signaling, Necroptosis, and Apoptosis - multiple species pathways. Further analysis revealed that the effects of these IRDEGs were closely linked to immune cells. Through LASSO regression analysis, ten key genes (Alox5ap, Chil1, Clec7a, Dysf, Fcgr3, Fgf1, H2-K1, Il10ra, Rgs1, and Tnnt2) were screened. Compared with normal group, the expression levels of nine key genes except for the Fgf1 gene were significantly higher in ARHL group. Although some of these key genes were detected to be associated with age-related diseases, they have not yet been reported in ARHL.
The Arachidonate 5-lipoxygenase activating protein (ALOX5AP) gene is responsible for producing the protein 5-lipoxygenase activating protein (FLAP), which promotes the first step in the leukotriene signaling pathway involving 5-lipoxygenase (5LOX)57. Leukotriene B4 (LTB4) is the most important product of this pathway and powerfully promotes the production of monocyte chemoattractant protein 1 (MCP1) in human monocytes58. Both MCP1 and 5LOX products recruit white blood cells to sites of inflammation59. They partake in the pathogenesis of obesity, atherosclerotic cardiovascular disease and coronary heart disease. This might establish a connection among fatty tissue, inflammation, and insulin resistance60,61. 5-LOX, expressed in the central nervous system, might contribute to neurodegenerative diseases. LTB4, one of the downstream products of 5-LOX, might be a pathogenic factor of Parkinson’s disease62. It has been reported that the expression of 5-LOX and FLAP is elevated in the brains of aged rats, with the highest expression levels detected in animals with cognitive impairment63.
CHI3L1 belongs to the glycoside hydrolase family 18 and is regulated by changes in the cytokines, growth factors, extracellular matrix, drugs, and stress64. CHI3L1 is synthesized and secreted by a variety of cells including synoviocytes, chondrocytes, neutrophils, macrophages, fibroblast-like cells, smooth muscle cells and tumor cells. It is crucial in the inflammatory and remodeling processes as it promotes the formation of endothelial tubules and the migration and adhesion of vascular smooth muscle cells. CHI3L1 is closely associated with diseases such as arthritis, asthma, diabetes, sepsis, liver fibrosis, and coronary artery disease64,65. CHI3L1 profoundly affects the differentiation of macrophages, the accumulation of DCs, and the balance between Th1 and Th2 cells66. CHI3L1 promotes the recruitment of macrophage through the MAPK signaling pathway and secretes inflammatory chemokines, mainly IL-8 and CCL267. CHI3L1 could encode YKL-40, which can prevent the activation of microglial phagocytes. This condition increases with age and the onset of Alzheimer’s disease68,69,70. The expression of CHI3L1 is also significantly increased in aging liver tissue71.
DYSF encodes Dysferlin, a seven-C2 β-sheet domain-rich protein that plays an important role in the regeneration and maintenance of calcium-dependent cell membrane72. DYSF was discovered to promote phagocytosis, migration, and adhesion of THP1 cells. Supportive studies indicate that DYSF is specifically overexpressed in Alzheimer disease and might be involved in the pathogenesis of AD73.
IL-10RA specifically binds IL-1 and mediates IL-10 function. Previous research has shown that IL-10RA could reduce inflammation74. IL-10RA is thought to influence macrophage infiltration and function in inflammatory responses75. It is also important in forming the intestinal epithelial barrier to prevent inflammation76.
HLA−A, a gene within the HLA gene complex located on the short arm of the chromosome 677, presents antigens to CD8+ T cells as the main arm of cellular immunity78. It was reported that HLA was associated with chronic infection by Herpes zoste79 and positively correlated with cytolytic activity80. Future in-depth experimental researches are warranted to explore the potential effects of these key genes on the pathogenesis of ARHL.
The KEGG enrichment analysis in our study revealed several significant signaling pathways, mainly including NF-kappa B, PI3K-Akt, JAK-STAT, MAPK, Hippo, Necroptosis, and Apoptosis-multiple species. It has been confirmed previously that the involvements of some of these signaling pathways contributes to various inflammation, which is associated with some age-related diseases. For instance, by inhibiting the NF-kappa B pathway, it is possible to reduce the associated inflammation and oxidative stress, alleviate damage to cochlear hair cell, and thus prevent the development of ARHL81. Several studies have shown that inhibiting the activity of the PI3K/Akt pathway could decrease the level of anti-apoptotic protein Bcl-2, thereby promoting the death of aging cells82,83. The classic MAPK signaling pathway involves in the pathogenesis of autoimmune disorders, inflammatory conditions, and cancer84,85. Hearing loss caused by neomycin or gentamicin can be prevented by blocking MAPK cell death signaling86,87,88. The Hippo pathway, consisting of a series of kinases, regulates cell proliferation, differentiation, and apoptosis89. Yes-associated protein (YAP), a crucial element of the Hippo pathway, impairs the polarization of M2 macrophage and alleviates inflammation90. Recent clinical research indicates that the Hippo pathway is related to tissue inflammation and associated immunopathology91. YAP overexpression decreased ROS accumulation and then reduced neomycin-induced cochlear hair cell loss in a Hippo pathway92. Inflammation is closely associated with apoptosis process93, while apoptosis contributes to several age-related degenerative diseases, including ARHL94. Aging in the cochlea is associated with increased expression of apoptosis-related genes such as Bak, Bax, and Bim from the Bcl-2 family10. The deletion of the gene Bak could prevent ARHL in mice.
Traditionally, the inner ear was thought to be devoid of immune cells due to the barrier between the cochlea and the circulation known as the BLB. However, emerging evidence indicates the presence of immune cells such as resident macrophages in the cochlea14,95,96,97,98 and the involvement of the inner ear in immune responses98,99,100,101,102. Previous researches have shown that plasma cells, NK cells, activated CD4+ T cells, and regulatory T cells are important in the development of presbycusis103,104,105. Our bioinformatics analysis revealed that the infiltration abundance of Macrophages M1, Dendritic cells resting, and T follicular helper (Tfh) cells was statistically significantly higher in ARHL group than in Normal group, which indicates that the 3 immune cells might involve in the development of ARHL. Macrophages are essential for tissue homeostasis and inflammation and for defending against pathogens. It has been demonstrated that cochlea-resident macrophages play a significant role in inner ear immunity106,107. Macrophages were categorized into naive or non-activated macrophages (M0), pro-inflammatory macrophages (M1) promoting inflammation, and anti-inflammatory macrophages (M2) supporting tissue repair108,109. Covarrubias et al. demonstrated the accumulation of pro-inflammatory M1-like macrophages in metabolic tissues such as liver and visceral white adipose tissue during ageing and responses to acute inflammation110. As the heart ages, inflammaging occurs resulting from the type transformation of macrophage to a proinflammatory subtype (M1)111, which is a process of a raise in the basic inflammatory response during aging112,113.
Similarly, it has been proposed that dysfunction of Tfh cells may play a role in specific pathological conditions associated with senescence such as reduced responses to vaccination114 and increased vulnerability to atherosclerosis115, cancer116, and autoimmunity117. The main function of Tfh cells, a subtype of CD4+ T cells, is to support B cells and regulate antibody production118,119. Zhou and colleagues showed that the frequencies of blood CXCR5+ CD4+ Tfh cells increased in the aged population compared with younger subjects120. Dendritic cells also play a role in inflammaging121 and were reported to be relevant to sudden sensorineural hearing loss54. Dendritic cells resting produce proinflammatory cytokines to regulate immune homeostasis. Related cytokines they secreted include IL-1, IL-6, IL-12, TNF-α, NF-κB, etc., which are associated with various inflammatory and autoimmune diseases in humans122,123.
Given the close association between immune cells and inflammatory cytokines, the relationship between the 10 key genes converted to human genes and the infiltration abundance of the three immune cells was further investigated. A certain positive correlation was observed between Dendritic cells resting and CLEC7A, Macrophages M1 and RGS1, T cells follicular help and FCGR2A, etc. CLEC7A is the gene encoding the dectin-1 receptor and is inversely correlated with the expression of dendritic cells. CLEC7A induces the expression of the chemokines CXCL10 and CCL20, both of which powerfully attract lymphocytes and other inflammatory cells124,125,126. It serves as a diagnostic and prognostic biomarker for many inflammatory conditions, including psoriasis, multiple sclerosis, and lupus127,128. RGS1, a regulatory component of the G protein signaling family, links G protein-coupled receptors and calcium signaling129. Previous studies have demonstrated that RGS1 could enhance the production of inflammatory molecules and the chemotaxis of macrophages130,131. Fc fragment of IgG receptor IIa (FCGR2A) has been reported to be overexpressed in the colonic mucosa of patients with active ulcerative colitis in comparison with non-inflammatory controls132. These newly identified immune cells and cytokines may offer novel directions for treating ARHL.
Although it has been manifested in our work that inflammation might involve in the development of ARHL, this study does have limitations. Initially, we conducted bioinformatics analysis and developed a theoretical diagnostic model, which based on secondary mining of public database. A preliminary investigation was conducted to study the expression levels of hub genes and pathway-related genes. The primary limitation of this study is the absence of an external validation cohort to substantiate our findings, despite our comprehensive analyses conducted across multiple datasets. Furthermore, the datasets utilized in this study may have limitations regarding sample representativeness, potentially impacting the generalizability of the results. To establish a specific regulatory relationship, further validation with a larger clinical sample size is necessary to enhance the clinical relevance of our diagnostic and prognostic model. Additionally, further in vitro and in vivo experiments, including RT-qPCR, western blotting, and immunohistochemical analysis, are needed to elucidate the detailed mechanisms of inflammation associated with ARHL. Resource and time constraints also prevent us from performing additional experiments for further validation of our model. Therefore, future research should aim to incorporate more diverse datasets and experimental verification to improve the reliability and generalizability of the findings.
In recent years, there has been progress in our knowledge of the molecular mechanisms associated with the degeneration of cochlear cells and hearing loss that comes with age. This study enhances comprehension of the pathogenesis of ARHL and offers fresh perspectives on potential therapeutic targets for presbycusis. Inflammation is now known to be associated with serious degenerative diseases in older adults133. Despite the unclear causes of the pro-inflammatory characteristics associated with aging and age-related diseases, several hub genes and molecular pathways have been identified. The results of this study will offer prospective guidance for the management of ARHL.
Data availability
All data generated or analysed during this study are included in this published article (and its Supplementary Information files).
References
Bowl, M. R. & Dawson, S. J. Age-related hearing loss. Cold Spring Harb Perspect. Med. 9(8). (2019).
Lin, F. R. et al. Hearing loss and incident dementia. Arch. Neurol. 68(2), 214–220 (2011).
Rutherford, B. R., Brewster, K., Golub, J. S., Kim, A. H. & Roose, S. P. Sensation and psychiatry: Linking age-related hearing loss to late-life depression and cognitive decline. Am. J. Psychiatry 175(3), 215–224 (2018).
Stucky, S. R., Wolf, K. E. & Kuo, T. The economic effect of age-related hearing loss: National, state, and local estimates, 2002 and 2030. J. Am. Geriatr. Soc. 58(3), 618–619 (2010).
James, S. L. et al. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 392(10159), 1789–1858 (2018).
Lyu, A. R. et al. Mitochondrial damage and necroptosis in aging cochlea.Int. J. Mol. Sci. 21(7). (2020).
Bermudez-Munoz, J. M. et al. G6PD overexpression protects from oxidative stress and age-related hearing loss. Aging Cell. 19(12), e13275 (2020).
Frisina, R. D., Ding, B., Zhu, X. & Walton, J. P. Age-related hearing loss: Prevention of threshold declines, cell loss and apoptosis in spiral ganglion neurons. Aging (Albany NY) 8(9), 2081–2099 (2016).
Li, N. et al. Curcumin protects against the age-related hearing loss by attenuating apoptosis and senescence via activating Nrf2 signaling in cochlear hair cells. Biochem. Pharmacol. 212, 115575 (2023).
Tadros, S. F., D’Souza, M., Zhu, X. & Frisina, R. D. Apoptosis-related genes change their expression with age and hearing loss in the mouse cochlea. Apoptosis 13(11), 1303–1321 (2008).
McCabe, B. F. Autoimmune inner ear disease: Therapy. Am. J. Otol 10(3), 196–197 (1989).
Frye, M. D., Yang, W., Zhang, C., Xiong, B. & Hu, B. H. Dynamic activation of basilar membrane macrophages in response to chronic sensory cell degeneration in aging mouse cochleae. Hear. Res. 344, 125–134 (2017).
Zhang, W. et al. Perivascular-resident macrophage-like melanocytes in the inner ear are essential for the integrity of the intrastrial fluid-blood barrier. Proc. Natl. Acad. Sci. USA. 109(26), 10388–10393 (2012).
Hirose, K., Discolo, C. M., Keasler, J. R. & Ransohoff, R. Mononuclear phagocytes migrate into the murine cochlea after acoustic trauma. J. Comp. Neurol. 489(2), 180–194 (2005).
Fujioka, M., Okano, H. & Ogawa, K. Inflammatory and immune responses in the cochlea: Potential therapeutic targets for sensorineural hearing loss. Front. Pharmacol. 5, 287 (2014).
Lowthian, J. A. et al. Slowing the progression of age-related hearing loss: Rationale and study design of the ASPIRIN in HEARING, retinal vessels imaging and neurocognition in older generations (ASPREE-HEARING) trial. Contemp. Clin. Trials 46, 60–66 (2016).
Franceschi, C., Garagnani, P., Parini, P., Giuliani, C. & Santoro, A. Inflammaging: A new immune-metabolic viewpoint for age-related diseases. Nat. Rev. Endocrinol. 14(10), 576–590 (2018).
Uchida, Y. et al. Age-related hearing loss and cognitive decline - The potential mechanisms linking the two. Auris Nasus Larynx 46(1), 1–9 (2019).
Shen, Y. et al. Cognitive decline, dementia, alzheimer’s disease and presbycusis: Examination of the possible molecular mechanism. Front. Neurosci. 12, 394 (2018).
Fortunato, S. et al. A review of new insights on the association between hearing loss and cognitive decline in ageing. Acta Otorhinolaryngol. Ital. 36(3), 155–166 (2016).
Mitchell, P. et al. Relationship of Type 2 diabetes to the prevalence, incidence and progression of age-related hearing loss. Diabet. Med. 26(5), 483–488 (2009).
Watson, N., Ding, B., Zhu, X. & Frisina, R. D. Chronic inflammation - inflammaging - in the ageing cochlea: A novel target for future presbycusis therapy. Ageing Res. Rev. 40, 142–148 (2017).
Li, P., Qian, T. & Sun, S. Spatial architecture of the cochlear immune microenvironment in noise-induced and age-related sensorineural hearing loss. Int. Immunopharmacol. 114, 109488 (2023).
Qiu, K. et al. The emerging roles and therapeutic implications of immunosenescence-mediated inflammaging in age-related hearing loss. Am. J. Stem Cells 13(2), 101–109 (2024).
Cheng, Y. et al. Genetic analysis of potential biomarkers and therapeutic targets in age-related hearing loss. Hear. Res. 439, 108894 (2023).
Paplou, V., Schubert, N. M. A. & Pyott, S. J. Age-related changes in the cochlea and vestibule: Shared patterns and processes. Front. Neurosci. 15, 680856 (2021).
Su, Z. et al. Endoplasmic reticulum stress-induced necroptosis promotes cochlear inflammation: Implications for age-related hearing loss. Exp. Gerontol. 189, 112401 (2024).
Zhang, X. et al. Caffeine ameliorates age-related hearing loss by downregulating the inflammatory pathway in mice. Otology & neurotology: Official publication of the American Otological Society, American Neurotology Society [and] European Academy of Otology and Neurotology. ;45(3):227 – 37. (2024).
Liu, J. et al. Vitamin C inhibits NLRP3 inflammasome activation and delays the development of age-related hearing loss in male C57BL/6 mice. Neurosci. Lett. 836, 137897 (2024).
D’Souza, M., Zhu, X. & Frisina, R. D. Novel approach to select genes from RMA normalized microarray data using functional hearing tests in aging mice. J. Neurosci. Methods 171(2), 279–287 (2008).
Someya, S., Yamasoba, T., Prolla, T. A. & Tanokura, M. Genes encoding mitochondrial respiratory chain components are profoundly down-regulated with aging in the cochlea of DBA/2J mice. Brain Res. 1182, 26–33 (2007).
Davis, S. & Meltzer, P. S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23(14), 1846–1847 (2007).
Fishilevich, S. et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford). 2017. (2017).
Zhai, W. Y. et al. A novel inflammatory-related gene signature based model for risk stratification and prognosis prediction in lung adenocarcinoma. Front. Genet. 12, 798131 (2021).
Chen, L. et al. Identification of inflammatory-related gene signatures to predict prognosis of endometrial carcinoma. BMC Genom Data 23(1), 74 (2022).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12), 550 (2014).
Li, S., Gao, K. & Yao, D. Comprehensive Analysis of angiogenesis associated genes and tumor microenvironment infiltration characterization in cervical cancer. Heliyon 10(12), e33277 (2024).
Gene Ontology, C. Gene Ontology Consortium: going forward. Nucleic Acids Res. 43 (Database issue), D1049–D1056 (2015).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30 (2000).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16(5), 284–287 (2012).
Subramanian, A. et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 102(43), 15545–15550 (2005).
Hanzelmann, S., Castelo, R. & Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 14, 7 (2013).
Kang, J. et al. LASSO-based machine learning algorithm for prediction of lymph node metastasis in T1 colorectal cancer. Cancer Res. Treat. 53(3), 773–783 (2021).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37(7), 773–782 (2019).
Lu, H., Wu, J., Liang, L., Wang, X. & Cai, H. Identifying a novel defined pyroptosis-associated long noncoding RNA signature contributes to predicting prognosis and tumor microenvironment of bladder cancer. Front. Immunol. 13, 803355 (2022).
Wang, W., Zhang, J., Wang, Y., Xu, Y. & Zhang, S. Identifies microtubule-binding protein CSPP1 as a novel cancer biomarker associated with ferroptosis and tumor microenvironment. Comput. Struct. Biotechnol. J. 20, 3322–3335 (2022).
Chen, Y. & Wang, X. miRDB: an online database for prediction of functional microRNA targets. Nucleic Acids Res. 48(D1), D127–D31 (2020).
Zhou, K. R. et al. ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Nucleic Acids Res. 45(D1), D43–D50 (2017).
Huang, Q. & Tang, J. Age-related hearing loss or presbycusis. Eur. archives oto-rhino-laryngology: official J. Eur. Federation Oto-Rhino-Laryngological Soc. 267(8), 1179–1191 (2010).
Ebert, T. et al. Inflammation and premature ageing in chronic kidney disease. Toxins (Basel) 12(4). (2020).
El Assar, M., Angulo, J. & Rodriguez-Manas, L. Oxidative stress and vascular inflammation in aging. Free Radic Biol. Med. 65, 380–401 (2013).
Newcombe, E. A. et al. Inflammation: The link between comorbidities, genetics, and Alzheimer’s disease. J. Neuroinflamm. 15(1), 276 (2018).
Mao, H. & Chen, Y. Noise-induced hearing loss: Updates on molecular targets and potential interventions. Neural Plast. 2021, 4784385 (2021).
Yoon, S. H., Kim, M. E., Kim, H. Y., Lee, J. S. & Jang, C. H. Inflammatory cytokines and mononuclear cells in sudden sensorineural hearing loss. J. Laryngol Otol 133(2), 95–101 (2019).
Shi, X. et al. NLRP3-inflammasomes are triggered by age-related hearing loss in the inner ear of mice. Am. J. Transl Res. 9(12), 5611–5618 (2017).
Verschuur, C. A. et al. Markers of inflammatory status are associated with hearing threshold in older people: Findings from the hertfordshire ageing study. Age Ageing 41(1), 92–97 (2012).
Dixon, R. A. et al. Requirement of a 5-lipoxygenase-activating protein for leukotriene synthesis. Nature 343(6255), 282–284 (1990).
Huang, L. et al. Leukotriene B4 strongly increases monocyte chemoattractant protein-1 in human monocytes. Arterioscler. Thromb. Vasc. Biol. 24(10), 1783–1788 (2004).
Silva, A. R. et al. Monocyte chemoattractant protein-1 and 5-lipoxygenase products recruit leukocytes in response to platelet-activating factor-like lipids in oxidized low-density lipoprotein. J. Immunol. 168(8), 4112–4120 (2002).
Kaaman, M. et al. ALOX5AP expression, but not gene haplotypes, is associated with obesity and insulin resistance. Int. J. Obes. (Lond) 30(3), 447–452 (2006).
van der Net, J. B. et al. Arachidonate 5-lipoxygenase-activating protein (ALOX5AP) gene and coronary heart disease risk in familial hypercholesterolemia. Atherosclerosis 203(2), 472–478 (2009).
Kang, K. H., Liou, H. H., Hour, M. J., Liou, H. C. & Fu, W. M. Protection of dopaminergic neurons by 5-lipoxygenase inhibitor. Neuropharmacology 73, 380–387 (2013).
Mrowetz, H. et al. Leukotriene signaling as molecular correlate for cognitive heterogeneity in aging: An exploratory study. Front. Aging Neurosci. 15, 1140708 (2023).
Zhao, T., Su, Z., Li, Y., Zhang, X. & You, Q. Chitinase-3 like-protein-1 function and its role in diseases. Signal. Transduct. Target. Ther. 5(1), 201 (2020).
Puthumana, J. et al. Biomarkers of inflammation and repair in kidney disease progression. J. Clin. Investig. 131(3). (2021).
He, C. H. et al. Chitinase 3-like 1 regulates cellular and tissue responses via IL-13 receptor alpha2. Cell. Rep. 4(4), 830–841 (2013).
Kawada, M. et al. Chitinase 3-like 1 promotes macrophage recruitment and angiogenesis in colorectal cancer. Oncogene 31(26), 3111–3123 (2012).
Lananna, B. V. et al. Chi3l1/YKL-40 is controlled by the astrocyte circadian clock and regulates neuroinflammation and Alzheimer’s disease pathogenesis. Sci. Transl. Med. 12(574). (2020).
Molina-Martinez, P. et al. Microglial hyperreactivity evolved to immunosuppression in the hippocampus of a mouse model of accelerated aging and alzheimer’s disease traits. Front. Aging Neurosci. 12, 622360 (2020).
Vasunilashorn, S. M. et al. Proteome-wide analysis using somascan identifies and validates chitinase-3-like protein 1 as a risk and disease marker of delirium among older adults undergoing major elective surgery. J. Gerontol. Biol. Sci. Med. Sci. 77(3), 484–493 (2022).
Nishimura, N. et al. Chitinase 3-like 1 is a profibrogenic factor overexpressed in the aging liver and in patients with liver cirrhosis. Proc. Natl. Acad. Sci. USA. 118(17). (2021).
Li, Q., Zheng, H. & Chen, B. Identification of macrophage-related genes in sepsis-induced ARDS using bioinformatics and machine learning. Sci. Rep. 13(1), 9876 (2023).
Chen, J. A. et al. A multiancestral genome-wide exome array study of Alzheimer disease, frontotemporal dementia, and progressive supranuclear palsy. JAMA Neurol. 72(4), 414–422 (2015).
Acuner-Ozbabacan, E. S. et al. The structural network of Interleukin-10 and its implications in inflammation and cancer. BMC Genom. 15(Suppl 4(Suppl 4), S2 (2014).
Ding, Y., Li, H., Xu, L., Wang, Y. & Yang, H. Identification and validation of prognostic biomarkers specifically expressed in macrophage in IgA nephropathy patients based on integrated bioinformatics analyses. Front. Mol. Biosci. 9, 884588 (2022).
Zheng, L. et al. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor-dependent repression of claudin-2. J. Immunol. 199(8), 2976–2984 (2017).
Klein, J. & Sato, A. The HLA system. First of two parts. N Engl. J. Med. 343(10), 702–709 (2000).
Blum, J. S., Wearsch, P. A. & Cresswell, P. Pathways of antigen processing. Annu. Rev. Immunol. 31, 443–473 (2013).
Meysman, P. et al. Varicella-zoster virus-derived major histocompatibility complex class I-restricted peptide affinity is a determining factor in the HLA risk profile for the development of postherpetic neuralgia. J. Virol. 89(2), 962–969 (2015).
Rooney, M. S., Shukla, S. A., Wu, C. J., Getz, G. & Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160(1–2), 48–61 (2015).
Li, L. et al. UCHL1 regulated by Sp1 ameliorates cochlear hair cell senescence and oxidative damage. Exp. Ther. Med. 25(2), 94 (2023).
Kumar, R. et al. Epigallocatechin gallate suppresses premature senescence of preadipocytes by inhibition of PI3K/Akt/mTOR pathway and induces senescent cell death by regulation of Bax/Bcl-2 pathway. Biogerontology 20(2), 171–189 (2019).
Zhang, Y. et al. PIN1 Protects hair cells and auditory HEI-OC1 cells against senescence by inhibiting the pI3K/Akt/mTOR pathway. Oxid. Med. Cell. Longev. 2021, 9980444 (2021).
Chen, Y. et al. Beneficial effects of natural flavonoids on neuroinflammation. Front. Immunol. 13, 1006434 (2022).
Arthur, J. S. & Ley, S. C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 13(9), 679–692 (2013).
Eshraghi, A. A. et al. Blocking c-Jun-N-terminal kinase signaling can prevent hearing loss induced by both electrode insertion trauma and neomycin ototoxicity. Hear. Res. 226(1–2), 168–177 (2007).
Kalinec, G. M. et al. Pivotal role of Harakiri in the induction and prevention of gentamicin-induced hearing loss. Proc. Natl. Acad. Sci. USA. 102(44), 16019–16024 (2005).
Wang, J. et al. A peptide inhibitor of c-Jun N-terminal kinase protects against both aminoglycoside and acoustic trauma-induced auditory hair cell death and hearing loss. J. neuroscience: official J. Soc. Neurosci. 23(24), 8596–8607 (2003).
Zheng, A., Chen, Q. & Zhang, L. The Hippo-YAP pathway in various cardiovascular diseases: Focusing on the inflammatory response. Front. Immunol. 13, 971416 (2022).
Zhou, X. et al. YAP Aggravates inflammatory bowel disease by regulating M1/M2 macrophage polarization and gut microbial homeostasis. Cell. Rep. 27(4), 1176–1189 (2019).
Mia, M. M. & Singh, M. K. Emerging roles of the Hippo signaling pathway in modulating immune response and inflammation-driven tissue repair and remodeling. FEBS J. 289(14), 4061–4081 (2022).
Wang, M. et al. Hippo/YAP signaling pathway protects against neomycin-induced hair cell damage in the mouse cochlea. Cell. Mol. Life Sci. 79(2), 79 (2022).
Felix, F. B. et al. Blocking the HGF-MET pathway induces resolution of neutrophilic inflammation by promoting neutrophil apoptosis and efferocytosis. Pharmacol. Res. 188, 106640 (2023).
Someya, S. et al. Age-related hearing loss in C57BL/6J mice is mediated by Bak-dependent mitochondrial apoptosis. Proc. Natl. Acad. Sci. USA 106(46), 19432–19437 (2009).
Yang, W., Vethanayagam, R. R., Dong, Y., Cai, Q. & Hu, B. H. Activation of the antigen presentation function of mononuclear phagocyte populations associated with the basilar membrane of the cochlea after acoustic overstimulation. Neuroscience 303, 1–15 (2015).
Okano, T. et al. Bone marrow-derived cells expressing Iba1 are constitutively present as resident tissue macrophages in the mouse cochlea. J. Neurosci. Res. 86(8), 1758–1767 (2008).
Dong, Y. et al. Differential fates of tissue macrophages in the cochlea during postnatal development. Hear. Res. 365, 110–126 (2018).
Okano, T. Immune system of the inner ear as a novel therapeutic target for sensorineural hearing loss. Front. Pharmacol. 5, 205 (2014).
Hirose, K., Rutherford, M. A. & Warchol, M. E. Two cell populations participate in clearance of damaged hair cells from the sensory epithelia of the inner ear. Hear. Res. 352, 70–81 (2017).
Warchol, M. E. Interactions between macrophages and the sensory cells of the inner ear. Cold Spring Harb Perspect. Med. 9(6). (2019).
Hough, K., Verschuur, C. A., Cunningham, C. & Newman, T. A. Macrophages in the cochlea; an immunological link between risk factors and progressive hearing loss. Glia 70(2), 219–238 (2022).
Zhang, Y. et al. The detrimental and beneficial functions of macrophages after cochlear injury. Front. Cell. Dev. Biol. 9, 631904 (2021).
Tarbell, K. V. et al. Dendritic cell-expanded, islet-specific CD4 + CD25 + CD62L + regulatory T cells restore normoglycemia in diabetic NOD mice. J. Exp. Med. 204(1), 191–201 (2007).
Menges, M. et al. Repetitive injections of dendritic cells matured with tumor necrosis factor alpha induce antigen-specific protection of mice from autoimmunity. J. Exp. Med. 195(1), 15–21 (2002).
Verginis, P., Li, H. S. & Carayanniotis, G. Tolerogenic semimature dendritic cells suppress experimental autoimmune thyroiditis by activation of thyroglobulin-specific CD4 + CD25 + T cells. J. Immunol. 174(11), 7433–7439 (2005).
Jones, J. E. & Corwin, J. T. Regeneration of sensory cells after laser ablation in the lateral line system: Hair cell lineage and macrophage behavior revealed by time-lapse video microscopy. J. neuroscience: official J. Soc. Neurosci. 16(2), 649–662 (1996).
Bhave, S. A., Oesterle, E. C. & Coltrera, M. D. Macrophage and microglia-like cells in the avian inner ear. J. Comp. Neurol. 398(2), 241–256 (1998).
Wynn, T. A., Chawla, A. & Pollard, J. W. Macrophage biology in development, homeostasis and disease. Nature 496(7446), 445–455 (2013).
van der Vlist, M. et al. Macrophages transfer mitochondria to sensory neurons to resolve inflammatory pain. Neuron 110(4), 613–626 (2022). e9.
Covarrubias, A. J. et al. Senescent cells promote tissue NAD(+) decline during ageing via the activation of CD38(+) macrophages. Nat. Metab. 2(11), 1265–1283 (2020).
Ma, Y. et al. Deriving a cardiac ageing signature to reveal MMP-9-dependent inflammatory signalling in senescence. Cardiovasc. Res. 106(3), 421–431 (2015).
Baylis, D., Bartlett, D. B., Patel, H. P. & Roberts, H. C. Understanding how we age: Insights into inflammaging. Longev. Healthspan 2(1), 8 (2013).
Wang, M. & Shah, A. M. Age-associated pro-inflammatory remodeling and functional phenotype in the heart and large arteries. J. Mol. Cell. Cardiol. 83, 101–111 (2015).
Pallikkuth, S. et al. Dysfunctional peripheral T follicular helper cells dominate in people with impaired influenza vaccine responses: Results from the FLORAH study. PLoS Biol. 17(5), e3000257 (2019).
Triposkiadis, F., Xanthopoulos, A. & Butler, J. Cardiovascular aging and heart failure: JACC review topic of the week. J. Am. Coll. Cardiol. 74(6), 804–813 (2019).
He, S. & Sharpless, N. E. Senescence in health and disease. Cell 169(6), 1000–1011 (2017).
Gensous, N. et al. T Follicular helper cells in autoimmune disorders. Front. Immunol. 9, 1637 (2018).
Murayama, K. et al. CD4(+)CD8(+) T follicular helper cells regulate humoral immunity in chronic inflammatory lesions. Front. Immunol. 13, 941385 (2022).
Dong, L. et al. Functional differentiation and regulation of follicular T helper cells in inflammation and autoimmunity. Immunology 163(1), 19–32 (2021).
Zhou, M. et al. The effect of aging on the frequency, phenotype and cytokine production of human blood CD4 + CXCR5 + T follicular helper cells: Comparison of aged and young subjects. Immun. Ageing. 11, 12 (2014).
Iske, J. et al. Senolytics prevent mt-DNA-induced inflammation and promote the survival of aged organs following transplantation. Nat. Commun. 11(1), 4289 (2020).
Blanco, P., Palucka, A. K., Pascual, V. & Banchereau, J. Dendritic cells and cytokines in human inflammatory and autoimmune diseases. Cytokine Growth Factor. Rev. 19(1), 41–52 (2008).
Park, H. et al. A distinct lineage of CD4 T cells regulates tissue inflammation by producing interleukin 17. Nat. Immunol. 6(11), 1133–1141 (2005).
Gudmundsdottir, A. B., Brynjolfsdottir, A., Olafsdottir, E. S., Hardardottir, I. & Freysdottir, J. Exopolysaccharides from Cyanobacterium aponinum induce a regulatory dendritic cell phenotype and inhibit SYK and CLEC7A expression in dendritic cells, T cells and keratinocytes. Int. Immunopharmacol. 69, 328–336 (2019).
Hang, L. et al. Downregulation of the syk signaling pathway in intestinal dendritic cells is sufficient to induce dendritic cells that inhibit colitis. J. Immunol. 197(7), 2948–2957 (2016).
Kalia, N., Kaur, M., Sharma, S. & Singh, J. A Comprehensive in silico analysis of regulatory SNPs of human CLEC7A gene and its validation as genotypic and phenotypic disease marker in recurrent vulvovaginal infections. Front. Cell. Infect. Microbiol. 8, 65 (2018).
Mocsai, A., Ruland, J. & Tybulewicz, V. L. The SYK tyrosine kinase: A crucial player in diverse biological functions. Nat. Rev. Immunol. 10(6), 387–402 (2010).
Turner, M., Schweighoffer, E., Colucci, F., Di Santo, J. P. & Tybulewicz, V. L. Tyrosine kinase SYK: Essential functions for immunoreceptor signalling. Immunol. Today 21(3), 148–154 (2000).
Squires, K. E., Montanez-Miranda, C., Pandya, R. R., Torres, M. P. & Hepler, J. R. Genetic analysis of rare human variants of regulators of G protein signaling proteins and their role in human physiology and disease. Pharmacol. Rev. 70(3), 446–474 (2018).
Feng, Z. et al. Epithelium- and endothelium-derived exosomes regulate the alveolar macrophages by targeting RGS1 mediated calcium signaling-dependent immune response. Cell Death Differ. 28(7), 2238–2256 (2021).
Patel, J. et al. RGS1 regulates myeloid cell accumulation in atherosclerosis and aortic aneurysm rupture through altered chemokine signalling. Nat. Commun. 6, 6614 (2015).
Camarillo, G. F. et al. Gene expression profiling of mediators associated with the inflammatory pathways in the intestinal tissue from patients with ulcerative colitis. Mediators Inflamm. 2020, 9238970 (2020).
Howcroft, T. K. et al. The role of inflammation in age-related disease. Aging (Albany NY) 5(1), 84–93 (2013).
Funding
This research received financial support from the following sources: the National Natural Science Foundation of China (Grant No. 82201272), the Natural Science Foundation of Fujian Province (Grant No. 2022J01217), the Young and Middle-aged Key Personnel Training Project of Fujian Provincial Health Commission (Grant No. 2021GGA024) and the Science and Technology Innovation Joint Funding Project of Fujian Province (Grant No. 2021Y9099).
Author information
Authors and Affiliations
Contributions
X.G., C.C., Y.C., and C.L. conceptualized and designed the study. X.G. conducted the integration of the publicly available ARHL GEO datasets, followed by further data processing and analysis. C.C. conducted the LASSO regression analysis and converted them into human genes. Y.C. evaluated immune infiltration variation between ARHL group and controls by CIBERSORT and applied the MCPcounter algorithm to evaluate the relationship between identified humanized key genes and immune infiltration levels. C.Z. conducted GSEA and GSVA analysis on both high and low-risk groups. Y.L. established interaction networks for mRNA-miRNA, mRNA-transcription factor, and mRNA-RNA-binding protein. R.G. and S.X. conducted the clinical evaluation of IRDEGs and drew the ROC curve. X.G., C.C., Y.C., and C.L. contributed to manuscript writing and revision. All authors reached a consensus and gave their approval for the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gu, X., Chen, C., Chen, Y. et al. Bioinformatics approach reveals the critical role of inflammation-related genes in age-related hearing loss. Sci Rep 15, 2687 (2025). https://doi.org/10.1038/s41598-024-83428-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-83428-x