Abstract
The challenge of early detection and stratification in Parkinson’s disease (PD) is urgent due to the current emergence of mechanism-based disease-modifying treatments. In here, metabolomic and lipidomic parameters obtained by a standardized and targeted in vitro diagnostic research (IVDr) platform have a significant potential to address therapy-related questions and generate improved biomarker panels. Our study aimed to use IVDr nuclear magnetic resonance (NMR) spectroscopy to quantify metabolites and lipoproteins in PD blood serum from different cohorts to stratify metabolically driven subtypes of idiopathic and genetic PD. Serum aliquots from three neurodegeneration biobank cohorts (287 samples in total, including 62 PD patient samples with GBA mutation, 98/43 PD patient samples of early/late stages of disease duration, 20 PD samples from patients with mutations in recessive PD genes and some smaller subgroups of mitochondrial and double mutation cases) were prepared and analyzed with IVDr NMR spectroscopy, covering 39 blood serum metabolites and 112 lipoprotein parameters. Uni- and multivariate statistics were used to identify metabolism-driven changes under consideration of typical confounders such as age, sex and disease duration and set into context with clinical biomarkers such as CSF concentrations of alpha-synuclein, neurofilament light chain, and tau protein. Based on the different PD subgroups we performed a total of eight different comparisons. Highlights from these comparisons include increased citrate and dimethylglycine with a decrease of creatinine and methionine in healthy controls and early PD group compared to GBA, PD late and recessive PD. We furthermore identified decreased HDL-3 free cholesterol in genetic PD cases compared to sporadic subject samples (sum of the PD early and PD late groups). Considering medication, we found that the levodopa equivalent daily dose (LEDD) is mostly positively correlated with tyrosine and citrate in sporadic PD compared to pyruvate and phenylalanine in genetic PD. Cerebrospinal fluid levels of alpha-synuclein were negatively correlated with alanine. Further metabolites and lipoproteins with discriminatory power for double mutation PD cases involved ornithine, 2-aminobutyrate and 2-hydroxybutyrate as well as for mitochondrial phenotypes via LDL phospholipid, apolipoprotein and cholesterol subfractions. Quantitative IVDr NMR serum spectroscopy is able to stratify PD patient samples of different etiology and can contribute to a wider understanding of the underlying metabolism-driven alterations e.g. in energy, amino acid, and lipoprotein metabolism. Though our overall cohort was large, major confounders such as age, sex and medication have a strong impact. That is why absolute quantification and detailed patient knowledge about metabolic confounders, is a premise for future translation of NMR serum spectroscopy to routine PD diagnostics.
Similar content being viewed by others
Introduction
Parkinson’s disease (PD) is characterized by a unique combination of motor and non-motor symptoms1. While preclinical PD models suggest new targets for therapy2, there is huge lack to develop new robust biomarkers for early phases of the disease3.
In recent years, omics techniques (genomics, proteomics, transcriptomics, and metabolomics) have entered the research field to study PD pathology and pathogenesis4. From the genetic standpoint, the beta-glucocerebrosidase gene (GBA) mutation, also causing Gaucher’s disease, is a critical genetic variable5, which needs to be considered in population-wide studies of PD. In a recent study6, the increasing importance of proteomics and transcriptomics was further demonstrated in network-building (including correlations between miRNAs and proteins) and highlighting the potential PD biomarker 14-3-3 protein zeta/delta (YWHAZ) protein. Overall, cooperation of omics methods of PD population screening (genomics – via genome-wide association studies (GWAS), transcriptomics, and proteomics) tend to give complimentary results and allow to investigate processes behind protein folding (aggregations)7.
Metabolomics research in PD has proven to help screen tissue or biofluid samples for low molecular mass compounds with polar and lipid characteristics8. Metabolomic studies can provide essential insights into pathologic alterations of metabolic pathways and may lead to discovering new PD biomarkers. Using metabolomics, two analytical techniques are used to study PD: mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy9,10. Unlike MS, NMR based metabolomics offers the benefit of a non-destructive sample analysis and precise absolute quantification11,12. On the other hand, NMR suffers from poor sensitivity and metabolites can only be identified at relatively high concentrations. For any metabolomics investigation, the acquiring large sets of patient material with associated metadata is essential to contribute to a better understanding of Parkinson’s disease.
The utilization of NMR-based metabolic and lipoprotein parameters in combination with clinical biomarkers for PD, neurodegeneration and aging has been previously used in PD research13,14 and such combinations may reach a certain level of stratification in a PD phenotype, as was shown in a study by Troisi et al. with the use of gas chromatography - mass spectrometry15. One major research field is to identify significant metabolic anomalies that allow to distinguish iPD from other PD types.
In our labs, we recently applied quantitative NMR spectroscopy to examine serum and CSF samples from AD (Alzheimer´s disease) and MCI (mild cognitive impairment, investigated with AD biomarkers in CSF) patients16,17. Thus, the aim of our study was to elaborate how the NMR-based analysis of blood serum and lipoprotein parameters of different PD subtypes could be linked to genotypes and existing clinical indicators to expand the diagnostic PD toolbox. Such investigations may, at some point in the future, lead to novel screening programs or improved patient care e.g. using this technology as monitoring tool within curative approaches18.
Results
Analysis 1: the comparison of main PD groups elucidates imbalances in blood metabolites
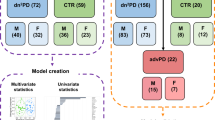
The overall cohort is the sum of three subproject cohorts that were measured at different time spans between 2021 and 2023 and whose samples derived from two different biobanks, as illustrated in Table 1; Figs. 1 and 2. Serum samples for cohorts 1 and 3 were provided by a local biobank in Tübingen, Germany, whereas the subproject cohort 2 samples derived from a biobank at the DZNE in Bonn, Germany. The pooling of these cohorts allowed to reach higher n-numbers which are mandatory for a metabolomics approach.
Full clinical metadata and CSF biomarkers are provided in Supplementary Table 1. The following ID numbers and attributes have been used within the Supplementary Table 1. The GBA genetic variations have been labeled additionally with the grade of Gaucher’s disease severity and their positive association with PD19,20 and is increasing as follows: GBA risk → GBA mild → GBA severe. VUS label means “variance of unclear significance”.
Our present study classified patients as having either sporadic PD early or sporadic PD late. Those with PD duration parameter within five years were classified as having sporadic PD early entry, while those with more than five years were classified as having sporadic PD late group label.
The supplementary material also includes entries for comorbidities in non-PD controls (from all three cohorts of the current study). Five individuals, in our investigation, are marked as “Mito” because these patients were born with recessive mitochondrial diseases. It is also important to highlight another small group of participants diagnosed with dementia with Lewy bodies (DLB). The DLB condition was in included in the sporadic analysis group.
Overview of the cohorts, sample numbers and plan of study. (A) Descriptive statistics plot (mean ± SEM, SEM - Standard Error of the Mean) on age, showing the significantly higher age of the Sporadic PD late group (main 5 groups); (B) Descriptive statistics plot on sex in which a balance corridor of 40% ±10% among groups is observed (main 5 groups); (C) Descriptive statistics plot (mean ± SEM, SEM - Standard Error of the Mean) on age (remaining groups); (D) Descriptive statistics plot on sex (remaining groups); (E) Overall study plan– Created in BioRender. Berezhnoy, G. (2025) https://BioRender.com/m96r807.
Comparing the 5 main groups (Analysis 1 in Fig. 1E), methionine had a significant decrease (FDR < 0.05) in recessive PD, GBA, and late PD patient groups (Fig. 2). The non-wildtype PD groups (PD recessive, GBA) displayed lower creatinine levels and higher dimethylglycine (DMGly). Further significant metabolites were higher citric acid (citrate) levels in late and recessive PD and ethanol, which had many missing values (below limit of detection) as displayed in the violin plot. We observed that the highest blood formate levels were found in the group of control samples. After testing for FDR significant ANOVA variables in these five main patient groups, we could locate covariant dependencies between the main checked factors (PD disease duration, age, and sex) and ethanol (Supplementary Table 2).
Significant metabolite alterations within the five main studied groups. Heatmap of FDR significant findings checked via ANOVA (p < 0.05). The six significantly changed metabolites are creatinine, methionine, formate, ethanol, dimethylglycine (DMGly) and citrate (shown as dot plots, mean ± SEM). For Ethanol plot 9 dots were not plotted.
Analysis 2: male PD subjects are characterized by higher levels of VLDL and valine while females show increased LDL and HDL lipoproteins
Performing Analysis 2 in Fig. 1E and applying a stricter FDR-adjustment (p < 0.001, Supplementary Fig. 2) filter, numerous variables changed, when separately analyzing male and female subgroups (using Ward clustering method). Doing so, we spotted in males (but interestingly also PD recessive females, Table 1) higher levels of very low-density lipoprotein (VLDL) parameters. Males on average had higher blood creatinine and valine in comparison to females. Furthermore, the BCAAs leucine and isoleucine as well as Glyc/SPC were elevated in male cohorts. Finally, we identified citrate and DMGly being higher in some but not all male groups. In the subset of female groups, we found increased LDL subfractions LDL1-LDL3, creatine, HDL subfractions HDL1-HDL3, overall HDL content, and the SPC peak (supramolecular phospholipid composite) in comparison to males.
Analysis 3: sporadic PD early and sporadic PD late patients can be distinguished by their blood citrate levels
Comparing NMR data of Sporadic PD early with Sporadic PD late (Analysis 3 in Fig. 1E) did not reveal any significantly changed lipoproteins; however, FDR-significantly changed citrate (Supplementary Fig. 2) in the Sporadic PD late group. We must note that the Sporadic PD late group is skewed toward higher patient ages, and therefore FDR significant results of citrate from the two-group t-test could be due to the aging factor. However, after analysis of covariance performance check, we found that citrate is not biased towards the PD disease duration variable, age, and sex; and its final covariate p value is significant (Supplementary Table 2).
It was determined, for our cohorts of patient samples, that citrate portrayed the strongest t-test significance result (with FDR-rate considered) at the level of pFDR < 0.0125, while another TCA cycle-related metabolite, 2-oxoglutarate, was having pFDR of ~ 0.125 reported value of discrimination between the PD (early) v (late) groups. Of course, when the FDR-adjustment is dropped, t-statistics becomes significant for both metabolites. Succinate, another TCA-related metabolite that is measurable by our analytical assay applied, was neither raw p-value-wise nor after an FDR-adjustment even considerable for noteworthy changes between the two patient samples groups.
Analysis 4: genetic and sporadic PD patients are discriminated by HDL free cholesterol particles, creatinine, methionine and dimethylglycine
Comparing patient samples with sporadic (combination of PD early and late groups) against genetic PD (Analysis 4 in Fig. 1E, see Table 1, including double mutation carriers, PD recessive, PD GBA, and PD LRRK2 individuals’ samples; LRRK2 – leucine-rich repeat kinase 2) with two non-PD control groups (age- and sex-matched control group and group with subset of female participants due to age and sex matching; see Supplementary Table 2 for main statistics output), we were able to determine lowered histidine and methionine levels (Fig. 3).
Furthermore, creatinine was significantly lower in female controls and the genetic group. Of note, creatine was proven to be in covariance dependent on PD disease duration variable, or either to age or sex characteristics, therefore, the p value after covariance check is no longer significant, Supplementary Table 2). There were also technically variable changes of ethanol and asparagine (due to low n numbers). The HDL lipoprotein variable of HDL-3 free cholesterol was lowered in the PD genetic group, whereas dimethylglycine was elevated.
We also ran a significance t-test evaluation for only two diagnostic groups - genetic and sporadic PD. This observation of methionine and dimethylglycine was also proven to be FDR-significant (pFDR = 0.006 – Methionine; pFDR = 0.009 – DMGly). Overall, this additional two-group-based comparison highlighted six FDR-significant variables (Fig. 3), including two lipoprotein parameters from HDL subfractions of free cholesterol (HDL-3 and HDL-4). Characteristics of changes are the same also for the two-group comparison for those repeated parameters of the two analyses.
Analysis 5: correlation of NMR parameters with CSF dementia biomarkers, α-synuclein, BMI index, Levodopa equivalent dosage daily, cognition and depression scores, and other relevant data
We additionally performed correlation analyses based on existing neurodegeneration biomarkers and clinical metadata (Analysis 5 in Fig. 1E). As not all metadata variables were available for all patients, the respective group sizes changed accordingly and the updated numbers are provided in the Methods section and Supplementary Table 3.
Figure 4 and Supplementary Fig. 3 summarize our findings: First, we would like to highlight the negative correlation (r = −0.29, p = 0.01) between CSF total alpha-synuclein and blood alanine (which is lower in serum of late PD patients, when compared to Sporadic PD early). Second, the body mass index (BMI) was positively correlated with the inflammatory glycoprotein parameter Glyc/SPC ratio (FDR range of significance; r = 0.45, PFDR < 10−7). Third, the correlation between HDL-4 apolipoprotein Apo-A2 and CSF-measured human total tau protein was significantly negative (r = −0.22, P < 0.01). Furthermore, dimethylglycine and citrate were positively correlated (citrate – r = 0.25, PFDR < 0.01; DMGly – r = 0.25, PFDR < 0.01) with the calculated LEDD dosage.
Overall, there were no clear results from any correlations checked for datasets based on values of MoCA. We can report that the two-group selection of non-PD controls and the PD recessive group provided a positive correlation with 3-hydroxybutyrate, a ketone body metabolite, (r = 0.39, P value not significant) and MoCA. Finally, the clinical depression score BDI-II showed a positive correlation with the apolipoprotein ratio ApoB100/ApoA1 (r = 0.20, P < 0.01; Fig. 4).
Besides the parameters listed in Fig. 3, Nfl plasma and other parameters (Aβ1–42, Aβ1–40, Aβ1–38, P-tau-181, weight, height, all available MoCa values, sex, age, aao – age at onset, PDdd – PD disease duration, CSF NfL levels, MDS UPDRS III score) were checked via correlation analysis as well. However, correlation for those parameters were only marginal (Supplementary Fig. 3, Supplementary Table 3).
In here, we also had to overcome the issue of missing entries with some sub-group merging and summarize the main challenges and how we addressed them. One patient with mitochondrial disease (labeled as ‘Mito’) was diagnosed also PD, so their PDdd value (of PD disease duration) was applied due to their positive PD status. For metadata preparation of tables in correlation analysis, we merged some groups to preserve all available metadata entries and reach the highest n-number. Female controls (control_F) were merged with Mito and PD LRRK2 groups for alpha-synuclein data. For BMI data, PD LRRK2 and PD recessive groups were merged. For h-t-tau (h-Tau) data, DLB and PD LRRK2 groups were merged. For LEDD data, DLB and PD LRRK2 groups were merged. For MoCA data, DLB and Mito groups were merged. For BDI-II data, non-PD controls were merged with the extra female non-PD controls (Control_F). For Aβ1–40 data, Control_F and DLB groups were merged. For Aβ1–42 and also p-tau-181 data, DLB, Mito, and PD LRRK2 groups were merged. PD LRRK2 and PD recessive groups were merged for weight and height characteristics. For PDaao data, the PD LRRK2 group and the PD-Mito individual were merged. For Aβ1–38 data, Control_F and DLB groups were merged. For Nfl plasma data, Control_F, PD double mutation carrier, and PD LRRK2 groups were merged. For CSF NfL data, Control_F and Mito groups were merged. For MDS UPDRS-III data, Control_F, DLB, and PD LRRK2 groups were merged.
To further elaborate how NMR parameters have a potential to distinguish between Sporadic PD early and Sporadic PD late, we performed an area under the curve receiver operating characteristic AUC-ROC analysis on the LEDD dataset exclusively for both early and late PD duration groups. In this aspect, citrate was higher in the late PD group (Fig. 4). However, when checked directly, we could not find citrate as the closest metabolite parameter positively correlated with LEDD. By contrast, blood tyrosine showed a stronger correlation (for early and late PD groups), likely due to their close metabolic positions, as tyrosine is a precursor of L-dopa. Furthermore, for PD genetic groups there was a high positive correlation between LEDD and blood pyruvate. Negative correlations were identified between LEDD in the PD genetic subgroup and several parameters of the HDL4 subfraction.
Regarding the LEDD-driven influence on metabolome, we briefly did a split of LEDD dataset of our study and reclassified with accordance of levels (zero, low, medium, high) and patient sex as possible factors driving our metabolic reported alterations (Supplementary Fig. 4). With an applied threshold of pFDR < 0.001, we saw that most metabolic driver variables were dimethylglycine and citrate. On the lipoprotein part, we can summarize that several groups (re-stratified) were different based on how much of HDL lipoprotein particles, including HDL cholesterol blood values; were there and also the SPC signal.
We furthermore saw abnormal elevation of HDL triglycerides on average in the PD recessive (females) samples. As the following correlation shows, HDLs and also SPC in the strongest way correlate with the female sex factor.
We also did an integrated (correlation) analysis of blood methionine, DMGly and formate levels to check for any abnormalities in one-carbon metabolism in people with PD. We also included some extra parameters and metabolites as of described for the one-carbon metabolism, that we can detect with our methodology, as reviewed in21. We also did a correlation analysis to see if there was a connection between DMGly and LEDD in both GBA mutation positive and sporadic PD groups, to work out if DMGly changes can be affected by dopaminergic treatment.
Of note, we were able to see that blood citrate levels follow (by correlation analysis) closer to the factor of PD disease duration – leading metabolite, while patient age factor was positively correlating stronger with other metabolites in places prior to citrate (choline, TMAO, tyrosine, acetate, and dimethylsulfone) and also lipoprotein variable of HDL-1 subfraction signal constitute for apolipoproteins A1 (Supplementary Fig. 3).
Citrate, as any other our NMR parameters, did not meaningfully correlate to the MDS UPDRS-III patient data. Our determined value of correlation coefficient is + 0.013554 (r; Spearman’s; Supplementary Table 3).
Correlations of clinical biomarkers and metadata with metabolites and lipoprotein alterations. (A) A negative correlation was found between blood alanine and CSF alpha-synuclein, (B) the Glyc/SPC ratio parameter shows a significant positive correlation with BMI index, (C) lipoprotein characteristic HDL-4 apolipoprotein Apo-A2 had a negative correlation with human total tau levels, (D) DMGly and citrate metabolites have positive correlations with LEDD, (E) blood 3-HB levels show a nonsignificant tendency to correlate to MoCA score (within non-PD Controls and PD recessive groups of individuals’ samples only), (F) blood ApoB100/ApoA1 ratio correlate positively with BDI-II, (G) area-under-curve analysis (receiver operating curve building) demonstrates a good statistical differentiation of Sporadic PD early and Sporadic PD late patient groups (n total = 139) for the LEDD available entries only, (H) area-under-curve analysis plot with a logarithmically normed box plot of blood citrate indicating of Sporadic PD early and Sporadic PD late patient groups (n total = 139) for the significant discriminatory characteristics tested separately based on the LEDD available entries only patient samples, (I) tyrosine shows a positive correlation with LEDD for PD sporadic (early, late; n = 139), (J) pyruvate has a positive correlation with LEDD for PD genetic groups (GBA, recessive, LRRK2, and double mutation carriers; n = 92). The software used places LEDD in the number one position, with a correlation of value plus one with itself (segments I and J). The rates shown on a (“classical”) Receiver Operating Characteristic (ROC) curve, often so, are also plotting the true positive rate (the sensitivity) against the false positive rate (1-specifcity) for different threshold settings of a diagnostic test, as performed in MetaboAnalyst22. The red separator line is representing an “optimal cutoff” that is according to located point furthest from the diagonal and according to the Youden’s method23.
Analysis 6: methionine is significantly decreased in subtypes of GBA driven PD
Further discrimination within the PD GBA group may be of particular clinical interest (Analysis 6 in Fig. 1E), and we identified lowered methionine levels in GBA groups (ANOVA, Fig. 5). Unfortunately, the GBA VUS group could not be analyzed in Analysis 6 due to the low n-number limitation of the number of analyzed groups. The mentioned group is further checked in Analysis 7.
Metabolites that were distinct in this comparison were creatine, formate, asparagine, citrate, creatinine, ornithine, and DMGly. We also identified the significance of the VLDL-5 subtraction parameter of free cholesterol. It is essential to mention that PD duration covariate (and also sex- and age- based covariates) is helpful for further investigations. Doing so, we found creatine as such dependent metabolite (Supplementary Table 2). The data entries from asparagine have not been analyzed (too many zeros).
Significant metabolite and lipoprotein alterations in eight-group comparison (non-PD Control, PD genetic phenotype with GBA groups excluded, PD sporadic, PD GBA groups divided by severity grade). Heatmap of p-value FDR-significant findings checked via ANOVA statistical testing. Dot plot of methionine shows lowest level of this metabolite, shown as means and SEMs, in GBA groups.
Analysis 7: methionine and ornithine levels differ in diagnosed PD GBA patient groups
Interestingly we also found decreased methionine for GBA VUS in addition to the previous analysis. Alterations in methionine can be connected to the gut microbiota24,25,26. Concerning case-control discrimination of GBA PD patient samples and risk stratification of the GBA mutation (GBA risk, mild, severe), methionine was identified as significant over analyzed groups.
It was lower in comparison to non-PD controls (Fig. 6). Furthermore, a substancial increase in ornithine levels was shown in GBA patient groups. Overall, the PD GBA severity grade was significantly (on average) discriminated by ornithine elevation levels. Testing covariance resulted in identification of none of the two metabolites being dependent of PD disease duration time (or age, or sex) rather than discrimination of the analyzed patient groups (Supplementary Table 2).
Metabolomic and lipoprotein significant alterations in five-group comparison (non-PD Control, PD GBA groups separated by a severity grade). Heatmap of p-value FDR-significant findings checked via ANOVA statistical testing. Cohort groups providing dot plots of ornithine and methionine, shown as means ± SEM.
Analysis 8: A potential interplay of 2-aminobutyrate in PD recessive patients
We performed additional analyses in Analysis 8 (Fig. 1E) to find potential discriminating metabolites between non-PD controls and PD recessive mutation carriers, including some small patient groups (Table 1). The groups in question here are as follows: PD patient samples with double mutations (carriers), PD LRRK2 mutation-positive, PD PRKN biallelic, PD PRKN monoallelic, non-PD controls, non-PD controls the female surplus group, DLB individuals, and the Mito group.
In this analysis, we could not find significant differences according to univariate statistics. Thus, to further explore, we applied a regression analysis (PLS-DA) (Fig. 7). In here LDL lipoproteins were shown to be decreased in PD mitochondrial diseases-positive individuals and the DLB phenotype group. Higher 2-aminobutyrate (2-AB) were observed in PD PRKN. 2-AB was found at zero levels for the patient groups of PD LRRK2 positive and the double mutation carrier individuals’ samples (Supplementary Table 1).
Metabolomic and lipoprotein alterations in the comparison of smaller groups (non-PD Controls, DLB, Mitochondrial diseases, PD double mutation carriers, PD LRRK2 positive, PD PRKN biallelic, and PD PRKN monoallelic). Using regression analysis (PLS-DA) 2-aminobutyrate (2-AB, shown as means ± SEM) and, on the other panel, LDL-3 and LDL-4 lipoprotein particles showed strong discriminatory power. PLS-DA – partial least squares discriminant analysis.
Discussion
In clinical routine, patients and individuals with symptoms of Parkinson’s disease are usually stratified based on genetic testing, imaging, established CSF/blood biomarkers and clinical history. In recent years, scores of clinical questionnaires and tests have been used (e.g., the Unified Parkinson’s Disease Rating Scale (UPDRS) for clinical severity of the main PD symptoms, the Beck Depression Inventory (BDI) for mood assessment and the Parkinson’s Disease Questionnaire (PDQ-39) for subjective disease symptoms)27,28,29,30,31,32,33. In here, the Montreal Cognitive Assessment (MoCA) and the Mini Mental Status Examination (MMSE) are the tests most widely used for clinical research on PD dementia34,35, and could be likely used to diagnose (and improve accuracy in diagnosing) cognitive impairment and dementia in PD.
Of note, it has been shown that various rare mutations in genes causing familial Parkinson’s disease are linked to PD pathogenesis, having led to a broader understanding of PD as a whole, including idiopathic PD (iPD)36,37. Thus, to better understand iPD, some studies provide map-like pathological observations of clinical diagnostic data to better understand PD heterogeneity38,39,40,41. Furthermore, towards the impact of biological sex, the prevalence of PD is higher in males than females42,43. This is an additional challenge when aiming for stratification and mapping phenotypic features towards a refined and improved management of PD.
An overall analysis of CSF and blood serum biomarkers in a large (n = 453) cohort of PD patients revealed a panel of inflammatory markers and sex-specific differences44. Within DJ-1 parkinsonism, an important role is played by altered mitochondrial functions and oxidative stress45,46. Furthermore, the PARK7 mutation is associated with primary parkinsonism and can be disrupted by DJ-1 activity45,47,48. Both oxidative stress and energy metabolism are strongly impacted by mitochondria, so the features of these organelles must be considered when assessing imbalanced states in neurodegeneration phenotypes49,50,51,51. Here, quantitative metabolites levels could help to assess the magnitude of oxidative stress, aging, dietary effects, and overall neurodegeneration.
To evaluate the progress, prognosis and onset of PD, biomarker-based investigations reflecting the intensity of nervous system degeneration are highly relevant. Our present study used a quantitative in vitro diagnostics research (IVDr) NMR spectroscopy approach to measure a targeted panel of blood metabolites and lipoproteins that has to technical potential to be readily applied during routine clinical visits or within extensive population screening program.
Having a set of different PD subgroups, we performed a total of eight different statistical comparisons to assess how NMR parameters can stratify these different patients and to learn what major confounders have to be considered. Patients with recessive PD forms were studied to gain insights into the genetic basis of metabolomic findings derived from blood samples. The analysis focused on mutations associated with the parkin (PRKN, PARK2), PINK1 (PARK6), and PARK7 (DJ-1) genes. The respective proteins from all three are helping to maintain mitochondrial wellbeing and are crucial for the optimal functioning of mitochondria52,53,54,55.
In the forthcoming discussion sections, we will concentrate on the results that were identified as FDR-rate significant.
Methionine is among the strongest differentiating metabolites overall
The non-wildtype PD groups (genetic group) demonstrated lowered levels of methionine and histidine. This is in line with another recent PD blood metabolomics study, where the two metabolites were lower on average for advanced stage of PD patient group in contrast to controls13. Histidine provides antioxidative protection in the metabolism56,57. Methionine is protective against higher levels of ROS58 in a model of PD.
Besides methionine, creatinine was another important metabolite in Analysis 1, comparing the five major patient groups of our study. Both compounds were significantly altered in amyotrophic lateral sclerosis (ALS)59. The study showed that these two metabolites together with asymmetric dimethylarginine and some phospholipids were machine learning modeled to highlight a trend of metabolically relevant ALS pathogenic alterations in contrast to controls.
Importance of biomarkers and meta-analysis in PD research
Starting from the methodology of seed amplification assays (SAA), there were indications of alpha-synuclein SAA was checked as the most accurate way to make a diagnosis60,61,62,63. Xylaki et al. 2023 have reported a significant reviewing work to identify extracellular vesicles (EV) proteins and ribonucleic acids; and EV-derived biomarkers for diagnosing PD and assess their clinical significance64. Furthermore, Xie et al. 2023 did a multimodal meta-analysis of the changes of neuroimaging parameters in PD compared with normal controls through whole-brain meta-analysis65. There were no signs of significance via multimodal analysis among brain areas of PD patients, as they had series of spontaneous brain activity abnormalities65. Yet, a meta-analysis performed by Xu et al. 2023 showed a consistent increase of iron metabolism markers in the brain of PD patients66.
Ornithine alterations might serve as readout for oxidative stress in PD
Evaluating the impact of GBA, methionine and ornithine levels were changed along with the GBA mutation risk status. As for ornithine, in PD, it was found to be elevated in urine67. A recently published meta-analysis, however, did show PD-related reports on increased ornithine metabolite levels (blood, urine, CSF) compared to controls68. Ornithine changes (blood, urine) could indicate increased liver and kidney activity in PD and changes within the urea cycle metabolizing nitrogen.
However, according to one study of L-Ornithine-L-Aspartate69, it may prevent neuronal dysfunction in Parkinson’s disease in in-vitro models due to the capability of ornithine to cross the blood-brain barrier. As ornithine is a compound that participates in polyamine metabolism. Polyamines can participate in gene expression and antioxidative processes70,71,72, and ornithine was elevated in serum of PD patients73,74. On the other hand, it was proposed that ornithine causes longitudinal osmotic pressure in brain regions and induces damage74,75. Elevated ornithine levels were reported in aged (advanced stage) PD patients76. Therefore, ornithine could play a significant role in the blood-based screening for PD individuals.
It may also suggest poor digestive function77,78 in Parkinson’s disease patients (e.g., resulting in constipation). But, in this case, blood ornithine is reported as lowered in patients with functional constipation79. Finally, a urine of idiopathic PD patients’ screening study showed elevated ornithine also in that matrix67. Authors of the publication67 associated their ornithine finding with patients’ bowel problems, such as constipation, and mitochondrial dysfunction73,77,80.
Significant alterations dimethylglycine could mirror dysbiosis in PD
Several of our comparisons (Analysis 1, Analysis 2, Analysis 6) we found an increase in DMGly. This metabolite is result of choline metabolism and could indicate changes of gut-related metabolism. An increase in DMGly in our study groups could be a distinctive change of gut-related metabolism, which did not follow, however, the previously reported DMGly decrease in PD patients13. DMGly contributes to one-carbon metabolism with formic acid and methionine. L-Dopa may be an altering factor in the one-carbon metabolism as well81. Regarding one-carbon metabolism, the liver as main entrance organ for most gut-derived metabolites must be considered82. Therefore, it would be beneficial to study and compare blood changes and more accurate or frequent data on the metabolites belonging to both the metabolic pathway and metabolic profiling83.
Citrate as a metabolite of aging
Further results Analysis 1 showed elevations of citrate and dimethylglycine in other groups (PD GBA, Sporadic PD late, and PD rec) compared to controls and Sporadic PD early participants’ samples. Further studies with larger n-numbers have to support these findings. Overall, higher blood citrate might indicate increased aging84,85,86. There may be a link also towards oxidative stress. Blood citrate was increased in blood of PD, progressive supranuclear palsy, and multiple system atrophy patients when compared to controls87.
Changes of citrate refer to TCA cycle metabolism alteration, which (on par with other metabolic pathways) is one of several features in mitochondrial dysfunction and discrimination of Sporadic PD early and Sporadic PD late88. Regarding that, our findings of higher citric acid levels in Sporadic PD late are not in line with another NMR-based study of PD patients89 and do not meet the direction of changes compared to the de novo drug-naive and advanced stage PD individuals13. So, findings upon citrate remaining contradictory.
Our analysis found that there was no significant correlation between the blood citrate levels and the MDS UPDRS-III scores. Citrate and isocitrate did shown a certain tendency to be higher in aged population90. This same preprint also found that citrate was higher in Dopa-negative idiopathic PD rather than controls, also there citrate correlated itself in a negative way correlation to the factor of PD severity90. Interestingly, one paper suggested to collectively view PD-related evidence of citrate and also tyrosine metabolism alterations91 and linking into92 towards earlier investigations of PD in cerebrospinal fluid, blood, and saliva93,94.
Biological sex is a major confounder for NMR based serum metabolites and lipoproteins
Studying the impact of biological sex upon NMR-based parameters, VLDL particles were elevated overall in the men of our study, which is a typical trend as before95,96,97,98,99. Furthermore, levels of BCAAs were elevated in male subjects100,101. This is interesting since we discovered sex-related changes, including creatinine and valine, in our cohorts. Sex-specific differences have been underappreciated, that should no longer happen102,103,104,105. Multiomics approaches in human tissue research study of ALS and animal model investigations highlighted therapeutically related evidences and sex-related differentiations106. We believe that future research of PD neurodegeneration could benefit from further approaches in securing evidences in both therapy- and sex-related alterations.
Sex-specific and PD-related changes in lipoproteins in this study may be explained by starvation rates, insulin resistance, dyslipidemia, and possibly sarcopenia. Sarcopenia has a wide range of associations with patients’ comorbidities, yet specific links have been found how insulin resistance is associated with obesity and sarcopenia107.
On another hand of PD study field that is about sex hormones108, estrogen use after menopause was associated with a lower risk of developing Parkinson’s disease as reported in109. A study110 in monkeys showed111 that estradiol altered dopamine metabolism112 and transporter uptake in the brain after surgically induced menopause.
Measurement of dihydrotestosterone would have allowed better characterization of the metabolism of androgens and reveal more involvements about PD, as was pointed out in113. There is some evidence to suggest that drinking alcohol and smoking might reduce the risk of PD for men more than for women114. For example, there are differences in how men and women’s bodies react to alcohol and smoking, and also in how much nicotine they can take in114. A healthy alternative, physical exercise is a way to reduce PD-associated risks for men and women with also reflections towards their metabolic and lipid alterations114.
NMR parameters show rather week correlations with clinical PD biomarkers
Matching NMR parameters with clinical biomarkers, several correlations provided some interesting findings. Correlation analysis revealed a notable correlation between BMI and the NMR-based Glyc/SPC ratio. This ratio is an NMR-based metric widely during COVID-19 research. Both Glyc and SPC parameters, parts of the ratio, were correlated with the BMI index in a study of COVID-19 patients115.
We found that with higher LEDD, slightly higher DMGly levels are observed. This correlation could be linked to gut dysbiosis. Yet, DMGly is also part of the folate cycle metabolism and close to homocysteine via a remethylating pathway116. This report rose the idea of high homocysteine in blood may be due to the adverse effects of L-DOPA treatment. The one-carbon metabolism was reported to be connected for PD dementia patients cohort in the direction of higher homocysteine amounts in the brain due to L-DOPA117. In some additional clinical context, patients with autism were reported to have a significant increase in DMGly due to oxytocin118.
We noted another correlation of LEDD with blood citrate. Moreover, we found blood citrate to well discriminate LEDD-provided datasets of Sporadic PD early and late disease duration groups in the AUC analysis. Overall, other researchers showed via correlation analysis some significant changes in metabolomics that are potentially applicable for further research of idiopathic PD, as for example correlations between UPDRS III motor score and lactate and BCAAs (valine and isoleucine)123.
Evaluation of quantitative IVDr NMR spectroscopy to serve as diagnostic add-on in PD research
One strength of NMR spectroscopy is it is high-throughput and excellent experimental reproducibility when investigating large cohorts of human samples allowing to combine data from different batches. Furthermore, the absolute quantitation allows a direction comparison with clinical metadata and lab parameters or protein-based biomarkers. Blood research, in this approach, may provide further insights of PD pathology119. Furthermore, the combination of NMR-based data sets of multiple biofluids from PD patients with knowledge of the underlying genetic PD predispositions (including familial dominant and recessive forms of PD), may provide further context to previous findings in PD patients with PINK1 mutations120.
A recent NMR-based PD blood metabolomics study reported elevated plasma pyruvate levels121. Additional results from the same study showed a significant decrease in TCA cycle metabolites, suggesting the importance of underlying mitochondrial dysfunction. However, a limitation of that study was the relatively small number of participants. Another study identified elevated plasma levels of the branched-chain amino acids (BCAAs) valine and isoleucine87. BCAAs may reflect pathophysiological changes in the blood of individuals with neurodegenerative diseases that have been reported as well for patients with dementia16,122. Another metabolite that was elevated in PD blood was glutamate87 which is part of the alanine-aspartate-glutamate metabolism pathway. Comparing iPD patients with healthy controls123, higher serum levels of alanine, citrate, acetate, glycerophosphocholine, glycine, and trimethylamine-N-oxide (TMAO) were found in iPD. Interestingly, negative correlations were reported between BCAAs (and lactate) and UPDRS III motor score next to another negative correlation between TMAO and disease duration123. Alanine is involved in muscle metabolism. Which is interesting, as loss of muscle tissue (sarcopenia) is an aging parameter also in Parkinson’s disease124,125,126.
Amino acids transported to the brain (via the blood-brain barrier) are necessary to sustain essential brain functions and BCAAs contribute to a vital part of our brain energy metabolism87. In a study from 2020, patients with PD were investigated by NMR metabolomics127. It was demonstrated that acetate, ketone bodies (acetone and 3-hydroxybutyrate (3-HB)), lysine, glutamine, tyrosine, and phenylalanine were increased in PD with a decrease of glutamate127. In a later study of 2022, an NMR standard operation procedure (SOP128,129,130 solution was used to profile metabolites and lipoproteins in blood samples of de novo drug-naive PD patients, healthy controls, and advanced PD patients with dopaminergic treatment13. However, the evidence of PD progressive patients’ alternated blood phenotype was drawn to oxidative stress factors in Meoni et al. 2022 42. Later, the study researchers created a biological age prediction model in PD patients to investigate the discrepancies observed between age estimation in control individuals and age prediction in patients via NMR-based metabolomics14.
In recent years, metabolomics and lipidomics have emerged as promising analytical tools to measure human biofluids upon health and disease status. IVDr NMR spectroscopy is a method of choice as it is highly reproducible, quantitative, and high throughput. IVDr NMR-based studies have been successfully applied, especially for metabolic disorders such as diabetes, cardiovascular disease131, or hypertension132. Under the hypothesis that the duration and severity of PD are impacted also by metabolism, we gathered 287 blood serum samples within three different subprojects.
We merged these cohorts for a total of 8 different analysis. Though we estimated that statistical power would be sufficient, only a handful of metabolites and lipoproteins met significance threshold for multiple hypothesis testing correction (FDR adjustment). Still, our results demonstrate a first step how IVDr NMR could contribute to future PD screening, progress monitoring or treatment efficacy testing programs. Future investigations should include higher n-numbers for the respective subgroups as within many of our statistical tests we found clear trends of changing metabolites, however p-values after FDR correction remained > 0.05. This is mainly for blood serum metabolite and lipoprotein analysis, which is crucial as these parameters can quickly change to diet and exercise, and a perfect standardized collection is almost impossible. In conclusion, we could give evidence for several significantly changed metabolites and lipoproteins in different PD patient groups, which shows that the impact of metabolism should not be neglected in PD research and that to do so, IVDr NMR serum spectroscopy is a suited tool.
Limitations of the study
Study design: This project was designed as a metabolomics study. It lacks a holistic clinical metadata knowledge, such as imaging experiments or further clinical laboratory results, which may either not be available or not yet completed, or which otherwise can only be analyzed in a different - time-dependent - type of patients’ analysis. We were also not able to locate all the clinical data from the measured samples as a form of complete matrix. Therefore, several important PD and dementia parameters were represented with a much lower n-number due to the aforementioned data unavailability.
NMR technology: Despite all the recent advantages of NMR spectroscopy, it is well known that the use of complex mixtures such as biofluids comes at the cost of low sensitivity of the method, despite its accuracy. Indeed, the range of reference proton resonance signal concentrations is in the millimolar range. Additionally, overlapping signals during an analysis comes at the cost of some larger error discrepancies.
Sample collection: Next, the use of patient samples for metabolomics studies always is threatened by delayed collection and deep-freezing which could result in the degradation of certain metabolites. Unfortunately, there is no easy way to quickly take a sample of cerebrospinal fluid or any other fluid close to the human brain. Additionally, when a blood or CSF sample is taken, disinfectant alcohols may be introduced into the sample.
Confounder analysis: Age matching between groups was not fully achieved. This is due to the specific nature of the study involving an older population with PD (PD_late). Efforts were made to minimize sudden age differences and to extract statistical data analysis that was fully relevant to the present study. Yet, to make sure our findings are reflective of the disease progression, we ran generalized linear model (GLM) in R (version 4.2.1) and R-studio (2022.2.3.492) with covariates, such as sex, age, and PD disease duration (PDdd).
Methods
Cohort description
The present investigation was performed in accordance with the Declaration of Helsinki. All participants gave their written informed consent. Samples for the current NMR study were obtained from the Neuro-Biobank Tuebingen and the DZNE Bonn. The code reference of the study is TUEPAC133, Tübingen data, and Bonn reference codes are DESCRIBE and DANCER134. CSF amyloid and tau data are also referenced in135,136.
Patients were classified, in our present study, as having sporadic PD early if PD endured within five years, or sporadic PD late if endured for more than five years.
Clinical parameters were received in form of an incomplete matrix (datasets with missing measurements) and considered for separate analysis: age at onset and disease duration (analyzed as full years, Supplementary Table 1, Table 1), weight (total n = 174 complete records, Cohorts 2 & 3), height and body mass index (BMI, total n = 173 complete records, Cohorts 2 & 3), Montreal Cognitive Assessment Index Score137 (MoCA, total n = 240, Cohorts 1, 2, & 3), MDS-Unified Parkinson’s Disease Rating Scale33 section III (UPDRS-III, total n = 221, non-MDS ratings138 (n = 14) were not used, Cohorts 1, 2, & 3), Beck Depression Inventory–II139 (BDI-II, total n = 172, Cohorts 1, 2, & 3), L-Dopa equivalent daily dose (LEDD, total n = 259, Cohorts 1, 2, & 3), CSF-measured neurofilament light chain protein level (Nfl, total n = 111, Cohorts 1, 2, & 3), plasma-measured Nfl (total n = 38, Cohort 2), CSF-measured α-synuclein (total n = 77, Cohorts 1 & 3), CSF-measured Aβ38 (total n = 35, Cohort 2), CSF-measured Aβ40 (total n = 36, Cohorts 2 & 3), CSF-measured Aβ42 and human total tau (total n = 151, Cohorts 1, 2, & 3), p-tau-181 (total n = 150, Cohorts 1, 2, & 3). In other words, with each parameter measured for a certain n number of individuals’ samples statistical analysis was measured based on the finalized table with the n number of parameter entries and measured NMR data.
Additionally, biological replicate samples (for the same patient at another timepoint, n = 16 patients, 32 samples, Subproject Cohort 3, Supplementary Table 1) were taken and cross-cohort biological replicates from Cohort 2 (marked with #, n = 5 patients, 10 samples) as well as from Subproject Cohort 1 (marked with @, n = 2 patients, 5 samples). Therefore, the part of samples corresponding to a same individual but at different time point taken were used in the present study.
To exclude sex as a confounding factor, surplus female controls were randomly selected to be excluded from the Controls group. The resulting female control were labeled as “Control_F” group (Fig. 1).
Quantitative NMR-based metabolite and lipoprotein analysis
Blood aliquots were stored at − 80 °C and transported on dry ice until preparation for NMR analysis. All samples were prepared according to a commercial SOP for blood plasma and serum (AVANCE IVDr Methods, Bruker BioSpin GmbH & Co. KG, Ettlingen, Germany). On the day of preparation, the samples were thawed to room temperature and prepared. The annotation and quantification of serum spectra are based on server-based service from Bruker BioSpin GmbH & Co. KG (Ettlingen, Germany). Nuclear magnetic resonance experiments were accomplished on a Bruker Avance III HD 600 MHz NMR spectrometer (Bruker BioSpin AG, Fällanden, Switzerland). Samples were measured with a 5 mm TXI probe, using Bruker TopSpin, including additionally required IVDr SOP experiments and software plug-ins as provided by Bruker BioSpin GmbH & Co. KG, Ettlingen, Germany.
The data obtained contained a total number of 39 compounds (incl., ethanol and glycerol) and metabolites (via Bruker IVDr Quantification In Plasma/Serum, B.I.Quant-PS™, analysis package), and a total of 112 lipoprotein parameters (via Bruker IVDr Lipoprotein Subclass Analysis B.I. LISA™ (Bruker BioSpin GmbH & Co. KG, Ettlingen, Germany). We also had utilized data on glycoprotein signals from NMR and also supramolecular phospholipid composite signal (SPC) as provided by a Bruker service called PhenoRisk PACS™ RuO (intented for research use only; Bruker BioSpin GmbH & Co. KG, Ettlingen, Germany).
More information on the used Bruker solutions, NMR metabolomics and some applications in research was recently published140.
Statistical analysis
Data was analyzed with the MetaboAnalyst 6.0 (online-based) package for statistical analysis and meta-analysis22. Logarithmic scaling was applied. GraphPad Prism software version 10.1.1 (323) was used for plot visualization and age-based comparisons after age-matching of the merged clinical cohorts (see also Supplementary Fig. 5). For each comparison, the following set of parameters was determined: p-values (student’s non-parametric t-testing (via Wilcoxon rank-sum testing method) and non-parametric ANOVA (via Kruskal Wallis Testing method) analysis of variance), and Spearman correlation coefficients (used in PatternHunter). For the t testing, all of the described results were significant via unequal n-numbers check. Graphs (mean ± SEM format) were produced based on input values and generated in GraphPad Prism 10.1.1 (323) for displaying metabolomics data.
Additionally, covariance analysis was done with replacing zeros as missing values and subsequent imputation. We used a generalized linear model (GLM) in R (version 4.2.1) and R-studio (2022.2.3.492) in order to determine whether the covariates, such as sex, age, and PDdd (PD disease duration) acted as confounders. We used GLM with Gaussian link and gamma log link function for normally distributed and skewed data, respectively. Of note, covariate adjusted p-value less than 0.05 is considered significant and such covariates did not confound the association between the group and dependent variable. The test recalculated zeros as missing values and no logarithmic scaling. The primary study statistics are not.
A regression model (PLS-DA, partial least squares discriminant analysis) was used, in short, to establish ranking (via VIP values, variable importance in projection) as discriminatory features of the first component of the regression model score performed for two or more patient groups. The operated threshold for VIP values was > 1.50, and these values were considerably important for reporting.
Several figures in the current work were created with BioRender.com online-based service.
Data availability
Raw NMR quantitation data (metabolites, lipoproteins and inflammation markers) and basic clinical metadata is included in Sup. Table 1. Further clinical data beyond the information in the manuscript and supplement files is available upon request to: thomas.gasser@uni-tuebingen.de.
References
Tansey, M. G. et al. Inflammation and immune dysfunction in Parkinson disease. Nat. Rev. Immunol. 22, 657–673 (2022).
Richter, F., Stanojlovic, M., Käufer, C., Gericke, B. & Feja, M. A mouse model to test novel therapeutics for Parkinson’s disease: an update on the Thy1-aSyn (line 61) mice. Neurotherapeutics 20, 97–116 (2023).
Mollenhauer, B. & Zhang, J. Biochemical premotor biomarkers for Parkinson’s disease. Mov. Disord. 27, 644–650 (2012).
Ren, R., Sun, Y., Zhao, X. & Pu, X. Recent advances in biomarkers for Parkinson’s disease focusing on biochemicals, omics and neuroimaging. Clin. Chem. Lab. Med. (CCLM). 53, 1495–1506 (2015).
Beavan, M. et al. Evolution of prodromal clinical markers of Parkinson disease in a GBA Mutation–Positive cohort. JAMA Neurol. 72, 201–208 (2015).
Yadav, S. K. et al. Transcriptomics and proteomics approach for the identification of altered blood MicroRNAs and plasma proteins in Parkinson’s disease. Cell. Mol. Neurobiol. 43, 3527–3553 (2023).
Dumitriu, A. et al. Integrative analyses of proteomics and RNA transcriptomics implicate mitochondrial processes, protein folding pathways and GWAS loci in Parkinson disease. BMC Med. Genom. 9, 5 (2016).
Gątarek, P., Pawełczyk, M., Jastrzębski, K., Głąbiński, A. & Kałużna-Czaplińska, J. Analytical methods used in the study of Parkinson’s disease. TRAC Trends Anal. Chem. 118, 292–302 (2019).
Shao, Y. & Le, W. Recent advances and perspectives of metabolomics-based investigations in Parkinson’s disease. Mol. Neurodegeneration. 14, 3 (2019).
Luan, H., Wang, X. & Cai, Z. Mass spectrometry-based metabolomics: targeting the crosstalk between gut microbiota and brain in neurodegenerative disorders. Mass. Spectrom. Rev. 38, 22–33 (2019).
Smolinska, A., Blanchet, L., Buydens, L. M. C. & Wijmenga, S. S. NMR and pattern recognition methods in metabolomics: from data acquisition to biomarker discovery: A review. Anal. Chim. Acta. 750, 82–97 (2012).
Dumas, M. E. et al. Assessment of analytical reproducibility of 1H NMR spectroscopy based metabonomics for Large-Scale epidemiological research: the INTERMAP study. Anal. Chem. 78, 2199–2208 (2006).
Meoni, G. et al. Metabolite and lipoprotein profiles reveal sex-related oxidative stress imbalance in de Novo drug-naive Parkinson’s disease patients. Npj Parkinson’s Disease. 8, 14 (2022).
Dimitri, G. M., Meoni, G., Tenori, L., Luchinat, C. & Lió, P. NMR spectroscopy combined with machine learning approaches for age prediction in healthy and Parkinson’s disease cohorts through metabolomic fingerprints. Appl. Sci. 12, 8954 (2022).
Troisi, J. et al. A metabolomic signature of treated and drug-naïve patients with Parkinson’s disease: a pilot study. Metabolomics 15, 90 (2019).
Berezhnoy, G., Laske, C. & Trautwein, C. Metabolomic profiling of CSF and blood serum elucidates general and sex-specific patterns for mild cognitive impairment and Alzheimer’s disease patients. Front. Aging Neurosci. 15, 1219718 (2023).
Berezhnoy, G., Laske, C. & Trautwein, C. Quantitative NMR-Based Lipoprotein Analysis Identifies Elevated HDL-4 and Triglycerides in the Serum of Alzheimer’s Disease Patients. Int. J. Mol. Sci. 23, 12472 (2022).
Jia, L. et al. A metabolite panel that differentiates Alzheimer’s disease from other dementia types. Alzheimer’s Dement. 18, 1345–1356 (2022).
Gan-Or, Z. et al. Differential effects of severe vs mild GBA mutations on Parkinson disease. Neurology 84, 880–887 (2015).
Huh, Y. E. et al. Glucosylceramide in cerebrospinal fluid of patients with GBA-associated and idiopathic Parkinson’s disease enrolled in PPMI. Npj Parkinson’s Disease. 7, 102 (2021).
Konno, M. et al. The one-carbon metabolism pathway highlights therapeutic targets for Gastrointestinal cancer (Review). Int. J. Oncol. 50, 1057–1063 (2017).
Pang, Z. et al. MetaboAnalyst 6.0: towards a unified platform for metabolomics data processing, analysis and interpretation. Nucleic Acids Res. 52, W398–W406 (2024).
Youden, W. J. Index for rating diagnostic tests. Cancer 3, 32–35 (1950).
Andréasson, M., Brodin, L., Laffita-Mesa, J. M. & Svenningsson, P. Correlations between methionine cycle metabolism, COMT genotype, and polyneuropathy in L-Dopa treated Parkinson’s disease: A preliminary Cross-Sectional study. J. Parkinson’s Disease. 7, 619–628 (2017).
Zhou, L. et al. High methionine intake alters gut microbiota and lipid profile and leads to liver steatosis in mice. Food Funct. 15, 8053–8069 (2024).
Yang, Y. et al. Dietary methionine restriction improves the gut microbiota and reduces intestinal permeability and inflammation in high-fat-fed mice. Food Funct. 10, 5952–5968 (2019).
Peto, V., Jenkinson, C., Fitzpatrick, R. & Greenhall, R. The development and validation of a short measure of functioning and well being for individuals with Parkinson’s disease. Qual. Life Res. 4, 241–248 (1995).
Hagell, P. & Nygren, C. The 39 item Parkinson’s disease questionnaire (PDQ-39) revisited: implications for evidence based medicine. J. Neurol. Neurosurg. Psychiatry. 78, 1191–1198 (2007).
Visser, M., Leentjens, A. F. G., Marinus, J., Stiggelbout, A. M. & van Hilten, J. J. Reliability and validity of the Beck depression inventory in patients with Parkinson’s disease. Mov. Disord. 21, 668–672 (2006).
BECK, A. T., WARD, C. H. & MENDELSON, M. ERBAUGH, J. An inventory for measuring depression. Arch. Gen. Psychiatry. 4, 561–571 (1961).
Martínez-Martín, P. et al. Parkinson’s disease severity levels and MDS-Unified Parkinson’s disease rating scale. Parkinsonism Relat. Disord. 21, 50–54 (2015).
Disease, M. D. S.T.F.o.R.S.f.P.s. The unified Parkinson’s disease rating scale (UPDRS): status and recommendations. Mov. Disord. 18, 738–750 (2003).
Goetz, C. G. et al. Movement disorder Society-sponsored revision of the unified Parkinson’s disease rating scale (MDS-UPDRS): scale presentation and clinimetric testing results. Mov. Disord. 23, 2129–2170 (2008).
Hoops, S. et al. Validity of the MoCA and MMSE in the detection of MCI and dementia in Parkinson disease. Neurology 73, 1738–1745 (2009).
Nasreddine, Z. S. et al. The Montreal cognitive assessment, MoCA: A brief screening tool for mild cognitive impairment. J. Am. Geriatr. Soc. 53, 695–699 (2005).
Mok, K. Y. et al. Deletions at 22q11.2 in idiopathic Parkinson’s disease: a combined analysis of genome-wide association data. Lancet Neurol. 15, 585–596 (2016).
Lees, A. J., Hardy, J. & Revesz, T. Parkinson’s disease. Lancet 373, 2055–2066 (2009).
Tolosa, E., Garrido, A., Scholz, S. W. & Poewe, W. Challenges in the diagnosis of Parkinson’s disease. Lancet Neurol. 20, 385–397 (2021).
Berg, D. et al. Prodromal Parkinson disease subtypes — key to Understanding heterogeneity. Nat. Rev. Neurol. 17, 349–361 (2021).
Cova, I. & Priori, A. Diagnostic biomarkers for Parkinson’s disease at a glance: where are we? J. Neural Transm. 125, 1417–1432 (2018).
Teymourian, H. et al. Closing the loop for patients with Parkinson disease: where are we? Nat. Rev. Neurol. 18, 497–507 (2022).
San Luciano, M. et al. Sex differences in LRRK2 G2019S and idiopathic Parkinson’s disease. Ann. Clin. Transl Neurol. 4, 801–810 (2017).
Saunders-Pullman, R., Wang, C., Stanley, K. & Bressman, S. B. Diagnosis and referral delay in women with Parkinson’s disease. Gend. Med. 8, 209–217 (2011).
Lerche, S. et al. CSF and Serum Levels of Inflammatory Markers in PD: Sparse Correlation, Sex Differences and Association With Neurodegenerative Biomarkers. Front. Neurol. 13, 834580 (2022).
Dolgacheva, L. P., Berezhnov, A. V., Fedotova, E. I., Zinchenko, V. P. & Abramov, A. Y. Role of DJ-1 in the mechanism of pathogenesis of Parkinson’s disease. J. Bioenerg. Biomembr. 51, 175–188 (2019).
Krebiehl, G. et al. Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s Disease-Associated protein DJ-1. PLoS ONE. 5, e9367 (2010).
Shendelman, S., Jonason, A., Martinat, C., Leete, T. & Abeliovich, A. DJ-1 is a Redox-Dependent molecular chaperone that inhibits α-Synuclein aggregate formation. PLoS Biol. 2, e362 (2004).
Skou, L. D., Johansen, S. K., Okarmus, J. & Meyer, M. Pathogenesis of DJ-1/PARK7-Mediated Parkinson’s disease. Cells 13, 296 (2024).
Cid-Castro, C., Hernández-Espinosa, D. R. & Morán, J. ROS as regulators of mitochondrial dynamics in neurons. Cell. Mol. Neurobiol. 38, 995–1007 (2018).
Burté, F., Carelli, V. & Chinnery, P. F. Yu-Wai-Man, P. Disturbed mitochondrial dynamics and neurodegenerative disorders. Nat. Rev. Neurol. 11, 11–24 (2015).
Ryan, B. J., Hoek, S., Fon, E. A. & Wade-Martins, R. Mitochondrial dysfunction and mitophagy in Parkinson’s: from Familial to sporadic disease. Trends Biochem. Sci. 40, 200–210 (2015).
Hauser, D. N. et al. Hexokinases link DJ-1 to the PINK1/parkin pathway. Mol. Neurodegener. 12, 70 (2017).
Sun, M. E. & Zheng, Q. The Tale of DJ-1 (PARK7): A Swiss army knife in biomedical and psychological research. IJMS 24, 7409 (2023).
Hayashi, T. et al. DJ-1 binds to mitochondrial complex I and maintains its activity. Biochem. Biophys. Res. Commun. 390, 667–672 (2009).
Junn, E., Jang, W. H., Zhao, X., Jeong, B. S. & Mouradian, M. M. Mitochondrial localization of DJ-1 leads to enhanced neuroprotection. J. Neurosci. Res. 87, 123–129 (2009).
Wade, A. M. & Tucker, H. N. Antioxidant characteristics of L-histidine. J. Nutr. Biochem. 9, 308–315 (1998).
Holeček, M. Histidine in health and disease: metabolism, physiological importance, and use as a supplement. Nutrients 12, 848 (2020).
Catanesi, M. et al. L-Methionine protects against oxidative stress and mitochondrial dysfunction in an in vitro model of Parkinson’s disease. Antioxidants 10, 1467 (2021).
Chang, K. H. et al. Altered metabolic profiles of the plasma of patients with amyotrophic lateral sclerosis. Biomedicines 9, 1944 (2021).
Zheng, Y. et al. Comparison of biospecimens for α-synuclein seed amplification assays in Parkinson’s disease: A systematic review and network meta-analysis. Eur. J. Neurol. 30, 3949–3967 (2023).
Santacruz, C. A. et al. Association of cerebrospinal fluid protein biomarkers with outcomes in patients with traumatic and non-traumatic acute brain injury: systematic review of the literature. Crit. Care. 25, 278 (2021).
Okuzumi, A. et al. Propagative α-synuclein seeds as serum biomarkers for synucleinopathies. Nat. Med. 29, 1448–1455 (2023).
Kluge, A. et al. Detection of neuron-derived pathological α-synuclein in blood. Brain 145, 3058–3071 (2022).
Xylaki, M. et al. Extracellular vesicles for the diagnosis of Parkinson’s disease: systematic review and Meta-Analysis. Mov. Disord. 38, 1585–1597 (2023).
Xie, H. et al. Abnormalities of cerebral blood flow and the regional brain function in Parkinson’s disease: a systematic review and multimodal neuroimaging meta-analysis. Front. Neurol. 14, 1289934 (2023).
Xu, Y., Huang, X., Geng, X. & Wang, F. Meta-analysis of iron metabolism markers levels of Parkinson’s disease patients determined by fluid and MRI measurements. J. Trace Elem. Med Biol. 78, 127190 (2023).
Kumari, S. et al. Identification of potential urine biomarkers in idiopathic Parkinson’s disease using NMR. Clin. Chim. Acta. 510, 442–449 (2020).
Luo, X., Liu, Y., Balck, A., Klein, C. & Fleming, R. M. T. Identification of metabolites reproducibly associated with Parkinson’s disease via meta-analysis and computational modelling. Npj Parkinson’s Disease. 10, 126 (2024).
Sisalli, M. J. et al. L-Ornithine L-Aspartate restores mitochondrial function and modulates intracellular calcium homeostasis in Parkinson’s disease models. Cells 11, 2909 (2022).
Schuber, F. Influence of polyamines on membrane functions. Biochem. J. 260, 1–10 (1989).
Løvaas, E., Carlin, G. & Spermine An anti-oxidant and anti-inflammatory agent. Free Radic. Biol. Med. 11, 455–461 (1991).
Fiori, L. M. & Turecki, G. Implication of the polyamine system in mental disorders. J. Psychiatry Neurosci. 33, 102–110 (2008).
Hatano, T., Saiki, S., Okuzumi, A., Mohney, R. P. & Hattori, N. Identification of novel biomarkers for Parkinson’s disease by metabolomic technologies. J. Neurol. Neurosurg. Psychiatry. 87, 295–301 (2016).
Plewa, S. et al. The metabolomic approach reveals the alteration in human serum and cerebrospinal fluid composition in Parkinson’s disease patients. Pharmaceuticals 14, 935 (2021).
Çelik, V., Çiğdem, B., Kapancik, S., Kiliçgün, H. & Bolayir, E. The importance of increased serum ornithine levels in the pathogenesıs of Alzheımer and Parkınson’s dıseases. Asian J. Res. Rep. Neurol. 1, 1–8 (2018).
Chang, K. H. et al. Alterations of sphingolipid and phospholipid pathways and ornithine level in the plasma as biomarkers of Parkinson’s disease. Cells 11, 395 (2022).
Gabrielli, M. et al. Prevalence of small intestinal bacterial overgrowth in Parkinson’s disease. Mov. Disord. 26, 889–892 (2011).
Barone, P. et al. The PRIAMO study: A multicenter assessment of nonmotor symptoms and their impact on quality of life in Parkinson’s disease. Mov. Disord. 24, 1641–1649 (2009).
Wang, J. et al. Characteristics of the gut Microbiome and serum metabolome in patients with functional constipation. Nutrients 15, 1779 (2023).
Perier, C. & Vila, M. Mitochondrial biology and Parkinson’s disease. Cold Spring Harbor Perspect. Med. 2, a009332 (2012).
Murray, L. K. & Jadavji, N. M. The role of one-carbon metabolism and homocysteine in Parkinson’s disease onset, pathology and mechanisms. Nutr. Res. Rev. 32, 218–230 (2019).
Kim, J. H., Jin, S., Eo, H., Oh, M. S. & Lim, Y. Two faces of Catechol-O-Methyltransferase inhibitor on One-Carbon metabolism in Parkinson’s disease: A Meta-Analysis. Nutrients 15, 901 (2023).
Havelund, J., Heegaard, N., Færgeman, N. & Gramsbergen, J. Biomarker research in Parkinson’s disease using metabolite profiling. Metabolites 7, 42 (2017).
Mycielska, M. E., James, E. N. & Parkinson, E. K. Metabolic alterations in cellular senescence: the role of citrate in ageing and Age-Related disease. IJMS 23, 3652 (2022).
Auro, K. et al. A metabolic view on menopause and ageing. Nat. Commun. 5, 4708 (2014).
Menni, C. et al. Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol. 42, 1111–1119 (2013).
Babu, G. N. et al. Serum metabolomics study in a group of Parkinson’s disease patients from Northern India. Clin. Chim. Acta. 480, 214–219 (2018).
Toomey, C. E. et al. Mitochondrial dysfunction is a key pathological driver of early stage Parkinson’s. Acta Neuropathol. Commun. 10, 134 (2022).
Ahmed, S. S. S. J., Santosh, W., Kumar, S. & Christlet, H. T. T. Metabolic profiling of Parkinson’s disease: evidence of biomarker from gene expression analysis and rapid neural network detection. J. Biomed. Sci. 16, 63 (2009).
Balck, A. et al. The role of dopaminergic medication and specific pathway alterations in idiopathic and PRKN/PINK1-mediated Parkinson’s disease. Sci. Adv. 11(20), eadp7063 (2025)
Otto, C. et al. Comprehensive analysis of the cerebrospinal fluid and serum metabolome in neurological diseases. J. Neuroinflamm. 21, 234 (2024).
Li, X., Fan, X. Y., Yang, H. T. & Liu, Y. F. Review of Metabolomics-Based biomarker research for Parkinson’s disease. Mol. Neurobiol. 59, 1041–1057 (2022).
Jiménez-Jiménez, F. J., Alonso-Navarro, H., García-Martín, E. & Agúndez, J. A. G. Cerebrospinal and blood levels of amino acids as potential biomarkers for Parkinson’s disease: review and meta-analysis. Eur. J. Neurol. 27, 2336–2347 (2020).
Kumari, S. et al. Quantitative metabolomics of saliva using proton NMR spectroscopy in patients with Parkinson’s disease and healthy controls. Neurol. Sci. 41, 1201–1210 (2020).
Magkos, F., Patterson, B. W., Mohammed, B. S., Klein, S. & Mittendorfer, B. Women produce fewer but Triglyceride-Richer very Low-Density lipoproteins than men. J. Clin. Endocrinol. Metabolism. 92, 1311–1318 (2007).
Contois, J. H. et al. Reference intervals for plasma Apolipoprotein B determined with a standardized commercial immunoturbidimetric assay: results from the Framingham offspring study. Clin. Chem. 42, 515–523 (1996).
Freedman, D. S. et al. Sex and age differences in lipoprotein subclasses measured by nuclear magnetic resonance spectroscopy: the Framingham study. Clin. Chem. 50, 1189–1200 (2004).
Jungner, I. et al. Apolipoprotein B and A-I values in 147 576 Swedish males and females, standardized according to the world health Organization–International federation of clinical chemistry first international reference materials. Clin. Chem. 44, 1641–1649 (1998).
Johnson, J. L. et al. Gender and Racial differences in lipoprotein subclass distributions: the STRRIDE study. Atherosclerosis 176, 371–377 (2004).
Konz, T. et al. Sex-Specific Associations of Blood-Based Nutrient Profiling With Body Composition in the Elderly. Front. Physiol. 9, 01935 (2018).
Vignoli, A., Tenori, L., Luchinat, C. & Saccenti, E. Age and sex effects on plasma metabolite association networks in healthy subjects. J. Proteome Res. 17, 97–107 (2018).
Vaidya, B., Dhamija, K., Guru, P. & Sharma, S. S. Parkinson’s disease in women: mechanisms underlying sex differences. Eur. J. Pharmacol. 895, 173862 (2021).
Galea, L. A. M., Choleris, E., Albert, A. Y. K., McCarthy, M. M. & Sohrabji, F. The promises and pitfalls of sex difference research. Front. Neuroendocrinol. 56, 100817 (2020).
Sellau, J. et al. Immunological clues to sex differences in parasitic diseases. Trends Parasitol. 40, 1029–1041 (2024).
Westerman, S. & Wenger, N. K. Women and heart disease, the underrecognized burden: sex differences, biases, and unmet clinical and research challenges. Clin. Sci. 130, 551–563 (2016).
Caldi Gomes, L. et al. Multiomic ALS signatures highlight subclusters and sex differences suggesting the MAPK pathway as therapeutic target. Nat. Commun. 15, 4893 (2024).
Cleasby, M. E., Jamieson, P. M. & Atherton, P. J. Insulin resistance and sarcopenia: mechanistic links between common co-morbidities. J. Endocrinol. 229, R67–R81 (2016).
Jurado-Coronel, J. C. et al. Sex differences in Parkinson’s disease: features on clinical symptoms, treatment outcome, sexual hormones and genetics. Front. Neuroendocrinol. 50, 18–30 (2018).
Liu, B. & Dluzen, D. E. Oestrogen and nigrostriatal dopaminergic neurodegeneration: animal models and clinical reports of Parkinson’s disease. Clin. Experimental Pharmacol. Physiol. 34, 555–565 (2007).
Morissette, M. & Di Paolo, T. Effect of estradiol on striatal dopamine activity of female Hemiparkinsonian monkeys. J. Neurosci. Res. 87, 1634–1644 (2009).
Smith, K. M. & Dahodwala, N. Sex differences in Parkinson’s disease and other movement disorders. Exp. Neurol. 259, 44–56 (2014).
LeWitt, P. A. et al. Markers of dopamine metabolism in Parkinson’s disease. Neurology 42, 2111–2111 (1992).
Bovenzi, R. et al. Sex hormones differentially contribute to Parkinson disease in males: A multimodal biomarker study. Eur. J. Neurol. 30, 1983–1990 (2023).
Schaffner, S. L., Tosefsky, K. N., Inskter, A. M., Appel-Cresswell, S. & Schulze-Hentrich, J. M. Sex and gender differences in the molecular etiology of Parkinson’s disease: considerations for study design and data analysis. Biology Sex. Differences. 16, 7 (2025).
Rössler, T. et al. Quantitative serum NMR spectroscopy stratifies COVID-19 patients and sheds light on interfaces of host metabolism and the immune response with cytokines and clinical parameters. Metabolites 12, 1277 (2022).
Paul, R. & Borah, A. L-DOPA-induced hyperhomocysteinemia in Parkinson’s disease: elephant in the room. Biochim. Et Biophys. Acta (BBA) - Gen. Subj. 1860, 1989–1997 (2016).
Kalecký, K., Ashcraft, P. & Bottiglieri, T. One-Carbon metabolism in Alzheimer’s disease and Parkinson’s disease brain tissue. Nutrients 14, 599 (2022).
Kato, Y. et al. Oxytocin-induced increase in N,N-dimethylglycine and time course of changes in Oxytocin efficacy for autism social core symptoms. Mol. Autism. 12, 15 (2021).
Dimitri, G. M., Meoni, G., Tenori, L., Luchinat, C. & Lió, P. NMR Spectroscopy Combined with Machine Learning Approaches for Age Prediction in Healthy and Parkinson’s Disease Cohorts through Metabolomic Fingerprints. Appl. Sci. 12, 8954 (2022).
Prestel, J. et al. Clinical and molecular characterisation of a Parkinson family with a novel PINK1 mutation. J. Neurol. 255, 643–648 (2008).
Ahmed, S. S. S. J., Santosh, W., Kumar, S. & Christlet, H. Metabolic profiling of Parkinson’s disease: evidence of biomarker from gene expression analysis and rapid neural network detection. J. Biomed. Sci. 16, 63 (2009).
Conde, R. et al. NMR analysis seeking for cognitive decline and dementia metabolic markers in plasma from aged individuals. J. Pharm. Biomed. Anal. 238, 115815 (2024).
Kumari, S. et al. Metabolomic analysis of serum using proton NMR in 6-OHDA experimental PD model and patients with PD. Neurochem. Int. 134, 104670 (2020).
Sakuma, K. & Yamaguchi, A. Recent advances in Pharmacological, hormonal, and nutritional intervention for sarcopenia. Pflugers Arch. - Eur. J. Physiol. 470, 449–460 (2018).
Tan, A. H. et al. Altered body composition, sarcopenia, frailty, and their clinico-biological correlates, in Parkinson’s disease. Parkinsonism Relat. Disord. 56, 58–64 (2018).
da Luz, M. C. L., Bezerra, G. K. A., Asano, A. G. C., de Chaves, M. C. & Cabral, P. C. Determinant factors of sarcopenia in individuals with Parkinson’s disease. Neurol. Sci. 42, 979–985 (2021).
Toczylowska, B., Zieminska, E., Michałowska, M., Chalimoniuk, M. & Fiszer, U. Changes in the metabolic profiles of the serum and putamen in Parkinson’s disease patients – In vitro and in vivo NMR spectroscopy studies. Brain Res. 1748, 147118 (2020).
Vignoli, A. et al., High-Throughput Metabolomics by 1D NMR. Angew. Chem. Int. Ed. 58, 968–994 (2019).
Ghini, V., Quaglio, D., Luchinat, C. & Turano, P. NMR for sample quality assessment in metabolomics. New Biotechnol. 52, 25–34 (2019).
Takis, P. G., Ghini, V., Tenori, L., Turano, P. & Luchinat, C. Uniqueness of the NMR approach to metabolomics. TRAC Trends Anal. Chem. 120, 115300 (2019).
Gil-Redondo, R. et al. MetSCORE: a molecular metric to evaluate the risk of metabolic syndrome based on serum NMR metabolomics. Cardiovasc. Diabetol. 23, 272 (2024).
Kazenwadel, J. et al. Stratification of hypertension and SARS-CoV-2 infection by quantitative NMR spectroscopy of human blood serum. Commun. Med. (Lond). 3, 145 (2023).
Lerche, S. et al. CSF NFL in a longitudinally assessed PD cohort: age effects and cognitive trajectories. Mov. Disord. 35, 1138–1144 (2020).
Upadhyay, N. et al. Fronto-striatal alterations correlate with apathy severity in behavioral variant frontotemporal dementia. Brain Imaging Behav. 18, 66–72 (2024).
Jessen, F. et al. Design and first baseline data of the DZNE multicenter observational study on predementia Alzheimer’s disease (DELCODE). Alzheimers Res. Ther. 10, 15 (2018).
Jessen, F. et al. Subjective cognitive decline and stage 2 of alzheimer disease in patients from memory centers. Alzheimer’s Dement. 19, 487–497 (2023).
Julayanont, P., Brousseau, M., Chertkow, H., Phillips, N. & Nasreddine, Z. S. Montreal cognitive assessment memory index score (MoCA-MIS) as a predictor of conversion from mild cognitive impairment to Alzheimer’s disease. J. Am. Geriatr. Soc. 62, 679–684 (2014).
Tison, F. et al. Parkinsonism in multiple system atrophy: natural history, severity (UPDRS-III), and disability assessment compared with Parkinson’s disease. Mov. Disord. 17, 701–709 (2002).
Dozois, D. J. A., Dobson, K. S. & Ahnberg, J. L. A psychometric evaluation of the Beck depression Inventory–II. Psychol. Assess. 10, 83–89 (1998).
Trautwein, C. Quantitative Blood Serum IVDr NMR Spectroscopy in Clinical Metabolomics of Cancer, Neurodegeneration, and Internal Medicine. in Clinical Metabolomics: Methods and Protocols (eds. Giera, M. & Sánchez-López, E.) 427–443Springer US, New York, NY, (2025).
Acknowledgements
The authors would like to thank Prof. Bernd Pichler from the Werner Siemens Imaging Center, University of Tübingen, Germany. We thank Daniele Bucci for excellent technical assistance.We are furthermore grateful to Tony Reinsperger and Manfred Spraul from Bruker BioSpin GmbH & Co. KG, Ettlingen, Germany. Samples and data were obtained from the biobank at the German Center for Neurodgenerative Diseases (DZNE) Bonn and the Neuro-Biobank of the University of Tuebingen, Germany (https://www.hih-tuebingen.de/en/about-us/core-facilities/biobank/). This biobank is supported by the local University, the Hertie Institute and the DZNE.We also thank several institutions for their help for the current study: Max Delbrück Center for Molecular Medicine in the Helmholtz Association (Max Delbrück Center) – MDC, the Center for Cognitive Neuroscience Berlin (CCNB) at the Freie Universität Berlin, Clinic and Polyclinic for Nuclear Medicine at the University Hospital Bonn (Venusberg Campus 1), Clinic and Polyclinic for Nuclear Medicine at the University Hospital Cologne, Department of Radiology and Nuclear Medicine of the University Hospital Magdeburg, Clinic and Polyclinic for Nuclear Medicine at the University of Munich – LMU Hospital Munich, Department of Nuclear Medicine at the Rostock University Hospital (Universitätsmedizin Rostock), Nuclear Medicine and Clinical Molecular Imaging – Department of Radiology – University Hospital Tübingen, Bernstein Center for Computational Neuroscience Berlin, MR-Research in Neurosciences scientific core facility of the Department of Cognitive Neurology of the University Medical Center Göttingen, Clinic and Polyclinic for Radiology of the Department of Radiology – LMU Hospital Munich, MR Research Division of the Department of Diagnostic and Interventional Neuroradiology – Department of Radiology – University Hospital Tübingen.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
CT and TG created and planned the research. GeB, GB, LW, and CS gathered laboratory data and medical records of patients for the metabolomics research. Coauthors CS, DP, MZ, FB, PL, IW, AJ, EJS, JHR, SR, WG, FM, FH, IK, ED, OK, AG, JL, JP, OP, MW, AS, EDü, CVR, UW, ST, BF, MB, IZ, GCP, ASp, and PR were either contributing by means of clinical samples or/and by providing extensive clinical records. CW and HS provided technical and analytical support. GeB, GB, LW, CS, and CT performed data analyses. GeB wrote the original draft of the paper. GeB, CT, CS, GB, DP, MZ, FB, PL, and TG edited the draft.
Corresponding authors
Ethics declarations
Competing interests
GeB and CT report a research grant by Bruker BioSpin GmbH & Co. KG. CW and HS are employed at Bruker BioSpin GmbH & Co. KG, Ettlingen, Germany. All other authors declare no competing interests.
Ethics declarations
The project was carried out in compliance with the Helsinki Declaration, and all participants provided written informed consent. Sample collection and study design were approved by the local Ethics Committee of Eberhard Karls University of Tübingen (ethics numbers 226/2021B02 and 199/2011BO1), Germany. Regarding the Cohort 2 of the present study, the following ethic numbers are of reference – numbers 311/14 (status year 2021) and 312/14 (status year 2018). Harmonized voting was carried out at the Lead Ethics Bonn.
Metadata declaration
The data were provided by the study group of DESCRIBE and DANCER studies of the clinical research of the German Center for Neurodegenerative Diseases (DZNE). And data were provided by the study groups of TUEPAC study at the University Hospital of Tuebingen. The clinical data supporting the findings are available upon request to: thomas.gasser@uni-tuebingen.de.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Berezhnoy, G., Bae, G., Wüst, L. et al. Application of IVDr NMR spectroscopy to stratify Parkinson’s disease with absolute quantitation of blood serum metabolites and lipoproteins. Sci Rep 15, 17738 (2025). https://doi.org/10.1038/s41598-025-01352-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-01352-0