Abstract
Recent studies indicate that Secretogranin V (SCG5) is aberrantly expressed in various cancers and may be linked to tumor progression and prognosis. This study aims to evaluate the potential of SCG5 as a prognostic biomarker for non-small cell lung cancer (NSCLC). We employed a combination of bioinformatics analysis, Western blotting, and immunofluorescence techniques to investigate the role of SCG5 in NSCLC. A comprehensive analysis of TCGA and GEO pan-cancer datasets revealed a consistent upregulation of SCG5 across multiple cancer types. In NSCLC, SCG5 expression was significantly higher in tumor tissues compared to normal lung tissues (p < 0.001). Kaplan-Meier survival analysis demonstrated that patients with elevated SCG5 expression exhibited lower overall survival rates, suggesting a strong association with poor prognosis. Univariate and multivariate COX regression analyses, conducted on both TCGA cases and our collected patient data, confirmed SCG5 as an independent prognostic factor for NSCLC. Furthermore, immune infiltration analysis indicated a significant correlation between SCG5 expression and various immune cell subpopulations, underscoring its potential role as a biomarker for adverse outcomes. Western blot analysis further validated the elevated levels of SCG5 in NSCLC tissues and cell lines compared to their normal counterparts. Based on our findings, we hypothesize that SCG5 may serve as a valuable biomarker for predicting the prognosis of non-small cell lung cancer, thereby guiding future research in the fields of diagnosis, progression, therapy, and prognosis of NSCLC.
Similar content being viewed by others
Introduction
Lung cancer is the most common form of cancer and the leading cause of cancer death, accounting for 18.0% of all cancer deaths1. The prognosis for lung cancer is extremely poor, with a five-year survival rate of only 10–20% of patients diagnosed with lung cancer in most countries2. Non-small cell lung cancer is a highly heterogeneous tumor that accounts for approximately 85% of all newly diagnosed lung cancers3. Although existing biomarkers such as epidermal growth factor receptor EGFR mutations, ALK rearrangements, and PD-L1 expression have significantly advanced the diagnosis and treatment of non-small cell lung cancer, they still possess certain limitations in predicting patient prognosis and guiding personalized therapy. Therefore, the development of new biomarkers is of considerable importance for the early diagnosis, evaluation of disease prognosis, and decision-making regarding treatment for non-small cell lung cancer4. Therefore, it is crucial to investigate new NSCLC biomarkers in order to understand the mechanisms of NSCLC.
The SCG5 gene, also known as Secretogranin V, is situated in band 3 of region 1 on the long arm of human chromosome 15. Spanning approximately 55 kb in size, this gene comprises six exons5. Research suggests that the neuroendocrine protein 7B2, which is encoded by SCG5, functions as a molecular chaperone for the proteogen converting enzyme PC2. This role involves preventing the premature activation of PC2 in the regulatory secretory pathway6,7. Notably, abnormal expression of PC2 has been linked to the proliferation and differentiation of various tumor cells8. Studies on human pheochromocytoma have established a direct correlation between PC2 and endocrine gland tumors. Furthermore, PC2 expression has been observed in tumors such as breast cancer, colon cancer, and small cell lung cancer (SCLC)9,10. SCG5 is significantly linked to the prognosis of various cancers. Research has shown that SCG5 exhibits substantial differences in gene expression between brain metastases and lymph node metastases in breast cancer patients. Patients with elevated levels of SCG5 expression in the primary tumor had a lower rate of distant metastasis-free survival compared to patients with lower levels of SCG5 expression11. In another study on cutaneous squamous cell carcinoma (cSCC), SCG5 was identified as a hub gene associated with cSCC metastasis. Experimental validation revealed that increased expression of the hub gene SCG5 has a notable negative impact on the prognosis of cSCC12.
The role of the SCG5 gene in NSCLC remains unknown. This study conducted bioinformatics analysis to examine the molecular profile of SCG5, analyzing its expression in various cancers using public databases. The research also assessed the expression pattern and prognostic significance of SCG5 in lung cancer. Furthermore, differential expression and immune infiltration analyses were performed, and the expression of SCG5 in NSCLC was validated using clinical tissues. Overall, the findings indicate that SCG5 serves as a significant biomarker for non-small cell lung cancer.
Materials and methods
SCG5 gene information
The location of the SCG5 gene in human chromosomes and its subcellular localization were visualized using the GeneCards database13. The OPENTARGET platform14was utilized to investigate the association of the SCG5 gene with disease.
Differential and correlated expression analysis
Differential expression of the SCG5 gene across various cancer types was demonstrated using the TIMER database15 (http://timer.cistrome.org/). The analyses were conducted in R version 3.6.3 to investigate differences in SCG5 expression at the mRNA level in different disease states (tumor or normal). The results were visualized through box plots and paired sample line plots. Microarray datasets (GSE19804, GSE118370, GSE27262, GSE33532) were acquired from the Gene Expression Omnibus (GEO) database to identify genes implicated in NSCLC development. The GSE19804 dataset comprises 120 samples (T = 60, N = 60). The GSE118370 dataset contains 12 samples (T = 6, N = 6). The GSE27262 dataset includes 50 samples (T = 25, N = 25). The GSE33532 dataset is composed of 100 samples (T = 80, N = 20; these samples are derived from four distinct sites of a single primary tumor and their matched distant normal lung tissues from 20 patients). More detailed information can be found in the Supplementary Information. Differentially expressed genes (DEGs) were illustrated using volcano plots, and relevant genes were identified from the dataset and displayed as heat maps.
Kaplan-Meier survival analysis
The Kaplan-Meier curve16 (https://kmplot.com/analysis/), also known as the survival curve, is a widely used method in survival analysis to assess the impact of a single factor on survival rates. To enhance the assessment of how SCG5 gene expression influences overall survival (OS) and post-progression survival (PPS) in patients with non-small cell lung cancer, which encompasses both lung adenocarcinoma (LUAD) and lung squamous carcinoma (LUSC), we conducted an online analysis of the GSE and TCGA datasets using resources available on this site (data for the GSE and TCGA were sourced from the KM online analysis platform). Kaplan-Meier survival curves were generated using the survminer package in the R programming language, and the significance of differences in survival rates was evaluated employing the Log-rank test.
Correlation expression analysis and tumor immune infiltration analysis
The protein-protein interaction (PPI) network for SCG5 was established using data sourced from the STRING database17 (http://string-db.org) (Version: 12.0). Interactions with PPI scores exceeding − 0.40 were selected for visualization purposes. Immune infiltration analysis was performed using the single sample GSEA (ssGSEA) algorithm in the R package GSVA (v3.6). The final results showed the difference in 24 immune cell infiltration scores between the high and low SCG5 expression groups by comparing boxplots between groups.
Univariate and multivariate COX regression analyses
The relationship between clinicopathological characteristics and prognosis was analyzed through univariate and multivariate COX regression analyses to identify the independent impact of each variable on patient prognosis. Patient data from the TCGA database were utilized, with a study sample consisting of 1026 patients. Variables included in the analysis were pathological T stage, high and low SCG5 expression, pathological M stage, primary treatment outcome, sex, age, tumor location, tumor type, and number of pack-years of smoking. To further validate the independent prognostic role of SCG5 in patients with NSCLC, we collected clinical samples from 95 NSCLC patients. We then conducted univariate and multivariate COX regression analyses on the collected data and plotted Kaplan-Meier survival curves.
Nomogram construction
To further evaluate individual patients, we developed a prognostic nomogram statistical prediction model using the clinical data and results of COX regression analysis. Each variable’s impact on survival prognosis was quantified as a score in the nomogram, with a corresponding score interval for each variable. These scores were then summed to provide a total score, which was utilized to predict patient survival at one, three, and five years through the nomogram.
Plotting the ROC curve
Differential RNA-Seq levels of the SCG5 gene were employed as a biomarker to differentiate lung cancer tissue from healthy lung tissue. The performance of SCG5 expression in distinguishing lung cancer tissue from normal lung tissue was evaluated using ROC curves generated with the pROC package in R software.
Cell culture
The human lung cancer cell lines A549(ATCC number : CCL-185 ), H1975(ATCC number : CRL-5908 ), H23(ATCC number : CCL-185), H1299(ATCC number : CRL-5803 ), H460(ATCC number : HTB-177), and 4006(ATCC number : CRL-5800 ) utilized in the study were sourced from Procell, China. A549, H1975, H1299, H23 and 4006 cells were maintained at 37 °C in a 5% CO2 incubator with humidification. The culture medium consisted of 45 ml RMPI-1640 basal medium (Procell, China), supplemented with 5 ml 10% fetal bovine serum (Procell, China) and 1 ml penicillin-streptomycin solution (NCMbiotech, China).
Human normal lung epithelial cells BEAS-2B were procured from Procell, China. These cells were maintained at 37 °C in a humidified incubator with 5% CO2. The culture medium consisted of 45 ml of DMEM high-sugar basal medium (Procell, China), supplemented with 5 ml of 10% fetal bovine serum (Procell, China), and 1 ml of penicillin-streptomycin solution (NCMbiotech, China). Cell passaging was carried out using 0.25% Trypsin-EDTA (NCMbiotech, China) when cells reached 80% confluency.
Sources of human tissue specimens
This study analyzed 95 cases of non-small cell lung cancer admitted to Yancheng Third People’s Hospital (The Sixth Affiliated Hospital of Nantong University) from June 2014 to June 2022. The research parameters encompassed basic patient information (age, gender), clinical characteristics (smoking history, tumor location, pathological type), staging indicators (pathological T stage, pathological M stage, TNM staging, histological staging, lymph node metastasis status), treatment outcomes (initial treatment effect), molecular features (SCG5 gene expression level) and samples from peritumor and tumor tissue. Regarding sample processing, surgically removed tissue samples were promptly placed in ice boxes. Following aseptic sampling procedures, two preservation methods were employed: samples for Western blot detection were stored in a liquid nitrogen environment, while samples for immunohistochemical analysis were fixed with 10% formalin, made into 5 μm thick frozen sections, and ultimately preserved at -80 °C. The inclusion criteria were as follows: (1) histopathologically confirmed NSCLC; (2) age range from 18 to 90 years; (3) surgical indication without neoadjuvant radiotherapy; (4) no significant surgical contraindications; (5) exclusion of endocrine and metabolic diseases; (6) no history of mental illness. All subjects completed more than five years of follow-up. This study was approved by the Ethics Committee of the Third People’s Hospital of Yancheng City (Affiliated Hospital 6 of Nantong University) (Ethical Approval No: LS2023-46), and written informed consent was obtained from all participating patients.
Western blot
Proteins were initially extracted from the cells, washed twice with PBS (Procell, China), and then treated with RIPA lysate (Servicebio, China) and PMSF solution (Beyotime, China) in a ratio of 100:1. The mixture was resuspended on ice for 30 min. The pre-cooled centrifuge (15 min, 4 °C, 14,000 rpm) was used to separate the lysed and fragmented cells, retaining the supernatant for protein quantification analysis. Protein samples containing 50 µg were then separated by SDS-PAGE and transferred to PVDF membranes. Following blocking with 5% skimmed milk for two hours, the membranes were rinsed with TBST and incubated overnight at 4 °C with primary antibodies (Proteintech, China). Subsequently, the membranes were rinsed with TBST and incubated with a secondary antibody for one hour at room temperature in the absence of light. Immunoblotting was carried out using an enhanced chemiluminescence kit (NCMbiotech, China) and a fully automated chemiluminescence image analysis system (Tanon-5200Multi, China) was utilized. The SCG5 antibody (Proteintech, Cat No. 10761-1-AP) was diluted to a ratio of 1:1000, while GAPDH (Proteintech, Cat No. 10494-1-AP) was diluted to 1:10000 and served as the reference protein for standardizing protein expression levels.
Immunohistochemistry and Immunofluorescence
Paraffin was removed using xylene, followed by hydration through a series of alcohol concentration gradients. Next, the sections were treated with a 3% hydrogen peroxide solution to inhibit endogenous peroxidase activity and embedded in 5% BSA. Subsequently, the sections were incubated overnight at 4 °C with a dilution of primary antibody (rabbit polyclonal antibody, dilution of 1:500, Proteintech, China), washed with PBST solution, and underwent secondary antibody incubation for 30 min at 37 °C. After another wash with PBST solution, the sections were stained with DAB. Dropwise addition of DAB color development solution was done for 2 min, followed by rinsing with distilled water. The sections were then restained with hematoxylin for 3 min, rinsed with PBS until satisfactory staining was achieved, dehydrated, and embedded in neutral resin. Finally, the sections were observed and photographed under a microscope.
A549 cells were cultured in 12-well plates and processed for cell slides. The medium was aspirated and rinsed with a PBS solution. The cells were fixed with 4% paraformaldehyde for 10 min at room temperature, followed by three washes with ice-cold PBS solution. Subsequently, the cells were incubated with PBS containing 0.1-0.25% Triton X-100 for 10 min and washed three times with PBS solution. Following this, the cells were treated with 5% BSA for 30 min at room temperature, then incubated with a primary antibody dilution (rabbit polyclonal antibody, dilution of 1:500, Proteintech, China) at 4 °C overnight. After three PBS washes, the cells were exposed to a fluorescence-labeled secondary antibody dilution for 1 h at room temperature in the dark. The cells were then washed three times with PBS for 5 min each under light protection. Subsequent to staining the nuclei with DAPI for 5 min in the dark, the cells were washed with PBS. Finally, the coverslips were sealed with a drop of anti-fluorescence quenching sealer, and the images were examined and captured using a fluorescence microscope.
Statistical analysis
We conducted a differential analysis of gene expression data utilizing the DESeq2 software package. This package employs a negative binomial distribution model to identify significantly differentially expressed genes while normalizing the sequencing depth across samples. To minimize false-positive results arising from the analysis of numerous gene differences, we applied the Benjamini-Hochberg method to correct all p-values for the false discovery rate (FDR). Only genes with an FDR of less than 0.05 were deemed statistically significantly different. Quantitative data were obtained by analyzing the grayscale values of the Western blot bands using Image J software. The grayscale values of the target protein and the internal reference protein were subsequently processed using GraphPad Prism software for statistical analysis.
To evaluate the relationship between SCG5 gene expression and patient prognosis, we conducted univariate and multivariate COX proportional hazards regression analyses. Additionally, Kaplan-Meier survival curves and Log-rank tests were employed to assess the differences in survival between patients with high and low SCG5 gene expression. The hazard ratio (HR) and its 95% confidence interval (CI) were utilized to quantify the impact of SCG5 gene expression on patient survival. All statistical analyses were carried out using R software, with a significance threshold set at p < 0.05.
Measurements in this study were presented as mean ± SD (standard deviation of the mean). Statistical significance between groups was assessed using Student’s t-test, with significance levels defined as: *P < 0.05; **P < 0.01; ***P < 0.001.
Results
SCG5 localization and associated disease networks
To investigate the functional characteristics and pathological relevance of the SCG5 gene, we performed a systematic search of the GeneCards database. The results show that the 7B2 protein, as the encoded product of SCG5, has a tissue-specific distribution and is mainly located in the extracellular matrix and nuclear regions. At the same time, its expression was also detected in cytoskeletal components (Fig. 1A). From the perspective of genomic localization, this gene was confirmed to be located on human chromosome 15 (Fig. 1B). Based on the gene-disease network construction and analysis, SCG5 shows significant associations with various types of diseases, covering the spectrum of hereditary/familial/congenital diseases, digestive system diseases, and various malignant tumors (Fig. 1C).
Bioinformatics analysis shows that SCG5 is highly expressed in lung cancer cells and tumor tissue
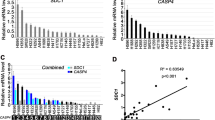
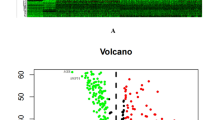
We conducted a pan-cancer analysis of SCG5 expression across multiple cancer types. Utilizing the online analysis data from the TIMER database, we observed that SCG5 generally exhibited an upward trend in various cancers, including invasive breast cancer, colon cancer, head and neck squamous cell carcinoma, and lung adenocarcinoma (Fig. 2A-B-C). Further analysis revealed that the expression level of SCG5 in these cancers was significantly higher than in normal tissues, suggesting that SCG5 may play a crucial role in cancer occurrence and development. By validating the data from the GTE database, we further examined the expression status of SCG5, which confirmed its high expression levels across multiple cancers (Fig. 2F). This finding aligns with the results from paired sample comparisons (Fig. 2E), further indicating the potential of SCG5 as a biomarker. Additionally, volcano plot analysis identified differentially expressed genes in lung cancer tissues, showing that SCG5 expression was up-regulated to a notable extent in these tissues (Fig. 2G). The heatmap illustrated the top 20 differentially expressed genes between normal lung tissues and lung cancer tissues, highlighting a significant difference in SCG5 expression between these two tissue types, particularly the relatively high levels in lung cancer tissues, which further supports the biological significance of SCG5 in lung cancer (Fig. 2D).
Comparison of SCG5 gene expression levels in normal and tumor tissues. (A–C) TIMER database showing SCG5 expression in pan-cancerous tissues; (D) Heatmap visualization of differential gene expression levels in normal lung tissue versus lung cancer tissue; (E, F) Differential expression of SCG5 in NSCLC and its paired adjacent tissue; (G) Volcano plot of differentially expressed genes.
PPI network analysis and immune cell infiltration analysis of the SCG5 gene
Utilizing the STRING database, the protein-protein interaction network was built, identifying the ten chaperone proteins with the highest predicted scores, which are listed as follows: Based on their STRING scores (Fig. 3A), the top ten chaperone proteins include: PCSK2 (0.983), SCG3 (0.925), CHGB (0.893), SCG2 (0.865), CHGA (0.800), PCSK1 (0.786), GREM1 (0.761), FMN1 (0.739), PCSK1N (0.693), and CPE (0.610). These proteins exhibit a significant correlation with SCG5 in the STRING database, suggesting they may hold crucial roles in the regulation and function of SCG5. Further experimental investigations are necessary to confirm the biological relevance of these interactions. Additionally, we analyzed the correlation between infiltration scores of 24 immune cells and SCG5 gene expression in lung cancer patients from the TCGA database. Our findings revealed a significant positive correlation between SCG5 expression in lung cancer and the infiltration of various immune cells, including CD8 T cells and Treg cells (Fig. 3B). By constructing a PPI network, we identified the interaction relationships between SCG5 and other proteins. Concurrently, we conducted an immune infiltration analysis to evaluate the correlation between the expression level of SCG5 and the degree of immune cell subset infiltration in lung cancer patients. This multi-dimensional analytical approach not only facilitates the exploration of the interaction mechanisms of SCG5 with other key proteins at the molecular level but also suggests that SCG5 may play a crucial role in the tumor microenvironment by modulating the infiltration patterns of immune cells, thereby influencing tumor progression.
Western blot and immunohistochemistry showed that SCG5 was highly expressed in lung cancer cells and tumor tissue
To validate the favorable results observed in the bioinformatics analysis, we examined the expression levels of SCG5 in human NSCLC tissues and NSCLC cell lines. Western blot analysis was employed to assess the expression levels of SCG5 across various lung cancer cell lines. The results demonstrated that SCG5 was expressed at significantly higher levels in these cell lines compared to the human normal lung epithelial BEAS-2B cells (Fig. 4A-B). Following this, the expression and localization of SCG5 in A549 cells were investigated using immunofluorescence imaging, which revealed a predominant localization of SCG5 in the nucleus (Fig. 4C), corroborating the findings presented in Fig. 1. Furthermore, the expression of SCG5 in NSCLC tissues and adjacent normal lung tissues was evaluated through immunohistochemistry, indicating a higher expression of SCG5 in NSCLC compared to normal tissues (Fig. 4D). Additionally, protein samples from six pairs of NSCLC tissues and adjacent normal tissues were collected and analyzed via western blot to further confirm the elevated expression of SCG5 in lung cancer tissues (Fig. 4E-F). In summary, the experimental findings support the idea that SCG5 expression is elevated in human NSCLC tissues compared to normal tissues. This observation aligns with the results derived from the bioinformatics analysis.
Western blot and immunohistochemistry showed that SCG5 was highly expressed in lung cancer cells and tumor tissues. (A, B) Western blot for the expression of SCG5 protein in various lung cancer cell lines and normal human lung epithelial cells (data are the results of three independent biological experiments); (C) immunofluorescence assay for the expression and localization of SCG5 in A549 cells; (D) immunohistochemistry assay for the expression of SCG5 in tumor tissues from non-small cell lung cancer patients and adjacent normal lung tissues; (E, F) Western blotting was employed to analyze SCG5 expression in protein samples extracted from six pairs of human lung cancer tissues and their adjacent normal tissues (data are the results of three independent biological experiments).
ROC curve and prognostic analysis of the SCG5 gene
Using the Kaplan-Meier (KM) online tool, we conducted an analysis of the association between SCG5 expression levels in microarray data (203889_at) and patient survival outcomes, which include overall survival and post-progression survival, among individuals diagnosed with lung adenocarcinoma and lung squamous cell carcinoma. The findings revealed that patients exhibiting higher SCG5 expression levels experienced significantly reduced overall survival and post-progression survival in comparison to those demonstrating lower SCG5 levels (Fig. 5A/B). Notably, in the cohort with LUSC, increased SCG5 expression was linked to diminished overall survival, implying that SCG5 might influence survival outcomes in patients with LUSC. However, it is noteworthy that after disease progression, we did not observe any statistically significant differences in survival between patients categorized as having high versus low SCG5 expression (Fig. 5C/D). This indicates that although SCG5 expression may affect overall survival in LUSC patients, it does not appear to impact survival once the disease has advanced.
ROC curves and survival prediction analysis of the SCG5 gene. (A, B) Analyzed the KM database to study the prognostic association of SCG5 expression with overall survival and post-progression survival in patients with LUAD; (C, D) Analyzed the KM database to study the prognostic association of SCG5 expression with overall survival and post-progression survival in patients with LUSC; (E) ROC curves; (F) Kaplan-Meier survival analysis of NSCLC patients with different SCG5 expression levels in immunohistochemistry; (G) Nomogram of predicted 1-3- and 5-year survival rates of lung cancer patients.
Additionally, the levels of differential RNA-Seq for SCG5 were utilized as a biomarker for differentiating normal lung tissue from lung cancer tissue. The analysis of the ROC curve demonstrated an area under the curve (AUC) value of 0.850 (95% CI: 0.811–0.890), suggesting that the expression levels of SCG5 can effectively separate normal lung tissue from lung cancer tissue (Fig. 5E). To improve the clinical usage of SCG5 as a prognostic biomarker for NSCLC, we created a prognostic nomogram model that includes SCG5 expression levels, pathological stage, and several other clinicopathological characteristics. This model synthesizes results from univariate and multivariate COX regression analyses, quantifying how each variable contributes to predicting survival rates at one, three, and five years. Key factors represented in the nomogram include SCG5 expression, tumor stage (T, N, M), age, smoking history, tumor type (either adenocarcinoma or squamous cell carcinoma), and tumor location (Fig. 5G). These results underscore the promise of SCG5 as a prognostic indicator for NSCLC patients, highlighting the necessity of tracking SCG5 expression within clinical settings to inform treatment approaches and enhance patient outcomes.
Univariate and multivariate COX regression analysis
To assess the potential independent prognostic value of SCG5 for pathological features related to lung cancer prognosis, we conducted univariate and multivariate COX regression analyses using patient data from the TCGA database. Our findings revealed a significant association between patient prognosis and variables such as tumor stage, presence of distant metastases, as well as high SCG5 expression and age (p < 0.05). Conversely, factors such as gender, tumor location, tumor type, and years of smoking did not show a significant association with prognosis (p > 0.05) (Table 1). After conducting a multivariate COX regression analysis, we were unable to establish a significant correlation between SCG5 and OS (Table 1). To mitigate the effects of data heterogeneity and potential errors in the initial data quality of the database, we re-collected clinical data from 95 patients with non-small cell lung cancer at our hospital and performed univariate and multivariate COX regression analyses again (Tables 2 and 3). The results indicated a significant difference in overall survival between the high and low expression groups of the SCG5 gene in both univariate and multivariate analyses. In the univariate analysis, high SCG5 expression was associated with a poorer prognosis (HR = 0.318, 95% CI: 0.163–0.621, P = 0.001). Furthermore, after controlling for other clinical covariates, the multivariate analysis continued to demonstrate that high SCG5 expression was significantly linked to a poorer prognosis (HR = 0.387, 95% CI: 0.155–0.967, P = 0.042). Additionally, Kaplan-Meier survival curves illustrated that the survival rate of the SCG5 low-expression group was significantly higher than that of the high-expression group, with a hazard ratio of 0.3240 (95% CI: 0.1791–0.5860, p = 0.0004)(Fig. 5F). These findings support the potential of the SCG5 gene as an independent prognostic factor.
Discussion
Non-small cell lung cancer is the most prevalent type of lung cancer, representing approximately 85% of all cases3. NSCLC has shown resistance to traditional radiotherapy and chemotherapy treatments18,19. With ongoing research into cancer development mechanisms, targeted therapies based on biomarkers have emerged as the primary treatment approach for advanced NSCLC patients. To enhance the efficacy of treatment and the overall quality of life for NSCLC patients, the exploration of new biomarkers is crucial for understanding the mechanisms underlying NSCLC development and improving the accuracy of early diagnosis and prognostic evaluation20.
In recent years, the SCG5 gene has been identified as playing a role in the development and progression of various cancers, such as small cell lung cancer21, squamous cell carcinoma of the skin12, and gastrointestinal tract tumors22. SCG5 functions as a molecular chaperone for the proteogen convertase PC2 and plays a role in the processing and maturation of various proteins6,7. It has been postulated that the functional relationship between SCG5 and PC2 suggests that their gene expressions may be coordinately regulated. The proximal promoter regions of these two genes contain binding sites for cAMP response element (CRE), specificity protein 1 (SP1), AP1, and AP2, yet they lack other apparent regulatory elements23,24. In an animal model of insulin-induced hypoglycemic shock, a significant decrease in mRNA levels of SCG5 and PC2 was observed after 6 h. However, over an extended period (up to 72 h), SCG5 mRNA levels continued to decline, while PC2 mRNA levels increased significantly and remained elevated. This differential expression may be linked to the reduced conversion of proPC2 to PC2, indicating that the downregulation of SCG5 may serve as a cellular mechanism that limits PC2 activity25.Co-transfection of SCG5-PC2 gene in human hepatocellular carcinoma cell line HepG2, mouse melanoma cell line B16, mouse breast carcinoma cell line 4T1, human lung carcinoma cell line A549, human breast carcinoma cell line MCF-7 and mouse mast cell tumor cell line P815 showed that with the increase of PC2 gene dose, these tumor cells inhibited their proliferation to different degrees and exhibited apoptosis26. In addition, SCG5 may be involved in pancreatic cancer metastasis through the interaction of the PPAR pathway, PI3K-Akt pathway and ECM receptor27. In the study of cutaneous squamous cell carcinoma (cSCC), SCG5 was identified as one of the hub genes associated with cSCC metastasis, and its high expression was correlated with cSCC invasion and migration12. It was also found that two important transcription factors, MYC and SPI1, may affect the signaling pathways associated with cSCC by targeting the expression of the hub genes. MYC, as a proto-oncogene, is involved in the transcriptional regulation of a variety of genes, including cell cycle, cell growth and cell metabolism28,29. In NSCLC, SPI1 induces SNHG6 up-regulation and regulates NSCLC cellular processes through the SNHG6/Mir-485-3p/VPS45 axis to exert oncogenic effects30.Despite this, the relationship between SCG5 and non-small cell lung cancer remains unclear in existing studies. Hence, our study aims to explore the potential cancer-promoting impact of SCG5 on NSCLC through bioinformatic analysis.
Our study systematically investigates the role and clinical significance of SCG5 in NSCLC through bioinformatics analysis, experimental validation, and clinical data evaluation. Our findings indicate that the expression level of SCG5 in NSCLC tissues and cell lines is significantly higher than that in normal controls, suggesting its potential involvement in the onset and progression of NSCLC. Kaplan-Meier survival analysis further revealed that the overall survival rate of patients with high SCG5 expression was significantly lower than that of the low-expression group. Additionally, univariate and multivariate COX regression analyses confirmed the potential of SCG5 as an independent prognostic factor in NSCLC patients. The results of ROC curve analysis demonstrated that the AUC for SCG5 was 0.850, indicating high sensitivity and specificity in differentiating NSCLC from normal tissue. Notably, the Nomogram model proposed in our study offers a reliable tool for individualized patient prognosis assessment based on SCG5 expression levels, pathological stage, and fundamental patient characteristics. This model holds potential for clinical application, as it integrates multiple factors to predict patient survival rates at 1, 3, and 5 years, thereby aiding in the development of personalized treatment strategies.
The tumor microenvironment (TME) is closely associated with tumorigenesis and metastasis31,32,33. T cells, including CD4 + T cells and CD8 + T cells, play a key role in the TME34,35. Immunotherapy is currently an important treatment for cancer and has made remarkable clinical progress. T cell-mediated immunotherapy plays an important role36. By immune infiltration analysis, we found that the expression of SCG5 in lung cancer was significantly and positively correlated with the infiltration of various immune cells. This result suggests that SCG5 may play an important role in the NSCLC tumor microenvironment and tumor immunoregulatory mechanism. In the subsequent phase of our research, we will quantify immune cell infiltration in tumor tissue using immunohistochemistry, flow cytometry, and single-cell RNA sequencing. This will allow us to assess the correlation between SCG5 gene expression and immune cell distribution. Additionally, we will investigate the regulatory effects of SCG5 on immune checkpoints, such as PD-1 and PD-L1, through RT-qPCR and Western blot analyses to explore its role in immune evasion. The methods outlined above may elucidate the potential regulatory role of SCG5 in tumor immunity. Consequently, targeted therapy against SCG5 could represent a promising avenue for immunotherapy in NSCLC.
Despite the preliminary results of our study, which reveal the potential role of SCG5 in NSCLC, our study still has some limitations. As mentioned above, SCG5 is closely related to the secretory function of neuroendocrine cells, and we speculate whether it also affects the biological behavior of NSCLC through a similar mechanism. Therefore, in the future, we will combine cellular and animal experiments to investigate the specific mechanism of SCG5 in NSCLC. For example, we can use knockdown or overexpression experiments to study the effects of SCG5 on tumor cell proliferation, invasion and migration. In addition, the present study was mainly based on the bioinformatics analysis of SCG5 gene and some simple cell line experiments, and did not explore the specific molecular mechanism between SCG5 and NSCLC through in vivo and in vitro experiments. In the future, we will investigate the signaling pathways and targets of SCG5, and use proteomics, genomics, metabolomics and other methods to comprehensively reveal the functional network of SCG5 on NSCLC. We will continue to search for molecules that specifically inhibit the activity of SCG5 using high-throughput screening and computer-aided drug design, and then use molecular docking to search for possible binding sites between small molecule drugs and SCG5. Transgenic mice may also be generated using gene editing techniques such as CRISPR/Cas9 to observe the effects of specifically silencing or inhibiting the expression of the SCG5 gene in mice and to study its in vivo and in vitro regulatory mechanisms in NSCLC. Finally, we will collect more information from large samples of NSCLC patients and compare it with our experimental results based on database analyses to better assess the potential of SCG5 as a prognostic marker for NSCLC.
Conclusions
Our findings indicate that SCG5 expression is significantly elevated in non-small cell lung cancer tissues compared to normal lung tissues. Furthermore, higher SCG5 levels correlate with poorer overall survival in patients. In conclusion, this research provides a reliable foundation for the utilization of SCG5 as a biomarker in the diagnosis and prognosis of NSCLC, while also highlighting avenues for future investigations into SCG5.
Data availability
The datasets generated during the present study are available for download from the Gene Expression Omnibus (GEO) database, accessible via the following accession numbers: GSE19804, GSE118370, GSE27262, and GSE33532.
References
Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021).
Allemani, C. et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 391, 1023–1075 (2018).
Gridelli, C. et al. Non-small-cell lung cancer. Nat. Rev. Dis. Primers. 1, 15009 (2015).
Herrera-Juárez, M., Serrano‐Gómez, C., Bote‐de‐Cabo, H. & Paz‐Ares, L. Targeted therapy for lung cancer: beyond EGFR and ALK. Cancer 129, 1803–1820 (2023).
Mbikay, M., Seidah, N. G. & Chrétien, M. Neuroendocrine secretory protein 7B2: structure, expression and functions. Biochem. J. 357, 329–342 (2001).
Muller, L., Zhu, X. & Lindberg, I. Mechanism of the facilitation of PC2 maturation by 7B2: involvement in ProPC2 transport and activation but not folding. J. Cell Biol. 139, 625–638 (1997).
Benjannet, S., Savaria, D., Chrétien, M. & Seidah, N. G. 7B2 is a specific intracellular binding protein of the prohormone convertase PC2. J. Neurochem. 64, 2303–2311 (1995).
Konoshita, T. et al. Expression of PC2 and PC1/PC3 in human pheochromocytomas. Mol. Cell. Endocrinol. 99, 307–314 (1994).
Khatib, A. M. et al. Inhibition of proprotein convertases is associated with loss of growth and tumorigenicity of HT-29 human Colon carcinoma cells. J. Biol. Chem. 276, 30686–30693 (2001).
Mbikay, M., Sirois, F., Yao, J., Seidah, N. & Chrétien, M. Comparative analysis of expression of the proprotein convertases Furin, PACE4, PC1 and PC2 in human lung tumours. Br. J. Cancer. 75, 1509–1514 (1997).
Mamoor, S. SCG5 is a differentially expressed gene in human metastatic breast cancer, in the brain and in the lymph nodes. (2021). https://doi.org/10.31219/osf.io/h5w4n
Chen, L. et al. Identification of Metastasis-Associated genes in cutaneous squamous cell carcinoma based on bioinformatics analysis and experimental validation. Adv. Ther. 39, 4594–4612 (2022).
Stelzer, G. et al. The genecards suite: from gene data mining to disease genome sequence analyses. CP Bioinf. 54, (2016).
Carvalho-Silva, D. et al. Open targets platform: new developments and updates two years on. Nucleic Acids Res. 47, D1056–D1065 (2019).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514 (2020).
Lánczky, A. & Győrffy, B. Web-Based survival analysis tool tailored for medical research (KMplot): development and implementation. J. Med. Internet Res. 23, e27633 (2021).
Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51, D638–D646 (2023).
Meyer, M. L. et al. New promises and challenges in the treatment of advanced non-small-cell lung cancer. Lancet 404, 803–822 (2024).
Brooks, E. D. Safe and effective systemic therapy for early-stage non-small-cell lung cancer. Lancet 402, 829–831 (2023).
Tostes, K., Siqueira, A. P., Reis, R. M., Leal, L. F. & Arantes, L. M. R. B. Biomarkers for immune checkpoint inhibitor response in NSCLC: current developments and applicability. Int. J. Mol. Sci. 24, 11887 (2023).
Iguchi, H. et al. Elevation of a novel pituitary protein (7B2) in the plasma in small cell carcinoma of the lung. Eur. J. Cancer Clin. Oncol. 25, 1225–1232 (1989).
Suzuki, H. et al. Production of pituitary protein 7B2 immunoreactivity by endocrine tumors and its possible diagnostic value. J. Clin. Endocrinol. Metabolism. 63, 758–765 (1986).
Ohagi, S. et al. Identification and analysis of the gene encoding human PC2, a prohormone convertase expressed in neuroendocrine tissues. Proc. Natl. Acad. Sci. U S A. 89, 4977–4981 (1992).
Braks, J. A. M., Broers, C. A. M., Danger, J. H. A. & Martens, G. J. M. Structural organization of the gene encoding the neuroendocrine chaperone 7B2. Eur. J. Biochem. 236, 60–67 (1996).
Seidel, B. et al. Neuroendocrine protein 7B2 is essential for proteolytic conversion and activation of proprotein convertase 2 in vivo. DNA Cell Biol. 17, 1017–1029 (1998).
LIu, C. et al. Effect of proprotein transferase 2 gene expression on tumour cell proliferation. J. Guangdong Pharm. Univ. 33, 117–122 (2017).
Xu, J., Liao, K., Wang, X., He, J. & Wang, X. Combining bioinformatics techniques to explore the molecular mechanisms involved in pancreatic cancer metastasis and prognosis. J. Cell. Mol. Medi. 24, 14128–14138 (2020).
Dong, Y., Tu, R., Liu, H. & Qing, G. Regulation of cancer cell metabolism: oncogenic MYC in the Driver’s seat. Sig Transduct. Target. Ther. 5, 124 (2020).
Jha, R. K., Kouzine, F. & Levens, D. MYC function and regulation in physiological perspective. Front. Cell. Dev. Biol. 11, 1268275 (2023).
Gao, N. & Ye, B. SPI1-induced upregulation of LncRNA SNHG6 promotes non-small cell lung cancer via miR-485-3p/VPS45 axis. Biomed. Pharmacother. 129, 110239 (2020).
Barkley, D. et al. Cancer cell States recur across tumor types and form specific interactions with the tumor microenvironment. Nat. Genet. 54, 1192–1201 (2022).
Guo, W. et al. Integrating microarray-based Spatial transcriptomics and single-cell RNA-sequencing reveals tissue architecture in esophageal squamous cell carcinoma. eBioMedicine 84, 104281 (2022).
De Visser, K. E. & Joyce, J. A. The evolving tumor microenvironment: from cancer initiation to metastatic outgrowth. Cancer Cell. 41, 374–403 (2023).
Zhou, W. et al. Stem-like progenitor and terminally differentiated TFH-like CD4 + T cell exhaustion in the tumor microenvironment. Cell. Rep. 43, 113797 (2024).
Hoekstra, M. E. et al. Distinct Spatiotemporal dynamics of CD8 + T cell-derived cytokines in the tumor microenvironment. Cancer Cell. 42, 157–167e9 (2024).
Chow, A., Perica, K., Klebanoff, C. A. & Wolchok, J. D. Clinical implications of T cell exhaustion for cancer immunotherapy. Nat. Rev. Clin. Oncol. 19, 775–790 (2022).
Acknowledgements
Declared none.
Funding
This research topic has been sponsored by the following fund projects: (1) Practice Innovation Program of Jiangsu Province (SJCX24_2060). (2) 2024 Yancheng City Key R&D Program (Social Development) Directive Project YCBE202421. (3) Nantong University’s 2023 Academic Level Research Project (Special Project of Yancheng. Third Institute) (YXY-Z2023008). (4) 2023 Nantong University Clinical Medicine Special Research Fund Project (2023JZ022). (5) 2021 Jiangsu Provincial Health and Health Commission Medical Research Guidance Project (Z2021087). (6) The Special Funds for Science Development of the Clinical Teaching Hospitals of Jiangsu Vocational College of Medicine (20219105).
Author information
Authors and Affiliations
Contributions
For this study, WZ conceived and designed the experiments, performed the experiments, analysed the data, prepared the graphs, wrote or reviewed the draft paper, and approved the final draft.WZ, RW, RG and YW analysed the data. HW and YL collected tissue samples. XL, JS conceived and designed the experiments, wrote or revised drafts of the article and approved the final version. All authors contributed to the article and approved the version submitted for publication.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical statement
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study was approved by the Ethics Committee of the Third People’s Hospital of Yancheng City ( Affiliated Hospital 6 of Nantong University) (Ethical Approval No: 2023-46).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, W., Wang, R., Guo, R. et al. The potential of secretogranin V as a prognostic biomarker in non-small cell lung cancer. Sci Rep 15, 16589 (2025). https://doi.org/10.1038/s41598-025-00747-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-00747-3