Abstract
Buruli ulcer (BU) a neglected disease induced by the bacterium Mycobacterium ulcerans, predominantly impacts tropical and subtropical areas with its pathophysiology ascribed to the Mycolactone protein. Current antibiotics frequently prove insufficient to manage advanced or chronic ulcers and the rise of drug resistance presents a considerable challenge. This work aims to address these challenges by employing computational methods to identify therapeutic candidates from organic compounds, which may be developed into more effective therapies for Buruli ulcer. The Gas-Chromatography Mass Spectrometry (GCMS) analysis of the Thymus Vulgaris identified the 29 bioactive compounds as potential drug candidates having different medicinal properties. Out of the 29 compounds against the mycolactone protein, 14 compounds demonstrated a binding affinity higher than − 6 kcal/mol predicted through PyRx. Among all compounds, gamma sitosterol and borneol showed the highest binding affinity − 7.7 kcal/mol. The ADMET analysis predicted that the compound borneol crosses the PGP + through the Blood Brain Barrier and gastrointestinal tract without violating Lipinski’s rule of 5 having high water solubility, and log p-value of 2.29. The molecular dynamic simulation was performed and showed the Eigenvalue of 1.332692e-04. The leads identified in the study have demonstrated encouraging outcomes with regard to their efficacy, toxicity, pharmacokinetics, and safety. Further experimental investigations can be conducted to evaluate their anti-bacterial activity, and their molecular frameworks could be utilized as a valuable foundation for designing new drugs for the treatment of Buruli ulcer.
Similar content being viewed by others
Introduction
Buruli ulcer (BU) is a severe, persistent skin illness caused by the bacteria Mycobacterium ulcerans (M. ulcerans). BU is endemic all over the world, with the majority of cases documented in Africa but also in Australia, Asia, and South America1. Research has revealed that the prevalence of Buruli ulcer is high in areas with slow-moving rivers, streams, and other bodies of water2. Additionally, Azumah and colleagues have conducted animal experiments that demonstrate the pathogenesis and transmission of the disease through Acanthamoeba3. For instance, in 2016, 1676 out of 1864 new cases worldwide were reported in this region4. The World Health Organization (WHO) recognized Buruli ulcer as a priority illness for research and development, urging enhanced efforts to provide new treatments and recommended the combination of either rifampicin and streptomycin or rifampicin and clarithromycin drugs for eight weeks of treatment5,6. However, injecting these drugs in the long term causes several side effects such as cochlear and vestibular damage7. Moreover, the long duration of treatment may cause non-compliance among those patients who live far away from healthcare departments8. In severe cases, surgery is the only option9, but it is expensive and can only be performed at well-equipped medical centers, which are not affordable for most patients. Therefore, new strategies for the treatment of BU with combinations of new drugs with low cost and minimal side effects are urgently needed to enhance the outcomes and well-being of the affected patients10. Mycolactone protein has played a role in the pathogenesis of the BU through multiple aspects such as, including cytotoxicity, inflammation, and immunological suppression11. This protein has been found to have direct cytotoxic effects on host cells, causing skin cells to die and necrotic ulcers to occur10. It hurts the human immune system, permitting M. ulcerans to further proliferate and cause infection12. Some potential advances in the hunt for novel therapies for BU have been made, including the use of antimicrobial peptides, immunomodulatory drugs, and phage therapy13.
In the field of biomedical sciences, the utilization of GCMS-derived plant chemicals for the treatment of Buruli ulcer is one potential area of investigation. Gas Chromatography Mass Spectrometry (GCMS), a technique that combines gas chromatography and mass spectrometry, was first elucidated by Dow Chemical scientists Fred McLafferty and Roland Gohlke in 1955-56. It is an effective analytical method to identify the chemical compounds of the mixtures14. Thymus vulgaris has been used as a medicinal plant to treat several respiratory diseases such as bronchitis, asthma and chronic cough. Depending on the bioactive compounds, vascular ailments such as tooth pain, indigestion and urinary tract diseases could be treatable15. Indian natives utilize T. vulgaris, a medicinal plant, due to its diverse range of beneficial properties. These properties encompass antioxidant, anti-inflammatory, rodenticidal, antibacterial, analgesic, insecticidal, diuretic, and antidiabetic effects.
The bioactive compounds such as flavonoids, alkaloids, and terpenoids exhibit antibacterial action against M. ulcerans. These chemicals might pave the way for the development of novel BU treatments16. In the current study, computer-aided drug design methodology was used to identify the best drug candidate against BU by using homology modeling, molecular docking, interaction analysis and ADMET profiling for the prediction of the toxicity of the drug compound. This study will provide a therapeutic approach for the treatment of the BU infection which is on the verge of becoming a pandemic in the future. Nevertheless, in-vivo, and in-vitro testing are yet to be required for the efficiency of the drug design and for more prediction of safety.
Materials and methods
Sample preparation
The Thymus vulgaris fresh plant (Not wild) was bought from the Islamabad Nursery, Islamabad. The whole leaf sample was washed and dried under shade at room temperature in compliance with the relevant institutional, national, and international guidelines and legislation. After drying, the leaves were made into fine powder in a grinder for 30 s.
Extraction method
The powder sample of T. vulgaris was soaked in the methanol (400 ml) for seven days for the preparation of the T. vulgaris methanolic extract using a Soxhlet extractor. In the process of extracting and evaporating, a semi-solid mass was formed that was amorphous and the extract was dried17.
Gas chromatography- mass spectrometry
The Gas Chromatography-Mass Spectrometry (GC-MS) analysis was performed for the identification of the bioactive compounds present in the methanolic extract of T. vulgaris. The Hewlett-Packard model no 6890 is used for the analysis with a DB-5 capillary tube of film thickness of 0.25 cm, diameter of 0.25 mm and 30 m in length. The flame ionization method was used in GC-MS analysis with a 30 atomic mass unit (amu) scan. The ionization energy was set to 75 eV and carrier gas such as helium was used with an air volume of 1.5 ml/min. The mass ranges between 10 and 2000 amu were detected using Joel Accu Time of Flight Analyzer (TOF) GCV at a resolution of 6000 Hz. Initially, the column was heated at 35 °C for five minutes and then increased up to 280 °C with a gradual rise of 8 °C temperature per minute. The components were separated in the column using a flow rate of 1 mL/min of the carrier gas. NIST MS 2.0 libraries were used to identify the peak area, retention time and name of the compound. This approach provides a faster, less laborious, and cost-effective method for discovering new drugs compared to conventional methods.
In-silico analysis of GCMS-derived compounds against mycolactone protein of BU
Retrieval of protein
The FASTA sequence of mycolactone protein was retrieved from NCBI (National Centre for Biotechnology Information), which is accessible at https://www.ncbi.nlm.nih.gov. The most reliable source for a complete record of nomenclature. It is possible to retrieve sequential references, maps, channels, variants, phenotypes, and genomic linkages.
Prediction of secondary structure of the protein
The self-Optimized Prediction Method with Alignment (SOPMA) server was used for the secondary structure prediction of the mycolactone protein. SOPMA is based on homolog technique and it has been found to considerably enhance the success rate of protein’s secondary structure prediction .
Protein tertiary structure prediction
The tertiary structure or 3D structure of the targeted mycolactone protein had been generated via Alpha Fold https://alphafold.ebi.ac.uk/. A DeepMind AI system called AlphaFold can infer a protein’s three-dimensional structure from its amino acid composition. It consistently delivers accuracy that is on par with experiment. This server’s goal is to make protein modeling available to all life science researchers freely available at science community18.
Retrieval of ligands
At https://pubchem.ncbi.nlm.nih.gov/, the largest database of publicly accessible chemical information, multiple ligands of the GC-MS-derived compounds had been retrieved. By using the name, structure, molecular formula, and other identifiers of the compounds, this database provides information about the toxicity profile, biological activities, chemical and physical properties, patents, citations in the literature, and more.
Mycolactone protein refinement
Refinement of protein mycolactone polyketide synthase has been done through discovery studio at https://discover.3ds.com/discovery-studio-visualizer-download. A special way to investigate biological and physicochemical processes right down to the atomic level is through modeling and simulation techniques. By opening the 3D structure in PDB format, the protein had been refined by deleting all the ligands and water molecules to obtain a purified, refined model19.
Protein validation and quality prediction
The refined protein structure was then validated by Ramachandran plot from PROCHECK which is available at this site SAVESv6.0 - Structure Validation Server (ucla.edu). The Ramachandran plot is a tool to analyze the conformation of amino acid residues in a protein structure to locate energetically favorable or unfavorable areas. The plot reveals the regions with the highest preference, as well as the regions that are moderately or barely acceptable, and the residues that are unacceptable20. Protein quality prediction was done through ProQ - Protein quality prediction ProQ - Protein quality prediction (bioinfo.se) and ProSA-Web (ProSA-web - Protein Structure Analysis (sbg.ac.at). ProQ21 and ProSA22 are web servers that use different methods to evaluate and predict protein structure and function quality. They are useful tools for protein modeling and analysis.
Protein’s active site prediction
The proteins Plus server (https://proteins.plus) was utilized to visualize, analyze, and manipulate the protein structure of the mycolactone. Proteins Plus server also provides tools for molecular modeling, protein engineering, and drug creation and is useful for predicting and refining protein structures. The ProteinsPlus server includes DoGSiteScorer, a tool that enables the identification of binding sites on a specific target of interest. This tool can analyze protein-ligand interactions and identify active regions within a protein structure23.
Screening of compounds using PYRX
Available at https://pyrx.sourceforge.io/downloads, PyRx is a program for virtual screening that can be used in computational drug discovery to compare libraries of compounds against potential drug targets. Pharmaceutical Chemists can use PyRx to conduct Virtual Screening from any platform, and the program assists users at every step of the process, including data preparation, job submission, and result analysis24.
Energy minimization
Energy minimization of ligands attaining the highest binding affinities had been done through discovery studio at https://discover.3ds.com/discovery-studio-visualizer-download. By removing all of the water molecules and ligands from the protein structure. Energy minimization refers to finding the lowest conformation of a molecule in order to stabilize the structure to its maximum level hence ligands and water molecules were removed to minimize the energy because only the minimized structures can provide the microscopic conformational changes, enthalpies of binding, inter-and intraresidue interactions24.
Molecular docking
Auto dock Vina is a program that simulates and evaluates how a small molecule binds to a protein. It helps to find potential drugs from large collections of chemicals. The ligand (Gamma-sitosterol and Borneol) and receptor (Maycolactone) files, typically in formats like PDB or MOL2, were prepared. Specific input parameters, including grid size, exhaustiveness, and scoring function options, were set to control the docking process. Auto dock Vina generated a grid map based on the specified grid size, defining the search space within the receptor for ligand binding evaluation. This software explores various ligand conformations and orientations within the receptor’s binding site to calculate binding affinities. A scoring function estimates the strength of ligand-receptor interactions. Upon completion, Auto dock Vina provided output files containing information about predicted ligand binding poses and their docked scores. Docking will be analyzed by the results to identify favorable ligand binding modes and evaluate binding affinities25.
Prediction of protein-ligand interaction
A computer program called LIGPLOT was used for the prediction of the protein-ligand interactions. It produces plain 2D diagrams of protein-ligand interactions using typical Protein Data Bank file input. Images for the PDBsum resource, which summarizes molecular structure, are created using the LIGPLOT. That had been available at https://www.ebi.ac.uk/thornton-srv/software/LigPlus/download.html26.
Toxicity prediction of the drug candidates
Following the virtual screening, the known ligands among which the top 2 compounds were analyzed by ADME/Tox studies and physio-chemically profiled. Drug toxicity had been predicted by using SWISSADME http://www.swissadme.ch/. SwissADME was developed to meet the needs of drug discovery and medicinal chemistry fields, where a crucial emphasis is placed on maintaining a balance between accuracy and speed to handle a substantial volume of molecules27.
Molecular dynamic simulations
The molecular dynamic simulations were performed through iMODs https://imods.iqfr.csic.es/. The docked complex of the mycolactone protein with the Top Scoring compound was submitted to the server and a simulation was performed. It permits the investigation of collective movements in proteins and nucleic acids utilizing internal coordinates (torsional space) with the aid of NMA (Normal Mode Analysis) by giving atomic coordinates in PDB format. N, CA, and C must be backbone atoms in order to define dihedral angles28.
Results
Retrieval of ligands
Multiple ligands have been retrieved from PubChem; the largest database of freely available chemical information available at https://pubchem.ncbi.nlm.nih.gov/.
Gas chromatography-mass spectrometry
The methanolic plant extract was subjected to GC-MS analysis, and the results are summarized in Table 1. The analysis identified 29 compounds, which accounted for 100% of the detected constituents. The GC-MS chromatogram is presented in Fig. 1.
In-silico analysis of GCMS-derived compounds against mycolactone protein of Buruli Ulcer (BU)
Retrieval of protein
The FASTA sequence of the mycolactone protein was retrieved from the NCBI with the accession ID: WP_096372707 consisting of 2041 amino acids.
Retrieval of ligands
The 29 compounds derived from the GC-MS analysis of the methanolic extract of T. vulgaris were retrieved from the PubChem database. All the compounds were retrieved and further analyzed as the drug candidate.
Prediction of protein secondary structure
Sopma online web server was used for the prediction of secondary structure. Alpha helices (Hh) were detected at about 39.79%, Random coils (Cc) were detected 39.83% and Beta-turns (Tt) were estimated 5.93%, whereas Extended strand (Ee) were also observed and predicted 14.44%. as shown in Fig. 2.
The prediction of the secondary structure of the mycolactone protein using the Sopma online tool. Different lines in different colors present a specific part of the protein in the secondary structure. Blue color represents the Helix, Red color presents the Sheets, Green color presents the Turns and Purple color is for Coils.
Prediction of tertiary structure
The 3D structure of mycolactone protein was generated using Alpha Fold. Model 01 was selected having GMQE (Global Model Quality Estimate) of about 0.71 and downloaded this structure in PDB format. The predicted 3D structure of mycolactone protein is shown in Fig. 3.
Protein validation and quality prediction
The protein underwent quality assessment using the ProQ server, which evaluated its quality based on two measures: The protein was evaluated by Maxsub score and LGscore. Maxsub score measures the number of residues with distances between the model and correct structure shorter than 3.5 Å. LGscore is a p-value score for the significance of a structural similarity match. The Predicted LGscore of the protein was 9.165 and the Maxsub score was 0.296, which indicates that it belongs to the range of a “very good model”. ERRAT value of the predicted mycolactone protein was “99.1456”. The value of ERRAT predicts that the structure of the protein is extremely good. On the basis of Phi (degrees) and Psi (degrees) plot statistics of Ramachandran plot was calculated 90.9%, 8.2%, 0.9% and 0.0% were the regions that were in most favorable areas, additional permitted/allowed regions, generously permitted/allowed regions and prohibited regions accordingly. The total number of mycolactone protein residues is 240 as shown in Fig. 4. The ProSA web showed that modeled protein has a z-score of -15.28, indicating that it lies in the range of X-ray assembled structures of protein as shown in Fig. 6b. A lower z-score indicates a better protein model (Fig. 5).
Pictorial presentation of mycolactone’s z-score, energy plot and predicted protein model by ProSA web server. Figure 6 (A): The mycolactone Z-score (shown by a dot “.”) X-ray crystallography and nuclear magnetic resonance spectroscopy evaluations of all protein chains in the Protein Data Bank revealed the observed range of sequence length. Figure 6 (B): Plot designed on the Energy of the mycolactone protein model.
Binding pockets of proteins
PROTEIN PLUS predicted 118 binding pockets. Selection of the active site was based on the pocket with the highest volume (V) and surface area, which was 665.15 nm3 and 945.4 nm2, respectively shown in Fig. 6. The selected active site consists of 52 residues. The active site contains a range of residues from 990, 1023–1034, 1056–1069, 1253–1260, 1458–1469, 1713–1725, 1982–1986 and 2010–2012.
Prediction of the selected Pockets or active sites with 52 residues. The active site is highlighted in different colors. Purple indicates highly conserved regions in the protein, while yellow represents less conserved regions, and green signifies regions where amino acids have significantly changed during evolution.
3.3.7. Screening of compounds using PYRX
In order to derive the best compound, 29 compounds had been selected and were docked against mycolactone protein using the multiple ligand tool PyRx. On the basis of their binding affinities, 29 compounds had been analyzed. Energy levels vary from − 4.9 to -7.7 kcal/mol. The compound borneol and gamma sitosterol had been chosen as the best candidate with the lowest binding affinity of -7.7 kcal/mol with toxin protein. The list of binding affinities of docked complexes is shown in Table 2.
Molecular docking analysis by auto dock vina
The result of the docking model for borneol with mycolactone protein has shown the highest binding affinity of -8.7 Å, compared to the binding affinity of -8 Å for gamma sitosterol. The docked complex of mycolactone with Gamma-sitosterol and borneol is shown in Fig. 7.
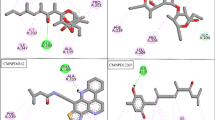
Represents the docked complex using discovery studio. (A) depicts the interaction of mycolactone protein with Borneol with hydrophobic interaction, showing the distance between ligand and receptor with bond types. (B) 2D structure of the docked complex with the aid of AutoDock Vina. (C) the docked complex of mycolactone with Gamma sitosterol showing hydrophobic interactions along with the distance and bond type. (D) 2D illustration of the docked complex of Gamma-sitosterol with mycolactone protein.
Ligplot analysis
The red figure displayed in Fig. 9 depicts the protein-ligand interaction and residues that are not directly involved in the interaction due to the greater bond angle. These depicted red interactions are hydrophobic in nature while blue bonds are hydrogen bonds. Ligplot analysis of docked Complex of mycolactone with Gamma-sitosterol and borneol is also shown in Fig. 8. Table 3 shows the interactions between the docked complexes.
The Protein-ligand interaction of the mycolactone protein with the gamma sitosterol and borneol using Ligplot. Most bondings are hydrophobic where; (A) shows the 2D diagram of Mycolactone interaction with gamma-sitosterol (B) shows the 2D interaction of mycolactone protein with borneol has been shown.
Toxicity prediction of the drug candidates
The top 2 compounds were further analyzed for the ADMET analysis. Gamma-sitosterol and Borneol were analyzed by SWISSADME. Best ADMET properties lie in the category where the log p-value is less than 4 and more soluble. Borneol showed the best ADMET properties and it did not violate any Lipinski’s rule and its log p value was less than 4 with more solubility (Tables 4 and 5). Boiled egg analysis from ADME suggested that if a compound has P-glycoprotein (PGP) -value, it shows that it has a strong bond with P-glycoprotein (PGP) and the drug will target the site after crossing the membrane. In the graphical representation, blue dots indicate molecules that are predicted to be expelled from the central nervous system via P-glycoprotein, whereas red dots represent molecules predicted to remain within the central nervous system without being expelled through P-glycoprotein.
In the provided visualization, the egg yolk points symbolize molecules that have the potential to passively traverse the Blood-Brain Barrier, while the egg white points represent molecules that may passively cross the gastrointestinal tract. The blue and red dots on the plot indicate molecules that are predicted to be expelled or retained by P-glycoprotein, respectively shown in Fig. 9.
Shows the boiled egg analysis by ADME, (A, B) give the best-boiled egg analysis (C) In the provided visualization, the egg yolk points symbolize molecules that have the potential to passively traverse the Blood-Brain Barrier, while the egg white points represent molecules that may passively cross the gastrointestinal tract. The blue and red dots on the plot indicate molecules that are predicted to be expelled or retained by P-glycoprotein, respectively.
Molecular dynamic simulation
In the provided visualization of Fig. 10 the Deformability and B-factors provide insight into the mobility profiles of docked proteins. In the Mycolactone protein-ligand borneol complex, the deformability and B-factors reveal peaks corresponding to regions within the protein structure where the highest peaks represent areas of high deformability. The B-factor graphs serve as a means to compare the Normal Mode Analysis (NMA) and the Protein Data Bank (PDB) fields of the complex.
Eigenvalues and variances are inversely related in each normal mode. Figure 10C displays an eigenvalue of 1.332692e-04 for the docked complex. Meanwhile, the variance graph of the borneol-target protein mycolactone complex depicts individual variances with purple-shaded bars and cumulative variance with green-shaded bars.
The covariance matrix of the complex provides information about correlations among residues. In the matrix, the red color illustrates significant correlations between residues, while white indicates uncorrelated motion, and blue signifies anticorrelations. Higher correlations suggest a more stable complex.
Elastic maps of the docked proteins visually represent associations among atoms, with darker-gray portions indicating stiffer regions, as illustrated in Fig. 10.
The Molecular Dynamics Simulation using IMODS of Mycolactone protein with ligand borneol (A) deformability graph, (B) B-factor, (C) eigenvalue, (D) variance map (with purple showing Individual variance and green showing total variance), (E) covariance map (with red representing correlated and blue representing anti-correlated motions), (F) elastic networking (with darker gray areas representing stiffer regions).
Discussion
Medicinal plants have played a crucial role in drug discovery due to the presence of bioactive compounds such as saponins, phenolics, steroids, lignans, alkaloids, terpenes and glycoside complexes29. Due to their biological and therapeutic properties, all these compounds have been used for the production of pharmaceutical drugs since ages. Compounds rich in polyphenols prove to be recommended for protection against ultraviolet-induced skin damage as T. vulgaris extract and thymol30. According to the Food and Drug Administration, the T. vulgaris plant is safe for human consumption and has no limitations on intake. For oral toxicity, oil showed a high lethal dose of 50 at 4 mg/kg and was recommended as non-toxic and safe31.
Antibiotic therapy is the primary treatment for BU, and the medicines suggested for eight weeks are rifampicin and streptomycin or rifampicin and clarithromycin. Furthermore, wound care, such as debridement and skin grafting, is critical for supporting healing and avoiding subsequent infections32. Surgery is often reserved for severe instances of BU that have failed to respond to antibiotics or wound care alone. However, these medications are not helpful in treating BU; hence, new and more effective treatments are needed18. The current study shows the docking analysis of the mycolactone protein of BU infection with the GC-MS-derived compounds from the medicinal plant known as Thymus vulgaris. The 3D structure of the protein was generated through SWISS-MODEL and quality structure prediction was performed through PROSA and PROCHECK33. The value of PROSA − 15.28 indicates that the mycolactone protein has fallen within the range of X-ray crystallographic structures, as the more negative z-score implies a better protein structure. The RAMACHANDRAN analysis shows 96.7% amino acids in allowed regions and 1.8% in disallowed regions which explains that the predicted protein is stable.
The screening of the compounds was performed and among all compounds, fourteen compounds showed a higher binding affinity towards mycolactone. These compounds show binding affinity greater than the threshold value which is considered effective against the target that is greater than − 6.0 kcal/mol. For a ligand to interact with its receptor effectively, it must dock to an active pocket and interact with specific amino acid residues that are crucial for protein-ligand interactions34. Among these 14 compounds borneol and gamma sitosterol showed the same docking energy of -7.7 which is further re-docked after energy minimization of compounds and energy increases up to -8.7 and − 8.0, respectively. Our results are similar to the study conducted that predicted the isocitrate lyase (ICL) protein of M. ulcerans and docked against natural compounds. The docking analysis showed the binding affinity of Euscaphic acid − 8.6 which had been reported as the drug-sensitive strain of Mycobacterium tuberculosis35. The study conducted by Sundarraj, S et al., 2012, provides the in vitro results, which support the ethnomedical use of γ-Sitosterol against cancer. The experimental results suggest that γ-Sitosterol exerts potential anticancer activity through cell cycle arrest, growth inhibition and apoptosis of cancerous cells36.
The toxicity analysis of the drug candidate is mandatory as it analyzes the lipophilicity, toxicity and solubility of the active compound37. The ADMET analysis of the top 5 predicted compounds was performed and results showed that borneol exhibits good ADMET properties while gamma sitosterol is poorly soluble. The lipophilicity iLogP value of borneol is 2.29 which is less than 5 which means that it does follow the five rule of Lipinski’s which is that the compound molecular weight should not be more than 500 Da, hydrogen bond acceptors not greater than 10 and hydrogen bond not greater than 5. Moreover, the log-p value should not be more than 538. An in-silico study conducted by Kwofie et al., evaluated the docking analysis and ADMET properties of against BU. The results show multiple ranges among which the top four compounds show affinities range in between 7.4 kcal/mol to − 8.0 kcal/mol with ZINC IDs ZINC000018185774, ZINC000095485921, ZINC000014417338 and ZINC000005357841. Their ADMET analysis shows that among these, two were accepted means showing good ADMET profiling and two were rejected the iron-dependent regulator (ideR) against 832 compounds for the potential drug designing39.
The current study shows that drug candidates derived from medicinal plant borneol have a high binding affinity with increased water solubility Log S -2.27 of soluble class, high GI absorption and it does cross the blood-brain barrier (BBB). These properties of borneol make it the best active compound and soluble drug candidate. The EGG-boiled analysis shows that the molecule may cross the blood-brain barrier and gastrointestinal tract as shown in Fig. 10f. Among different drug delivery systems used for the treatment of BU, hydrogels have been especially used for wound healing purposes and retain high water retention capability. However, these hydrogels could be easily applied and removed without affecting the wound but have poor water-soluble drug delivery systems due to their intrinsic hydrophobic nature40. In a work published in 2020, molecular docking simulations were used to find chemicals that can bind to and block the enzyme mycolactone, which is generated by M. ulcerans and contributes to the pathophysiology of Buruli ulcer41. Several substances, including quercetin and kaempferol, were shown to have potential inhibitory effects against mycolactone in recent research42.
A study conducted by Sen, S et al., shows the efficacy of streptomycin and rifampin drugs against BU. The oral administration of these drugs for 2 months causes potential side effects such as hearing loss43. Our study indicates the drug candidate borneol from a medicinal plant against mycolactone of BU shows no toxicity and side effects as shown from the ADMET analysis of the compounds and toxicity profiling. Further investigation, such as in-vivo studies, must be performed as drug detection and dose limits should be considered. This suggests that natural compounds borneol and gamma sitosterol derived from the Thymus vulgaris plant have therapeutic potential against BU. The combination of GC-MS-derived plant-based chemicals and in silico drug design offers great potential for the development of novel Buruli ulcer therapies. Researchers can speed up the drug development process and perhaps enhance treatment outcomes for this neglected disease by discovering and optimizing the most promising molecules from natural sources.
Conclusion
Mycolactone is a toxin secreted in the host tissue in response to the bacterial infection caused by the Buruli ulcer. The treatment available for BU such as rifampicin and streptomycin have adverse effects as long-term administration of these drugs causes hearing loss, nephrotoxicity, etc. The need for new strategies for the treatments of Buruli ulcer has increased based on natural products as they are having no side effects or toxicity. The bioactive compounds obtained from the GCMS analysis of the methanolic extract of the Thymus vulgaris plant were investigated as the drug candidates against the mycolactone protein of Buruli ulcer. The 3D structure of the protein was generated and docked against the 29 bioactive compounds via predicted binding sites. Among 29, two compounds, borneol and gamma sitosterol, showed the best binding affinity and were further analyzed for the protein-ligand interactions and ADMET analysis. Pharmacological profiling of borneol showed good water solubility, no toxicity and lipophilicity value less than five. Validating the efficacy of the lead compounds through biochemical assays could establish their significance as inhibitors of the mycolactone protein, which plays a vital role in the development of Buruli ulcer. The predicted leads can also be frameworks for developing more potent drugs to treat Buruli ulcers.
Data availability
All the data has been included in the manuscript.
References
Asiedu, K. & Etuaful, S. Socioeconomic implications of Buruli ulcer in Ghana: a three-year review. Am. J. Trop. Med. Hyg. 59 (6), 1015–1022 (1998).
Guarner, J. Buruli ulcer: review of a neglected skin mycobacterial disease. J. Clin. Microbiol. 56 (4), e01507–e01517 (2018).
Azumah, B. K. et al. Experimental demonstration of the possible role of Acanthamoeba polyphaga in the infection and disease progression in Buruli Ulcer (BU) using ICR mice. PLoS One. 12 (3), e0172843 (2017).
Tabah, E. N. Skin Neglected Tropical Diseases in Cameroon: The Need for Integrated Control and Elimination (University_of_Basel, 2018).
Geourjon, C. & Deléage., G. SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 11 (6), 681–684 (1995).
Ogbechi, J. Investigating the Mechanism behind the Tissue Necrosis in Mycobacterium ulcerans Infection (University of Surrey, 2017).
Klis, S. et al. Long term streptomycin toxicity in the treatment of Buruli Ulcer: follow-up of participants in the BURULICO Drug Trial. PLoS Negl. Trop. Dis. 8 (3), e2739 (2014).
Koka, E. Health seeking behaviour for Buruli Ulcer disease in the Obom sub-district of the Ga South Municipality of Ghana. Health 4, p43 (2020).
Leão et al. Tuberculosis, Leprosy, and other mycobacterioses, in Bioinformatics in Tropical Disease Research: A Practical and Case-Study Approach (Internet) (National Center for Biotechnology Information (US), 2007).
Röltgen, K. & Pluschke, G. Overview: Mycolactone, the macrolide toxin of Mycobacterium ulcerans. Methods Mol. Biol. 2387, 105–108 (2022).
Foulon, M. et al. Mycolactone toxin induces an inflammatory response by targeting the IL-1β pathway: mechanistic insight into Buruli ulcer pathophysiology. PLoS Pathog. 16 (12), e1009107 (2020).
Hall, B. S. et al. The pathogenic mechanism of the Mycobacterium ulcerans virulence factor, mycolactone, depends on blockade of protein translocation into the ER. PLoS Pathog. 10 (4), e1004061 (2014).
Lee, E. J. et al. A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1Fo ATP synthase. Cell 154 (1), 146–156 (2013).
Smirnov, A. et al. ADAP-GC 4.0: application of clustering-assisted multivariate curve resolution to spectral deconvolution of gas chromatography–mass spectrometry metabolomics data. Anal. Chem. 91 (14), 9069–9077 (2019).
Selvaraj, C. Molecular modeling and drug design techniques in microbial drug discovery. Essentials Bioinf. II (In Silico Life Sciences: Medicine), 185–231 (2019).
Tsouh Fokou, P. V. et al. Update on Medicinal Plants with Potency on < i > Mycobacterium ulcerans. BioMed. Res. Int. 2015, 917086. (2015).
Saleem, A. et al. HPLC, FTIR and GC-MS analyses of Thymus vulgaris phytochemicals executing in vitro and in vivo Biological activities and effects on COX-1, COX-2 and gastric Cancer genes computationally. Molecules 27 (23), 8512 (2022).
Waterhouse, A. et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46 (W1), W296–W303 (2018).
Yang, J. et al. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. 117 (3), 1496–1503 (2020).
Suhaibun, S. R. et al. Technology advance in drug design using computational biology tool. Malays J. Med. Health Sci. 16, 2636–9346 (2020).
Wallner, B. & i Elofsson, A. Pcons5: combining consensus, structural evaluation and fold recognition scores. 4248–4254 (2005).
Perera, D., Perera, K. M. L. & Peiris, D. C. A novel in silico benchmarked pipeline capable of complete protein analysis: a possible tool for potential drug discovery. Biology 10 (11), 1113 (2021).
Volkamer, A. et al. Analyzing the topology of active sites: on the prediction of pockets and subpockets. J. Chem. Inf. Model. 50 (11), 2041–2052 (2020).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J. Comput. Chem. 30 (16), 2785–2791 (2009).
Fährrolfes, R. et al. Proteins plus: a web portal for structure analysis of macromolecules. Nucleic Acids Res. 45 (W1), W337–W343 (2017).
Talele, T. T. et al. Successful applications of computer aided drug discovery: moving drugs from concept to the clinic. Curr. Top. Med. Chem. 10 (1), 127–141 (2010).
Daina et al. SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7 (1), 42717 (2017).
Lopéz-Blanco et al. iMod: multipurpose normal mode analysis in internal coordinates. Bioinformatics 27 (20), 2843–2850 (2011).
Rizwan, B. Therapeutic potential of Thymus vulgaris: A Review (2021).
Li, X. et al. Traditional uses, chemical constituents and biological activities of plants from the genus Thymus. Chem. Biodivers. 16 (9), e1900254 (2019).
Mothay, D. et al. Binding site analysis of potential protease inhibitors of COVID-19 using AutoDock. Virusdisease 31 (2), 194–199 (2020).
Yeboah-Manu, D. et al. Secondary bacterial infections of Buruli ulcer lesions before and after chemotherapy with streptomycin and rifampicin. PLoS Negl. Trop. Dis. 7 (5), e2191 (2013).
Hagar, M. et al. Investigation of some antiviral N-heterocycles as COVID 19 drug: molecular docking and DFT calculations. Int. J. Mol. Sci. 21 (11), 3922 (2020).
Mahanthesh, M. et al. Swiss ADME prediction of phytochemicals present in Butea monosperma (Lam.) Taub. J. Pharmacognosy Phytochemistry. 9 (3), 1799–1809 (2020).
Kwofie, S. K. et al. In silico screening of isocitrate lyase for novel anti-buruli ulcer natural products originating from Africa. Molecules 23 (7), 1550 (2008).
Sundarraj, S. et al. γ-Sitosterol from Acacia nilotica L. induces G2/M cell cycle arrest and apoptosis through c-Myc suppression in MCF-7 and A549 cells. J. Ethnopharmacol. 141 (3), 803–809 (2012).
Coutanceau, E. et al. Modulation of the host immune response by a transient intracellular stage of Mycobacterium ulcerans: the contribution of endogenous mycolactone toxin. Cell. Microbiol. 7 (8), 1187–1196 (2005).
Bakchi, B. et al. An overview on applications of SwissADME web tool in the design and development of anticancer, antitubercular and antimicrobial agents: A medicinal chemist’s perspective. J. Mol. Struct. 132712 (2022).
Kwofie, S. K. et al. Molecular informatics studies of the iron-dependent regulator (ideR) reveal potential novel anti-mycobacterium ulcerans natural product-derived compounds. Molecules 24 (12), 2299 (2019).
Hall, B. S. et al. The one that got away: how macrophage-derived IL-1β escapes the mycolactone-dependent Sect. 61 blockade in Buruli ulcer. Front. Immunol. 12, 5933 (2022).
Jeon, B. & Cizdziel, J. V. Determination of metals in tree rings by ICP-MS using ash from a direct mercury analyzer. Molecules 25 (9), 2126 (2020).
Röltgen, K. & Pluschke, G. Buruli ulcer: history and disease burden. Buruli ulcer: Mycobacterium ulcerans disease. 1–41 (2019).
Sen, S. & Chakraborty, R. Revival, modernization and integration of Indian traditional herbal medicine in clinical practice: importance, challenges and future. J. Traditional Complement. Med. 7 (2), 234–244 (2017).
Acknowledgements
Authors are thankful to the Researchers Supporting Project number (RSPD2025R568), King Saud University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Conceptualization, Muhammad Naveed; methodology, Imran Ali; software, Ayesha Saleem; validation, Mitub Al-harbi; formal analysis, Ayaz Ali Khan, investigation, Zeerwah Rajpoot; resources, Sameera Khalil.; data curation, Tariq Aziz.; writing—original draft preparation, Thamer H Albekairi.; writing—review and editing, Tariq Aziz; visualization, Ayaz Ali Khan supervision, Tariq Aziz and Muhammad Naveed.; project administration, Muhammad Naveed.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Naveed, M., Ali, I., Aziz, T. et al. Computational and GC-MS screening of bioactive compounds from Thymus Vulgaris targeting mycolactone protein associated with Buruli ulcer. Sci Rep 15, 131 (2025). https://doi.org/10.1038/s41598-024-83908-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-83908-0