Abstract
Reverse Vaccinology (RV) has revolutionized vaccine discovery, utilizing bioinformatics to surpass traditional methods in identifying genes and proteins. By analyzing pathogen genomic data, RV pinpoints proteins with key traits such as immunogenicity, surface localization, and conservation across strains. Despite its advantages, current RV tools face challenges like prediction accuracy, computational demands, and accessibility. To address these challenges, we introduce B-vac, an executable pipeline designed to streamline bacterial vaccine design. B-vac features a user-friendly interface and robust algorithms for high-throughput proteomics data analysis, covering modules like Localization, Non-host Homolog, Virulence Factor, and Epitope Mapping. It operates offline, enhancing accessibility for researchers with limited computational resources. B-vac is equipped with epitope libraries, bacterial proteomes and virulence factor database which helps the program process the protein sequences locally and feeds data back to users with the ability to set variables and toggles for cut-off and filter values. The B-vac pipeline uses a string-based matching approach to match proteomes supplied by users with the pipeline’s curated database. This approach aligns and compares pathogen protein sequences by string similarity and enables the researchers to easily identify motifs important for immunogenic function. Evaluation of the pipeline by employing the Helicobacter pylori proteome revealed B-vac’s effectiveness in identifying vaccine candidates. B-vac offers a user-friendly, standalone solution for bacterial vaccine development, eliminating the need for external libraries and enabling offline usability, addressing key gaps in convenience and accessibility compared to existing RV tools. B-vac can be downloaded from: https://mgbio.tech/tools/.
Similar content being viewed by others
Introduction
Bacterial infections and antibiotic resistance have now become one of the biggest global health challenges of the 21st century. The Centers for Disease Control and Prevention (CDC) reports that over two million people in the United States are affected by antibiotic-resistant infections annually, resulting in approximately 23,000 deaths. This alarming trend is compounded by the overuse and misuse of antibiotics, resulting to their ineffectiveness and thereby fueling multidrug resistance among bacterial pathogens1,2. Bacteria have evolved various mechanisms to resist antibiotics, such as genetic mutations, acquisition of resistance genes, and alterations in gene expression3,4. These mechanisms continuously evolve, posing critical challenges to existing treatment strategies5. Antimicrobial Resistance (AMR) has been identified as a high-priority public health concern by the World Health Organization since it causes several impacts on human health and the economy such as longer hospital stays and higher healthcare costs. Addressing Combating AMR requires cooperation across borders to rationalise antibiotic consumption, create new approaches to fighting infections, and promote equal access to potent medications6,7.
Vaccines are emerging as promising alternatives to antibiotics in the fight against bacterial infections. They reduce the need for antibiotics by preventing infections, and consequently slow down the development of antibiotic resistance8,9. Vaccines targeting bacterial pathogens are particularly vital in regions with limited healthcare resources, as they are designed to be affordable, stable without refrigeration, and administrable orally or intranasally. These features make them suitable for widespread global use10. Moreover, vaccines can prevent infections caused by multidrug-resistant (MDR) bacteria, which are hard to treat with existing antibiotics11,12. While vaccines for extracellular bacteria like tetanus and diphtheria have been successful, developing vaccines against intracellular bacteria remains a complex task requiring advanced technologies9. Innovative vaccine technologies, including reverse vaccinology and novel adjuvants are being explored to enhance vaccine efficacy against multidrug-resistant bacteria8.
Reverse vaccinology (RV) can be described as revolutionary approach to vaccine development, that uses pathogen’s genomic insights to identify potential vaccine candidates (PVCs) quickly and precisely as compared to traditional vaccinology methods. The approach that was initially introduced in the post-genomic era, started by sequencing the pathogen’s genome, which allowed researchers to analyze its whole antigenic repertoire. Unlike conventional methods which often required cultivation of the pathogen in vitro, RV relies on in silico methods for the analysis of pathogen’s genomic data. These tools look for genes that code for proteins with favorable characteristics for a vaccine and includes immunogenicity, exposure on the surface and/or conservation among different pathogens. This approach greatly accelerated and reduced the costs of identifying vaccine targets, making the journey from identifying a pathogen to developing a vaccine much faster13,14.
Traditionally, vaccine development was based on principles pioneered by Louis Pasteur, who introduced key techniques such as isolating, inactivating, and injecting pathogens to induce protective immunity. This approach resulted in production of vaccines for diseases such as rabies, typhoid, diphtheria, tetanus among others using attenuated pathogens, or simply components of microbes that can trigger immune response15,16. As time went on, advancements in molecular biology and biotechnology brought new techniques including genetic engineering, purification of microbial elements, and the use of live vectors to express vaccine proteins17. These improvements made the production of vaccines much more accurate and safer, however the use of these methods was limited by the amount of empirical testing that was still required. The advent of genomic technologies brought about a new era in vaccine development known as reverse vaccinology. This method not only overcame the challenges associated with traditional methods but also allowed the development of vaccines for pathogens that were previously considered intractable18,19.
The first successful application of reverse vaccinology was in developing a vaccine against serogroup B Neisseria meningitidis (MenB), a significant cause of sepsis and meningitis20. The 4CMenB vaccine, includes three recombinant antigens (fHbp, NadA, and NHBA) combined with outer membrane vesicles. This multicomponent vaccine has shown effectiveness in enhancing immune response across various age groups21,22,23. The 4CMenB vaccine underwent extensive clinical trials to evaluate its safety and efficacy. It was approved in Europe in 2013 and included in the UK’s National Immunization Program in 2015, showing an effectiveness of 83% against invasive MenB disease22,23. Research continues to refine MenB vaccines, exploring new antigens and formulations to enhance coverage and effectiveness. The use of reverse vaccinology remains a promising strategy for developing vaccines against other pathogens as well24,25,26.
Since then, several tools have been developed on principles of reverse vaccinology, each with unique features and methodologies. NERVE was designed to be user-friendly having integrated multiple algorithms for protein analysis. It ranks vaccine candidates and maintains comprehensive data for further analysis. NERVE is noted for its high recall of known protective antigens, making it efficient in identifying safe and experimentally viable candidates27. The authors of NERVE have since published an updated version, NERVE 2.0 (https://nerve-bio.org/home), which we have included in our benchmarking to evaluate its performance against other state-of-the-art tools28. Vaxign was the first web-based RV tool, and Vaxign2 enhances this with machine learning capabilities. Vaxign and Vaxign2 (https://violinet.org/vaxign2) offers comprehensive framework for vaccine design, including predictive and post-prediction analysis components29. Furthermore, known for its application in predicting vaccine candidates for various pathogens, VaxiJen (https://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) is widely used in RV30. It has been particularly applied to SARS-CoV-2, although experimental validation of its predictions is limited31. VacSol (https://sourceforge.net/projects/vacsol/) automates the prediction of vaccine candidates using a high-throughput approach. It efficiently screens bacterial proteomes and reduces false positives, making it a cost-effective tool for vaccine candidate identification32. Jenner-Predict focuses on host-pathogen interactions and pathogenesis, using functional domains to predict vaccine candidates. It has demonstrated better prediction accuracy compared to other tools, particularly in identifying non-cytosolic proteins involved in host-pathogen interactions33. Despite all of these pros, the above-mentioned current RV tools also face several technical and scientific limitations. Many RV tools, including VaxiJen and Jenner Predict, have low prediction accuracy, which limits their application in vaccine development. Only a small fraction of predicted candidates undergo experimental validation, which is crucial for confirming their potential as vaccines31,33,34. Some tools, such as NERVE, are designed to be user-friendly but still require significant expertise to install, run and interpret results effectively. This complexity can be a barrier for broader adoption27. Many tools focus on limited criteria, such as adhesin-likeliness, without considering other functional classes of proteins that may be involved in host-pathogen interactions and pathogenesis33. Tools like VacSol aim to reduce computational costs and time, but the efficiency of these processes can still be improved32. Moreover, most of the current RV tools like NERVE, Vaxign, and VacSol integrate various open-source bioinformatics tools and algorithms for protein analysis for screening of pathogen proteomes to identify potential vaccine candidates. Despite their utility, these tools often require internet access, local installations, and heavy computational resources, making them less accessible for researchers without advanced computational expertise or infrastructure.
To address these limitations, we developed B-vac, an executable program that integrates a series of internally designed algorithms for protein sequence processing, comparison and vaccine target analysis. Unlike existing tools described earlier, B-vac is designed to improve prediction accuracy by employing a streamlined, specialized approach to vaccine targets prediction and analysis, reducing reliance on broad, less accurate criteria. It also prioritizes ease of use, requiring no internet connection, command-line execution, or advanced computational expertise. B-vac’s self-contained architecture utilizing Python in its core framework, and user-friendly interface make it accessible to a broader range of researchers, including those without extensive bioinformatics experience. By focusing on practical, efficient workflows and eliminating the need for external dependencies, B-vac facilitates the identification of potential vaccine candidates with greater reliability and accessibility.
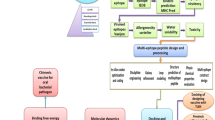
The predicted features in B-vac include protein subcellular localization, virulence factors, and epitope mapping among pathogen genomes, and sequence similarity to host (human) proteomes. Surface-exposed proteins, such as secreted proteins, fimbrial proteins, and outer membrane proteins, are crucial for vaccine development as they are accessible to the immune system. Studies have identified various surface proteins in pathogens like Streptococcus pneumoniae and Leptospira interrogans, which are promising vaccine targets due to their role in virulence and immune response elicitation35,36,37. In contrast, non-surface proteins are less suitable as they do not interact directly with host cells. Moreover, vaccine candidates should include virulence factors to elicit strong immune responses. Proteins that contribute to a pathogen’s virulence, such as adhesins, exoenzymes, and toxins, are essential for effective vaccines. These factors ensure a strong immune response, making them ideal candidates for vaccine development35,36,38. Additionally, effective vaccine targets should also avoid sequence similarity to host proteins to prevent autoimmunity. Identifying unique antigens that do not share homology with host proteins is critical to avoid autoimmunity. For instance, the Cp-P34 protein in Cryptosporidium is unique to the parasite and elicits immune responses, making it a potential vaccine candidate. These considerations are integral to the B-vac pipeline39. The overall architecture of B-vac pipeline is given in Fig. 1.
B-vac implementation
B-vac is written in Python v3.10.8, with its graphical user interface (GUI) developed using the Tkinter v8.6.12 library, which is a standard Python library for creating simple and user-friendly desktop interfaces. To ensure compatibility and ease of use on Windows and Linux (Ubuntu) platforms, it is compiled using PyInstaller v6.10.0, a tool that packages Python applications into standalone executables, allowing them to run without requiring a separate Python installation. The pipeline integrates extensive pre-saved datasets critical for reverse vaccinology. These datasets include protein FASTA files for each bacterial strain, specifically containing secreted, outer membrane, and fimbrial proteins, downloaded from the LocTree3 (http://www.rostlab.org/services/loctree3)40, for protein localization filtering, 916 CD4 + epitopes and 1659 CD8 + epitopes across multiple HLA alleles, stored in CSV format obtained from IEDB database v3 (accessed on March 13, 2025, https://www.iedb.org/), and 27,502 virulence factors obtained from the Virulence Factors Database (https://www.mgc.ac.cn/VFs/) with their corresponding IDs and protein fasta sequences (accessed on September 12, 2022)41,42,43. Additionally, it includes 67,297 B-cell linear epitopes in FASTA format obtained from IEDB and the human reference proteome downloaded from Uniprot (accessed on October 5, 2022, https://www.uniprot.org/) for non-host homologs analysis41,43.
B-vac is optimized for local execution without internet dependency. Testing was performed on two systems; an Intel i5-8350U CPU (1.70 GHz base / 1.90 GHz max) quadcore processor with 8 GB RAM running Windows 11, and an Intel i5-4570 CPU (3.20 GHz) quadcore processor with 4 GB RAM running Ubuntu 22.04.2 LTS. The pipeline supports batch processing of multiple protein sequences, with processing times averaging 20 min for 100 proteins under default parameters. B-vac’s architecture utilizes pre-saved datasets to enable local, resource-efficient processing of protein data. The GUI provides adjustable parameters (e.g., sequence identity thresholds, epitope lengths) and dynamically displays results, including filtered proteins, virulence factors, and mapped epitopes. By eliminating cloud dependencies and offering offline compatibility, B-vac streamlines strain-specific vaccine candidate identification while maintaining low memory overhead (< 1GB during runtime).
Graphical user interface of B-vac
The B-vac pipeline incorporates a user-friendly graphical user interface (GUI) optimized for rapid and effective vaccine target prediction and analysis, as illustrated in Fig. 2. This pipeline employs a string-based matching mechanism to compare the user’s provided proteome with a curated database. String-based matching mechanisms are fundamental in bioinformatics for aligning and comparing protein sequences based on their string similarity44,45. This approach is particularly helpful in recognizing conserved motifs or regions essential which might be important for protein function. Such statistically significant algorithms prioritize biologically relevant patterns, favoring conserved regions, and penalizing mismatches at key positions. This approach improves both the sensitivity and specificity of functional predictions of proteins44. Moreover, the user-defined identity percentage threshold in the pipeline acts as a filter, ensuring that only alignments with adequate sequence similarity are considered valid. This approach effectively balances sensitivity and specificity. These interconnected components synergistically contribute to a streamlined system of B-vac for precise and efficient vaccine candidates’ prediction, enabling researchers to focus on sequences that are most likely to provide useful immunogenic insights.
The user-friendly interface of B-vac enables users to upload proteome files in FASTA format (.faa or .fasta) for analysis. Users can customize their workflows by choosing from the available filters i.e. Localization, Non-Host Homologs, Virulence Factors, and Epitope Mapping through a well-organized layout. Key parameters like reliability score, identity percentage, and epitope lengths can be fine-tuned to meet the different analysis needs. The system also has the ability to handle dynamic processing, which is quite useful in display of results based on the given sequences and matching in the database. For example, when the Localization filter is selected and parameters like a 70% identity percentage and a reliability score of 50 are defined, the system immediately generates a list of proteins in the database that meet these criteria and displays the count of these proteins on the interface. Subsequently the Non-Host Homologs and Virulence Factors filters further refine the query dataset, by excluding the proteins having homology to the host and pinpointing important virulence factors respectively. The Epitope Mapping filter then identifies B-cell and T-cell epitopes according to user-specified lengths and identity percentages. Upon processing, the interface generates a summary which includes the lists of reliable proteins, predicted epitopes and the number of proteins filtered during each step of the process. The pipeline enables simultaneous and thorough analysis and is therefore suitable for high-throughput screening of vaccine candidates while minimizing manual intervention and errors.
Methods
B-vac is a comprehensive pipeline that integrates multiple internally developed algorithms with a clean graphical user interface with input fields and adjustable thresholds and filters for customizing analysis parameters to assist in RV.
B-vac algorithm for vaccine candidates filtering
The main input is a bacterial protein sequence, which is analyzed to filter and prioritize vaccine candidates. B-vac employs a custom string-based matching algorithm for sequence analysis, which calculates the percentage of matched residues between a submitted protein sequence and reference sequences from the dataset integrated in the software package. Sequences that meet or exceed the user-defined identity percentage threshold (e.g., 70%) are retained as potential candidates. Key adjustable parameters include:
Localization
This feature evaluates protein localization, a critical step in vaccine design. Proteins localized to the surface or secreted extracellularly are preferred candidates as they are accessible to the host immune system46,47,48. Localization will filter secreted, outer membrane and fimbrium proteins.
-
Select Bacteria Genus and Strain: Users can specify the genus and strain of interest, enabling strain-specific vaccine design.
-
Reliability Score: The reliability score used in the localization filter is based on the LocTree3, which predicts protein subcellular localization with a reliability index (RI) ranging from 0 (low confidence) to 100 (high confidence)40. B-vac incorporates these reliability scores to filter proteins, allowing users to set a threshold (e.g., 70) to retain only high-confidence predictions. These adjustable thresholds allow users to set confidence levels for protein localization evaluation, providing flexibility in stringency based on the organism being analyzed or project goals. The thresholds are based on common practices in reverse vaccinology, but users can modify them to suit their specific needs.
Non-host homologs
This section allows removal of proteins homologous to host proteins by setting thresholds for identity percentage and non-homology percentage, reducing the likelihood of autoimmune responses49,50,51,52. The threshold of 70% for non-host homology screening was chosen to balance sensitivity and specificity in identifying non-host proteins, but users can adjust this value to increase or decrease stringency based on their requirements.
Virulence factors
By incorporating virulence factors, the software identifies proteins essential for pathogen virulence, which are promising targets for subunit vaccine development53,54,55,56. Adjustable parameters include identity percentage to filter known virulence factors.
Epitope mapping
The B-vac pipeline extracts out antigenic epitopes recognized by B-cells (antibody-producing) and T-cells (CD4 + helper and CD8 + cytotoxic T-cells) from the input proteins. These epitopes are crucial for eliciting a robust immune response. B-cell epitopes are essential for the production of antibodies, which neutralize pathogens and prevent infection57. T-cell epitopes, on the other hand, are vital for the activation of CD4 + helper T-cells and CD8 + cytotoxic T-cells, which play key roles in coordination of immune response and directly killing infected cells58. The identification of these epitopes ensures that the vaccine can stimulate both humoral and cellular immunity and provide comprehensive protection against the pathogen59. Adjustable fields include length and identity thresholds for predicted epitopes.
Output metrics also display in right-hand panel of the interface, including:
-
Reliable Proteins: The number of candidate proteins that meet reliability criteria.
-
Proteins and PVCs: Total proteins analyzed and final vaccine candidates.
-
Epitope Predictions: Counts of epitopes mapped for B-cells, CD4+, and CD8 + T-cells.
Case study: Helicobacter pylori
To evaluate the functionality of the B-vac pipeline, the proteome of Helicobacter pylori, comprising 100 proteins, was downloaded from the NCBI database (https://www.ncbi.nlm.nih.gov/), and subsequently analyzed using the pipeline. The session initiated by browsing and uploading the proteome FASTA file (accepted formats .faa and .fasta), followed by saving the session in a user-defined directory to store the analysis results. The “Must Evaluate” option was checked to ensure all filters and methods; Localization, Non-Host Homologs, Virulence Factors, and Epitope Mapping, were applied without omission.
Within the Localization filter, parameters were adjusted to refine candidate proteins. The bacterial genus and species were selected, with the reliability score set to 50 and the identity percentage to 70. Upon applying these criteria, the right-hand panel of the interface displayed 192 reliable proteins from the pipeline’s dataset, which were subsequently matched against the query proteins. In the Non-Host Homologs filter, thresholds for identity and non-homology percentages were set at 35% and 70%, respectively, to exclude proteins homologous to the host genome, minimizing the risk of autoimmunity. The Virulence Factors filter was applied with an identity percentage threshold of 70%, to ensure that only proteins essential to pathogen virulence were retained for further analysis. Finally, Epitope Mapping was configured to assess antigenic epitopes. For B-cell epitopes, all lengths were included, while for T-cell epitopes, all CD8 + and CD4 + lengths were considered, with an identity percentage threshold set to 50%.
Upon submission, B-vac processed the protein dataset through all selected filters. The right-hand panel displayed the number of proteins passing all criteria and the counts of predicted epitopes for B-cells, CD8+, and CD4 + T-cells, providing a comprehensive overview for the analysis.
Results
Findings of H. pylori case study
Protein localization filter
Using the selected identity percentage thresholds, the analysis filters out five proteins which were saved in faa FASTA file format, with metadata embedded within the FASTA identifiers. These proteins showed a high identity match, ranging from 97 to 98%. Among them, four were categorized as secreted, indicating their potential accessibility to the host immune system, while one was classified as an outer membrane protein, supporting its suitability as a vaccine candidate.
Non-host homology filter
Using the non-host homologs filter, the analysis extracted four proteins from the five proteins that passed the localization filter. These proteins were also saved in .faa FASTA file format, with metadata embedded within the FASTA identifiers. The selected proteins showed non-homology identity percentage ranging from 71 to 90%, indicating their reduced similarity to host proteins and minimizing the risk of autoimmunity.
Virulence factors filter
Applying the virulence factors filter, the analysis identified two virulence factors among the four proteins that passed the non-host homology filter. These proteins were also saved in .faa FASTA file format, with information embedded in the FASTA identifiers. The selected virulence factors exhibited high identity percentages of 97% and 98%. Finally, two potential vaccine candidates (PVCs) with NCBI accession WP_000418838.1 and WP_000347746.1 were filtered out of the 100 proteins of Helicobacter pylori from the analysis. The detailed results of these analysis steps are given in Table 1.
Epitope mapping
Epitope analysis identified 434 total epitopes, with 17 B-cell epitopes on HLA-B07:02 with identity percentage ranging from 50 to 56% in one of the two PVCs WP_000418838.1, 36 CD4 + and 381 CD8 + T-cell epitopes with identity percentage ranging from 50 to 66% across the two potential vaccine candidates (PVCs). The detailed results of epitope analysis step are given in Supplementary Table S1a and S1b. The sequence fasta files of these results are given in Supplementary Files F1-F6.
Comparison of features and computational requirements
The comparative analysis of B-vac with other reverse vaccinology tools, including NERVE 2.0, Vaxign2, VaxiJen 2.0, VacSol, and Jenner-Predict, highlights the unique strengths and limitations of each tool (Table 2). B-vac stands out for its low computational requirements, ease of use, and self-contained architecture, requiring no internet connection, command-line execution, or advanced computational expertise. It integrates comprehensive datasets for localization (secreted, outer membrane, and fimbrial proteins from LocTree3), non-host homologs (human reference proteome), virulence factors (27,502 entries from VFDB), and epitope mapping, enabling filtering and dynamic results display on GUI. In contrast, NERVE 2.0 and Vaxign2 rely on web-based platforms with active internet connections, while VacSol requires moderate computational resources for high throughput screening. VaxiJen 2.0 and Jenner-Predict lack explicit focus on key filters like virulence factors and epitope mapping, with the latter having inaccessible URL. Notably, NERVE 2.0 failed to process our dataset with default parameters, succeeding only after disabling the virulent and loop-razor filters, completing predictions in 5 min. B-vac demonstrated superior efficiency, processing 100 proteins in 20 min with default parameters, outperforming Vaxign2 (3–4 h approx.) and matching VacSol (10 min). These results underscore B-vac’s potential as a reliable, user-friendly, and efficient tool for high-throughput vaccine candidate identification, addressing key limitations of existing tools.
Discussion
B-vac is a comprehensive software package for vaccine design of bacterial pathogens on principles of reverse vaccinology. B-vac integrates string-based matching algorithms to efficiently compare user-provided proteomic data against a manually curated database. This seamless pipeline enhances identification of immunogenic potential of proteins, offering a user-friendly platform for high-throughput vaccine target prediction. Our results indicate that B-vac can identify both known vaccine targets and potential novel candidates. However, additional validation across diverse datasets and experimental confirmations are required to evaluate its predictive accuracy and broader applicability. Possible directions for further development could include refining Bvac’s core algorithm to enhance the accuracy and efficiency of the matching and alignment processes. While deep learning and machine learning-based models offer potential improvements in performance, their integration would require careful consideration to maintain B-vac’s design principles of simplicity, offline usability, and minimal dependency on external resources. Algorithmic optimizations could also target existing filters i.e. Localization, Non-Host Homologs, Virulence Factors, and Epitope Mapping, to improve computational efficiency without compromising the tool’s lightweight architecture.
Beyond algorithmic refinements, the pipeline could be expanded to include new filters and criteria that support advanced reverse vaccinology workflows, such as prioritizing proteins based on immunogenicity scores, structural stability, or host-pathogen interaction networks. While B-vac currently focuses on providing customizable thresholds and filters to assist in reverse vaccinology, we acknowledge the importance of incorporating statistical significance metrics (e.g., P-values, confidence intervals, or ROC analysis) in future updates to further enhance the tool’s analytical capabilities. This approach would ensure that B-vac remains accessible and efficient for researchers without requiring complex hardware or external libraries. These enhancements would not only improve prediction reliability but also broaden the scope of vaccine target discovery.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Orenstein, W. et al. A call for greater consideration for the role of vaccines in National strategies to combat Antibiotic-Resistant bacteria: recommendations from the National vaccine advisory committee. Public Health Rep. 131, 11–16 (2016).
CDC. 2019 Antibiotic resistance threats report. Antimicrob. Resist. https://www.cdc.gov/antimicrobial-resistance/data-research/threats/index.html (2024).
Munita, J. & Arias, C. Mechanisms of antibiotic resistance. Microbiol. Spectr. 4, (2016).
Blair, J., Webber, M., Baylay, A., Ogbolu, D. & Piddock, L. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51 (2014).
MacLean, R. & Millán, S. The evolution of antibiotic resistance. Science 365, 1082–1083 (2019).
World Health Organization. Global Antimicrobial Resistance Surveillance System (GLASS) Report: Early Implementation 2017–2018 (World Health Organization, 2018).
Saeed, U. et al. Crisis averted: a world united against the menace of multiple drug-resistant superbugs -pioneering anti-AMR vaccines, RNA interference, nanomedicine, CRISPR-based antimicrobials, bacteriophage therapies, and clinical artificial intelligence strategies to safeguard global antimicrobial arsenal. Front. Microbiol. 14, (2023).
Baker, S., Payne, D., Rappuoli, R. & De Gregorio, E. Technologies to address antimicrobial resistance. Proc. Natl. Acad. Sci. 115, 12887–12895 (2018).
Osterloh, A. Vaccination against bacterial infections: Challenges, progress, and new approaches with a focus on intracellular bacteria. Vaccines 10, (2022).
Curtiss, R. Bacterial infectious disease control by vaccine development. J. Clin. Investig. 110 8, 1061–1066 (2002).
Mba, I. et al. Vaccine development for bacterial pathogens: advances, challenges and prospects. Tropical Med. Int. Health. 28, 275–299 (2023).
Khalid, K. & Poh, C. L. The promising potential of reverse Vaccinology-Based Next-Generation vaccine development over conventional vaccines against Antibiotic-Resistant Bacteria. Vaccines 11, 1264 (2023).
Rappuoli, R. Reverse vaccinology. Curr. Opin. Microbiol. 3 5, 445–450 (2000).
Goodswen, S., Kennedy, P. & Ellis, J. A guide to current methodology and usage of reverse vaccinology towards in Silico vaccine discovery. FEMS Microbiol. Rev. https://doi.org/10.1093/femsre/fuad004 (2023).
Schwartz, M. The pasteurian contribution to the history of vaccines. C.R. Biol. 345 (3), 93–107 (2022).
Giese, M. From pasteur to personalized vaccines. https://doi.org/10.1007/978-3-319-25832-4_1 3–24, (2016).
Plotkin, S. Vaccines: past, present and future. Nat. Med. 11, (2005).
Serruto, D. & Rappuoli, R. Post-genomic vaccine development. FEBS Lett. 580, (2006).
Lew-Tabor, A., Lew-Tabor, A. & Valle, M. A review of reverse vaccinology approaches for the development of vaccines against ticks and tick borne diseases. Ticks tick-borne Dis. 7 (4), 573–585 (2016).
Pizza, M. et al. Identification of vaccine candidates against serogroup B Meningococcus by Whole-Genome sequencing. Science 287, 1816–1820 (2000).
Serruto, D., Bottomley, M., Ram, S., Giuliani, M. & Rappuoli, R. The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: immunological, functional and structural characterization of the antigens. Vaccine 30 Suppl 2, (2012).
Masignani, V., Pizza, M. & Moxon, E. The development of a vaccine against Meningococcus B using reverse vaccinology. Front. Immunol. 10, (2019).
Andrews, S. & Pollard, A. A vaccine against serogroup B Neisseria meningitidis: dealing with uncertainty. Lancet Infect. Dis. 14 5, 426–434 (2014).
Martino, A. et al. Structural characterisation, stability and antibody recognition of chimeric NHBA-GNA1030: an investigational vaccine component against Neisseria meningitidis. Vaccine 30, 1330–1342 (2012).
Bi, H., Li, Y. B. & Jiang, S. D. The progress in research of Neisseria meningitidis serogroup B vaccine. 33, 158–161 (2010).
Hsu, C. A. et al. Immunoproteomic identification of the hypothetical protein NMB1468 as a novel lipoprotein ubiquitous in with vaccine potential. PROTEOMICS 8, 2115–2125 (2008).
Vivona, S., Bernante, F. & Filippini, F. N. E. R. V. E. New enhanced reverse vaccinology environment. BMC Biotechnol. 6, 35–35 (2006).
Conte, A. et al. NERVE 2.0: boosting the new enhanced reverse vaccinology environment via artificial intelligence and a user-friendly web interface. BMC Bioinform. 25, (2024).
Ong, E. et al. Vaxign2: the second generation of the first Web-based vaccine design program using reverse vaccinology and machine learning. Nucleic Acids Res. 49, W671–W678 (2021).
Doytchinova, I. A. & Flower, D. R. VaxiJen: a server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 8, 4 (2007).
Salod, Z. & Mahomed, O. Mapping potential vaccine candidates predicted by VaxiJen for different viral pathogens between 2017–2021—a scoping review. Vaccines 10, (2022).
Rizwan, M. et al. VacSol: a high throughput in silico pipeline to predict potential therapeutic targets in prokaryotic pathogens using subtractive reverse vaccinology. BMC Bioinform. 18, (2017).
Jaiswal, V., Chanumolu, S., Gupta, A., Chauhan, R. & Rout, C. Jenner-predict server: prediction of protein vaccine candidates (PVCs) in bacteria based on host-pathogen interactions. BMC Bioinform. 14, 211–211 (2013).
Dalsass, M., Brozzi, A., Medini, D. & Rappuoli, R. Comparison of open-source reverse vaccinology programs for bacterial vaccine antigen discovery. Front. Immunol. 10, (2019).
Aceil, J. & Avci, F. Pneumococcal surface proteins as virulence factors, immunogens, and conserved vaccine targets. Front. Cell. Infect. Microbiol. 12, (2022).
Zeng, L. B. et al. Comparative subproteome analysis of three representative Leptospira interrogans vaccine strains reveals cross-reactive antigens and novel virulence determinants. J. Proteom. 112, 27–37 (2015).
Wizemann, T. et al. Use of a whole genome approach to identify vaccine molecules affording protection against Streptococcus pneumoniae infection. Infect. Immun. 69, 1593–1598 (2001).
Cottom, C. O., Stephenson, R. & Wilson, L. & Noinaj, N. Targeting BAM for novel therapeutics against pathogenic gram-negative bacteria. Antibiotics 12, (2023).
Jaskiewicz, J., Tremblay, J., Tzipori, S. & Shoemaker, C. Identification and characterization of a new 34 kda MORN motif-containing sporozoite surface-exposed protein, Cp-P34, unique to Cryptosporidium. Int. J. Parasitol. https://doi.org/10.1016/j.ijpara.2021.01.003 (2021).
Goldberg, T. et al. LocTree3 prediction of localization. Nucleic Acids Res. 42, W350–W355 (2014).
The UniProt Consortium. UniProt: a hub for protein information. Nucleic Acids Res. 43, D204–D212 (2015).
Chen, L. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 33, D325–D328 (2004).
Kim, Y. et al. Immune epitope database analysis resource. Nucleic Acids Res. 40, W525–W530 (2012).
Ladunga, I., Wiese, B., Smith, R. & FASTA-SWAP FASTA-PAT: pattern database searches using combinations of aligned amino acids, and a novel scoring theory. J. Mol. Biol. 259 (4), 840–854 (1996).
Dandass, Y., Burgess, S., Lawrence, M. & Bridges, S. Accelerating string set matching in FPGA hardware for bioinformatics research. BMC Bioinform. 9, 197–197 (2008).
Arora, I. et al. mtx-COBRA: subcellular localization prediction for bacterial proteins. BioRxiv https://doi.org/10.1101/2023.10.04.560913 (2023).
Restrepo-Montoya, D. et al. Validating subcellular localization prediction tools with mycobacterial proteins. BMC Bioinform. 10, 134–134 (2009).
Vizcaíno, C. et al. Computational prediction and experimental assessment of secreted/surface proteins from mycobacterium tuberculosis H37Rv. PLoS Comput. Biol. 6, (2010).
Alshamrani, S. et al. Mining autoimmune-disorder-linked molecular-mimicry candidates in clostridioides difficile and prospects of mimic-based vaccine design: An in silico approach. Microorganisms 11, (2023).
Begum, S. et al. Molecular mimicry analyses unveiled the human herpes simplex and poxvirus epitopes as possible candidates to incite autoimmunity. Pathogens 11, (2022).
Fujinami, R. & Oldstone, M. Amino acid homology between the encephalitogenic site of Myelin basic protein and virus: mechanism for autoimmunity. Science 230 4729, 1043–1045 (1985).
Pahari, S. et al. Morbid sequences suggest molecular mimicry between microbial peptides and self-antigens: A possibility of inciting autoimmunity. Front. Microbiol. 8, (2017).
Markham, A. et al. Formulation and immunogenicity of a potential multivalent type III secretion system-based protein vaccine. J. Pharm. Sci. 99 11, 4497–4509 (2010).
Rawal, K. et al. Identification of vaccine targets in pathogens and design of a vaccine using computational approaches. Sci. Rep. 11, (2021).
Solanki, V. & Tiwari, V. Subtractive proteomics to identify novel drug targets and reverse vaccinology for the development of chimeric vaccine against acinetobacter baumannii. Sci. Rep. 8, (2018).
Sah, P. P., Bhattacharya, S., Banerjee, A. & Ray, S. Identification of novel therapeutic target and epitopes through proteome mining from essential hypothetical proteins in Salmonella strains: an in Silico approach towards antivirulence therapy and vaccine development. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 104315 https://doi.org/10.1016/j.meegid.2020.104315 (2020).
Farriol-Duran, R., López-Aladid, R., Porta-Pardo, E. & Torres, A. & Fernández-Barat, L. Brewpitopes: a pipeline to refine B-cell epitope predictions during public health emergencies. Front. Immunol. 14, (2023).
Shukla, P. et al. Immuno-informatics analysis predicts B and T cell consensus epitopes for designing peptide vaccine against SARS-CoV-2 with 99.82% global population coverage. Brief. Bioinform. 23, bbab496 (2022).
Zeng, Y. et al. Identifying B-cell epitopes using AlphaFold2 predicted structures and pretrained Language model. Bioinformatics 39, btad187 (2023).
Acknowledgements
The authors thank the Higher Education Commission (HEC) of Pakistan for funding this study under the NRPU project 16038.
Author information
Authors and Affiliations
Contributions
Amjad Ali led the conceptualization, funding acquisition, project administration, resources, supervision, and was equally involved in investigation, methodology, and review & editing of manuscript. Muhammad Hurrarah Hamid equally contributed to data curation, formal analysis, investigation, methodology, software, validation, visualization, and review & editing of manuscript. Samavi Nasir wrote the original manuscript draft and equally contributed to formal analysis, methodology, software, validation and revisions & editing of manuscript. Zaara Ishaq contributed to revisions and editing of manuscript equally, supported data curation, formal analysis, investigation, methodology, validation, visualization. Farha Anwer contributed to data curation equally, supported in formal analysis, investigation, methodology, software, validation, visualization, and review & editing of manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ali, A., Hamid, M.H.B., Nasir, S. et al. B-vac a robust software package for bacterial vaccine design. Sci Rep 15, 31745 (2025). https://doi.org/10.1038/s41598-025-01201-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-01201-0