Abstract
LUAD, a prevalent lung cancer with high mortality, has seen increased focus on molecular targeted therapies due to patient heterogeneity. Among these prospects, dystrophin-associated protein 2 (DRP2), a critical component of the dystrophin complex, underpins membrane-associated structures vital for intercellular interactions in vertebrates. Aberrations in DRP2 function have been linked to the occurrence and development of multiple diseases, prompting an inquiry into its potential link with LUAD progression. To delve into the potential roles of DRP2 in LUAD, we initiated a comprehensive investigation. First, we analyzed DRP2 expression patterns in LUAD using bioinformatics tools. This was subsequently validated through immunohistochemical staining, quantitative PCR, and Western blot analyses. Furthermore, we assessed the functional implications of DRP2 in LUAD cells, both in vitro and in vivo, utilizing assays such as cell cycle analysis, CCK-8 proliferation assay, Colony formation assay EdU incorporation, Transwell migration test, scratch wound healing assay, flow cytometry, and mouse models for tumor xenograft and metastasis. Results showed a strong correlation between high DRP2 expression in LUAD and poorer survival. Notably, DRP2 knockdown accelerated LUAD progression via the EMT pathway. These findings highlight DRP2’s crucial role in LUAD and its potential as a therapeutic target.
Similar content being viewed by others
Introduction
Despite notable advancements in lung cancer treatment in recent times, lung cancer continues to pose a formidable threat to human life and health, ranking as the leading cause of mortality globally1. In various lung cancer types, NSCLC comprises approximately 85% and represents the primary pathological subtype2. The intricate nature of NSCLC’s proliferation and metastasis mechanisms has hindered significant improvements in patient prognosis, resulting in a dismal five-year survival rate of merely 20%3. Notably, LUAD constitutes a significant majority of NSCLC cases. Although molecular targeted therapies and immunotherapies have significantly enhanced LUAD patient outcomes, not all LUAD patients harbor suitable molecular targets for these treatments3,4,5. Consequently, we hope that novel potential molecules capable of predicting LUAD prognosis, unravel the underlying molecular mechanisms regulating its progression, and devise effective, targeted therapeutic strategies.
DRP2, or Dystrophin Related Protein-2, belongs to the dystrophin gene family and is encoded by a 45-kilobase gene positioned on the Xp22 region. Its primary expression sites are the brain and spinal cord. Structurally akin to Dp116, DRP2 is suggested to execute analogous functions within the vertebrate central nervous system6. Beyond this, DRP2 assumes a vital part in the peripheral nervous system, collaborating with periaxin and gluconic anhydride, forming PDG. This complex contributes to the preservation of the Cajal band in myelin Schwann cells, emphasizing DRP2’s role7.Notably, Charcot-Marie-Tooth disease (CMT), a prevalent hereditary neurological condition with a frequency of around 1 in 2,500, arises from DRP2 deficiency, resulting in the disintegration of the Cajal band in affected individuals8,9.Moreover, DRP2 is a cornerstone molecule in the extracellular matrix-receptor interaction cascade and has garnered attention for its potential involvement in oncogenesis10. However, the exact expression patterns and functional contributions of DRP2 in LUAD have remained enigmatic. Hence, this study endeavors to clarify the expression status of DRP2 in LUAD and elucidate its pivotal role in cancer progression.
Our results demonstrate an elevated expression of DRP2 in LUAD samples, which significantly correlates with poorer patient outcomes. Furthermore, GSEA highlights the prominent enrichment of DRP2 in pathways regulating cell cycle progression and EMT, reinforcing DRP2’s substantial role in the biological mechanisms underlying LUAD.
Materials and methods
Data collection
The mRNA expression profiles for gene chip GSE31210 were retrieved from the GEO repository. RNA sequencing info & clinical specifics on LUAD patients were obtained from TCGA. To elucidate protein interactions, we leveraged the GeneMANIA online platform11 and the STRING database12 for constructing comprehensive interaction networks.
The cBioPortal analysis
The cBioPortal, an open-access online platform, streamlines the exploration of diverse cancer genomics databases. By bridging the gap between intricate genomic data and cancer researchers, it expedites the process of intuitively accessing high-quality molecular mapping and cancer genomics findings to clinical outcomes13. For our analysis, we carefully selected six LUAD datasets (https://www.cbioportal.org/).
Kaplan–Meier survival analysis
Using Kaplan Meier - plotter the following customizations: Automatically selecting the optimal cutoff for probes, and focusing the analysis on all histological types, particularly adenocarcinoma and squamous cell carcinoma, while maintaining default settings for all other parameters.
Cell culture and transfection
The cell lines BEAS-2B, A549, H1299, H226, and H1650 originate from the Chinese Academy of Sciences. Prior to the transfection process, the optimal growth density of A549 and H1299 cell lines reached about 40%. Lentiviral vectors specifically designed for DRP2 knockdown, along with their respective negative controls, were engineered and produced by Genechem (Shanghai, China). These vectors, carrying either non-targeting negative controls or short hairpin RNAs (shRNAs) directed against DRP2, were then introduced into the cell cultures. After purinomycin selection, a stable DRP2 knockdown cell line was successfully established. It is worth noting that the specific shRNA sequences targeting DRP2 used in this study are: 5’-GCATCAGACCAAGTGCTCTAT-3’ (sh-DRP2-1) and 5’-TAGAGCACTTGGTCTGATGC-3’ (sh-DRP2-2).
Reagents
Primary antibodies for Western blot: Rabbit DRP2 (#26631-1-AP, Proteintech, USA), mouse β-actin (#GB15001, Servicebio, China), mouse Snail (#L70G2, Cell Signaling, China), mouse Vimentin (#sc-6260, Santa Cruz, USA), mouse N-Cadherin (#610920, BD Lab, USA), mouse E-Cadherin (#610181, BD Lab, USA), along with anti-rabbit or anti-mouse secondary antibodies (#AS014,#AS017,ABclonal) were employed in dilutions of 1:1000–5000, while the secondary antibodies were used in a 1:5000–1:10,000 ratio.
Human LUAD tissues
From Soochow University’s First Affiliated Hospital, 31 lung adenocarcinomas and nearby non-cancerous tissues, each situated over 5 cm away from the tumor, were gathered. Notably, the patients involved had not undergone radiotherapy or chemotherapy prior to surgical intervention. Immediately post-surgery, the specimens were swiftly cryopreserved in liquid nitrogen to maintain their integrity. All tissue samples underwent rigorous pathological examination and confirmation to ensure accuracy. Moreover, the procurement of these specimens was granted approval by both the patients themselves and the Academic Advisory Committee of Soochow University, adhering to the highest ethical standards.
Immunohistochemical staining
After baking the tissue specimens embedded in paraffin blocks, a standard dewaxing and rehydration protocol was applied. The slides were then treated in 3% hydrogen peroxide (H2O2) for 25 min. The tissue sections were incubated with primary antibody at 4 °C overnight, specifically targeting DRP2, Ki67, and PCNA, at optimized dilutions (1:200, 1:1000, and 1:2000 respectively). For visualization, diaminobenzidine (DAB) from a commercial kit (#G1212, Servicebio, China) was utilized as the chromogenic substrate. After staining, the sections underwent counterstaining with hematoxylin to enhance contrast and were examined under an optical microscope.
The antibodies employed in these immunohistochemical analyses were sourced from reputable suppliers, including Anti-DRP2 antibody (#PAB20143, Abnova, China), Anti-Ki67 antibody (#GB111141, Servicebio, China), and Anti-PCNA antibody (#GB12010, Servicebio, China), all of which were diluted to their respective optimal concentrations.
The qRT-PCR assay
The qRT-PCR assay utilized the following primers: for DRP2, the forward primer was 5’-GAGACACATGCGGCCTTTATG-3’ and the reverse primer was 5’-CTCTAACTCCTCAAATGGGTGC-3’. β-actin was the reference gene.
Western blotting
During Western blot, cells were rinsed with chilled PBS, lysed in RIPA buffer (#YF369700, ThermoFisher, USA) containing inhibitors, and the extracted proteins were separated via SDS-PAGE. The proteins were then transferred onto a NC membrane (#1215471, Millipore, USA)。At room temperature, a protein-free solution (#PS108P, EpiZyme) was used to block membranes for 40 min。Following an overnight incubation with a specific primary antibody, the membrane was washed and subsequently incubated with a secondary antibody. The protein expression levels were finally detected and quantitatively evaluated using enhanced chemiluminescence (ECL) detection.
Migration and invasion assays
The cells undergo a digestion process, are then centrifuged and subsequently re-suspended in pure medium to achieve a density of 4 × 104 cells /100 µL. 100 µL of suspensions were distributed to each chamber. After incubation, the cells were fixed and then stained with crystal violet. After three thorough washes with PBS, gently wipe the room with a cotton swab and imaged under an inverted microscope. For the cell invasion assay, a 4:1 mixture of medium and matrix gel (Corning, Shanghai, China) was introduced into each chamber, and subsequent procedures remained unchanged.
Wound-healing assay
It was seeded in a sterile pipette parallel to a straight rule, scratched with a sterile pipette, rinsed with PBS three times, and imaged in six random fields of view. After 24 h, the cells were cleaned and imaged utilizing ImageJ Launcher software.
CCK-8 assay
A549 and H1299 cells were seeded into 96-well plates at 2 × 103 cells/well. CCK-8 solution (10 µL, APExBIO, USA) was added after incubation for 0, 24, 48, 72, or 96 h. After an additional 2 h of incubation, the optical density (OD450) at each time point was recorded using a microplate reader (CMax Plus, Shanghai, China). This process allowed for the quantification of cell proliferation over time.
Colony formation assay
Cells were seeded in 6-well plates (1.5 × 103 cells/well) and incubated for 8–10 days to form colonies. After washing with PBS, cells were fixed with methanol for 30 min and stained overnight with 0.1% crystal violet in 20% ethanol. The plates were washed, air-dried, and colonies counted. The experiment was repeated three times.
Animal experiments
Female BALB/c nude mice (4 weeks old) from Soochow University’s Lab Animal Center were implanted with Xenograft tumors. These tumors were prepared using 2 × 106 A549 cells in logarithmic growth, which were rinsed with PBS and re-suspended in serum-free medium mixed with matrix gel to form a 150µL suspension. The resulting tumors, once implanted in the axillary area of each mouse, were approximately-1mm14 in volume. Tumor size was measured every 5 days, and tumor volume was calculated. After 30 days, xenograft tumors were harvested for analysis. All procedures were ethically approved.
In the tail vein transfer experiment, 3 × 106 A549 cells were re-suspended in serum-free medium to prepare 200µL suspension, which was injected subcutaneously into each nude mouse. After transplantation for 8 weeks, the lungs of mice were taken, fixed with formaldehyde, embedded in paraffin, sliced, and stained by HE.
We ensure humane and ethical euthanasia/sacrifice of female BALB/c nude mice in our research, complying with all relevant guidelines. Trained personnel supervise the process, using anesthesia (0.3% pentobarbital sodium, dose based on weight) to ensure the mice are unconscious and pain-free. Once unconscious, cervical dislocation is performed for quick and effective euthanasia. We prioritize animal welfare and ethical treatment in all research activities.
Bioinformatics analysis
Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of differentially expressed genes was implemented using the clusterProfiler R package15,16,17.
Statistical analysis
All data and mappings were comprehensively analyzed utilizing the advanced capabilities of the Xiantao Academic Platform (www.xiantaozi.com). For statistical analysis and meticulous data processing, we employed the GraphPad Prism 9 software, ensuring rigor and precision. The outcomes are presented in a standardized format, as the mean accompanied by the standard deviation (SD), facilitating interpretation. Comparisons amongst datasets were facilitated through the application of Student’s t-test, with statistical significance designated to p-values falling below the threshold of 0.05.
Result.
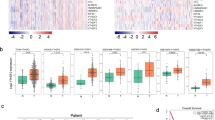
DRP2 expression was upregulated in LUAD tissues and correlated with aggressive clinical features
To investigate the function of DRP2 in pancarcinoma, we initiated by assessing DRP2 expression in the TCGA database. Prior research has demonstrated variable patterns, with decreased DRP2 expression noted in cancers like BLCA, READ, and GBM, whereas enhanced expression was observed in others, particularly LUAD and LUSC (Fig. 1A). To narrow our focus, we delved into the correlation between DRP2 levels and prognosis, utilizing Kaplan-Meier Plotter, an advanced online tool. Our analysis highlighted that heightened DRP2 expression in lung cancer patients is linked to poorer OS (Fig. 1B). Specifically, for LUAD, high DRP2 expression negatively impacted prognosis, with low DRP2 levels predictive of better outcomes and a hazard ratio of 1.37. Conversely, in LUSC, no significant survival difference was discernible. This steered our investigation towards elucidating DRP2’s role in LUAD.
DRP2 expression was upregulated in LUAD tissues and correlated with aggressive clinical features. (A) An investigation into the expression of DRP2 across various cancer types was conducted utilizing the TCGA database. (B) Kaplan-Meier survival curves show the effect of DRP2 on OS in LC, LUAD, and LUSC. (C,D) To further analyze DRP2 mRNA expression, the TCGA database was employed to compare unpaired (C) and paired (D) samples from LUAD and normal tissues. (E) The expression of DRP2 in LUAD was examined using GSE31210 database. (F–H) DRP2 mRNA expression (F) in tumor tissue (“ T “) and pairs of normal lung epithelial tissue (“N”) from 31 LUAD patients, and DRP2 protein expression and quantification (G,H) were assessed in twelve representative patient pairs. (I) DRP2 immunohistochemical staining of tumor tissue and paired normal tissue in two typical LUAD patients. (J–K) Association of DRP2 with clinical staging. *P < 0.05, **P < 0.01, ***P < 0.001 versus “N/normol” group. Scale bar = 50 μm.
Subsequently, leveraging both TCGA (Figs. 1C,D) and GSE31210 (Fig. 1E) datasets, we confirmed significant overexpression of DRP2 in LUAD tumors. To substantiate this finding, we performed qRT-PCR on 31 pairs of LUAD tissues and adjacent normals, conclusively showing a significant increase in DRP2 mRNA levels in tumor tissues (Fig. 1F). Furthermore, Western blot analysis on a subset of 12 paired tissue samples reinforced this observation, revealing a marked increase in DRP2 protein levels in cancerous lung tissues compared to their healthy counterparts (Fig. 1G, H). Furthermore, immunohistochemical staining from two representative patients specimen visually corroborated heightened DRP2 protein expression in LUAD versus adjacent normal lung epithelium (Fig. 1I). In summary, these comprehensive findings consistently point to upregulated DRP2 expression in human LUAD. To better understand the relationship between DRP2 expression and the progression of LUAD, 340 LUAD patients from TCGA with complete clinical data were categorized into high and low expression groups based on the expression level of DRP2. Their clinical data (age, gender), pathological characteristics, including pathological stage and TNM stage, were statistically analyzed (Table S1). The results revealed that DRP2 expression was not significantly associated with the T-stage, M-stage, age and gender of LUAD patients (P > 0.05). However, a significant correlation was observed between DRP2 expression and the N stage and pathological stage of the patients (P < 0.05), patients with elevated DRP2 levels had poorer clinical characteristics, with higher stage associated with higher DRP2 expression (Fig. 1J,K). Additionally, we performed immunohistochemical staining to compare the expression of EMT markers (E-cadherin and N-cadherin) in tumor tissues with high DRP2 expression versus adjacent normal lung epithelial tissues with low DRP2 expression (Figure S1). The results demonstrated a significant negative correlation between DRP2 and E-cadherin, while a significant positive correlation was observed between DRP2 and N-cadherin. These findings suggest that elevated DRP2 expression may play a role in the development and metastasis of LUAD, warranting further investigation into its underlying mechanisms. To determine whether DRP2 gene expression can independently predict the prognosis of LUAD, we employed various parameters from the clinical data downloaded from the TCGA database as independent variables and patient survival time as the dependent variable for Cox regression analysis. The results indicated that DRP2 gene expression is an independent risk factor affecting LUAD prognosis ( P < 0.05) (Figure S2A-B).Furthermore, our stratified analysis revealed that the prognostic value of DRP2 depends on the stage and is significant only in early-stage lung adenocarcinoma (LUAD) (Figures S2C–F). This nuanced finding highlights DRP2’s specific utility in early-stage disease, where its expression may serve as a critical biomarker for predicting clinical outcomes.
Genetic variation of DRP2 gene and PPI network in LUAD
Objective To investigate the mutation level of DRP2 gene in LUAD. Employing the online resource cBioPortal, we conducted an in-depth analysis of DRP2 mutations across six datasets, encompassing a cumulative total of 2276 LUAD samples (Fig. 2A). Our findings revealed that DRP2 exhibited an overall mutation rate of 2%, with nonsense mutations predominating as the most frequent mutation type (Fig. 2B). To assess the clinical relevance of DRP2 mutations, revealed no statistically significant differences in overall or disease-free survival between the mutant and non-mutant groups (Fig. 2C). While these mutations may imply a potential role in modulating tumor progression, our results do not conclusively establish a meaningful impact on the overall prognosis of LUAD patients.
Genetic variation of DRP2 gene and PPI network in LUAD. (A) The c-biopportal database shows the proportion of DRP2 mutations in 2276 LUAD samples. (B) Mutation types of DRP2 in 6 databases. (C) Prognostic correlations between DRP2 and mutations. (D) The PPI network of DRP2-interacting genes was visualized by GeneMANIA. (E) The PPI network of DRP2-interacting genes was visualized by STRING.
To gain further insights into the biological functions of DRP2, we constructed a PPI network. Initially, we leveraged the GeneMANIA database to identify the top 20 proteins that interact with DRP2 (Fig. 2D). Subsequently, using the STRING database, we narrowed our focus to the first 18 interacting proteins associated with SLIT2 (Fig. 2E), revealing potential interactions between DRP2 and key proteins such as SNTB1, SNTA1, SOX10, LAMC1, and EGR2.These findings suggest that DRP2 may play a pivotal role in diverse biological processes, including cell proliferation, maintenance of muscle membrane integrity and stability, cytoskeleton stability, and signal transduction, through its intricate interactions with these proteins. By participating in complex PPI networks, DRP2 appears to coordinate and regulate various cellular activities, highlighting its importance in the intricate machinery of cellular homeostasis.
In summary, while our study did not establish a direct link between DRP2 mutations and patient prognosis in LUAD, it underscores the multifunctional nature of DRP2 and its potential involvement in multiple biological pathways. Further investigations are warranted to fully elucidate the mechanisms underlying DRP2’s role in tumor biology and its potential therapeutic implications.
GO, KEGG and GSEA enrichment analysis for DRP2
We leveraged the TCGA database to discern genes that exhibit co-expression patterns with DRP2 in LUAD. Genes depicted in red dots signify a positive correlation with DRP2, whereas blue dots represent a negative correlation (P < 0.05) (Figure 3A). Figure 3B and C list the top 35 genes positively and negatively correlated with DRP2, respectively.
GO, KEGG and GSEA enrichment analysis for DRP2. (A) A Volcano plot of DRP2 co-expressed genes was made by TCGA LUAD data sets. (B,C) Top 35 positively and top 35 negatively correlated genes of DRP2. (D–F) Enrichment analysis of GO terms for DRP2 co-expression genes. (G) Enrichment analysis of KEGG terms for terms for DRP2 co-expression genes. (H–J) GSEA enrichment analyses of diferential expressed genes indicating an association of DRP2 with EMT and cell cycle.
Subsequently, we conducted GO and KEGG enrichment analyses on the top 500 genes positively correlated with DRP2 expression in the TCGA data set. Figure 3D showcases the enrichment of the foremost 20 biological processes, revealing that DRP2 is intricately linked to the regulation of organelle fission, nuclear division, and the transition of mitotic cell cycle phases. Figure 3E presents the cumulative analysis of the top 20 cellular components, indicating that DRP2 is primarily associated with chromosomal regions, spindles, and condensed chromosomes. Moreover, Fig. 3F depicts the enrichment of the first 20 molecular functions, emphasizing DRP2’s involvement in tubulin binding, ATP hydrolysis activity, and cation channel activity.
Furthermore, KEGG enrichment analysis of DRP2 underscored its primary association with the cell cycle (Figure 3G). To delve deeper into the potential functionality of the DRP2 gene, we employed GSEA on differentially expressed genes (Figs. 3H-J). Notably, we observed significant pathway enrichment in the G2/M checkpoint (FDR < 0.001, p < 0.001) and EMT signaling pathways (FDR < 0.001, p < 0.001), prompting us to further investigate the role of DRP2 in these crucial cellular processes.
DRP2 knockdown inhibited DRP2 knockdown inhibited EMT in A549 and H1299 cells
In alignment with GEO and TCGA cohorts, we noticed an elevated expression pattern of DRP2 in bronchial epithelial cells (exemplified by BEAS-2B) compared to NSCLC cell lines, including A549, H1299, H226, and H1650 (Figs. 4A-C). To elucidate the consequences of endogenous DRP2 downregulation, we devised two lentiviral constructs (sh-DRP2-1 and sh-DRP2-2) targeting DRP2, alongside a negative control lentivirus (sh-NC). These constructs were subsequently transfected into A549 and H1299 cells. Following transfection, we validated the knockdown of DRP2 and the expression of mesenchymal transition markers using qRT-PCR and Western blotting techniques (Figs. 4D-F). Our observations post-DRP2 knockdown revealed significant alterations in EMT markers, hinting at DRP2’s potential regulatory role in EMT. Notably, we found an upregulation of the epithelial marker E-cadherin, coupled with a downregulation of N-cadherin, Snail, and vimentin—all indicative of mesenchymal features—accompanied by reduced migratory potential. These findings underscore DRP2’s pivotal role in modulating LUAD cellular dynamics, specifically demonstrating that DRP2 silencing markedly impedes LUAD cell migration, and invasion. Although DRP2 knockdown altered baseline EMT marker expression, it did not suppress TGF-β-induced EMT (Figure S3A-B).
DRP2 knockdown inhibited EMT in A549 and H1299 cells. (A–C) The expression levels of DRP2 in bronchial epithelial cells (BEAS-2B) and various NSCLC cell lines (including A549, H1299, H1650, and H226) were analyzed through qRT-PCR (A) and Western blotting (B,C) methodologies, with β-actin protein utilized as the internal reference for standardization in these assessments. (D–F) The expression of EMT markers was detected by qRT-PCR (D) and western blotting (E,F). (G,H) The wound-healing assay was conducted utilizing DRP2-knockdown cells, alongside control cells. The extent of wound closure at 0 and 24 h post-initiation was quantitatively assessed. (I,J) Transwell assay was used to detect the migration and invasion ability of knockdown and control DRP2 cells. Count the number of cells in the field of view after crystal violet staining. Each experiment was performed three times and the data were expressed as mean ± standard deviation (SD, n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 versus “sh-NC” group. Scale bar = 100 μm.
Furthermore, our results emphasize DRP2’s significance in disrupting epithelial integrity and fostering cancer cell motility and invasion. Through wound healing and migration assays, we observed a notable decline in the migration capability of LUAD cell lines upon DRP2 knockdown (Figs. 4G-H). Furthermore, the migration and invasion potential of these cells were significantly attenuated upon DRP2 silencing (Figs. 4I-J). These findings collectively support the notion that silencing DRP2 can effectively curb the invasive and migratory properties of LUAD, highlighting its potential as a therapeutic target.
Knockdown of DRP2 inhibits proliferation in LUAD cells
Next, we delve deeper into the function of DRP2 in fostering the proliferation of LUAD cells. Initially, we employed flow cytometry to investigate how knocking down DRP2 impacts the cell cycle progression. Our findings revealed that, in contrast to the control group, the DRP2 knockdown cohort exhibited an elevated proportion of cells in the G1 phase and a corresponding decrease in the S phase (Fig. 5A-C). This observation hints that reduced DRP2 expression might impede the transition of cells from the G1 to the S phase, thereby suppressing cellular proliferation.
Knockdown of DRP2 inhibits proliferation in LUAD cells. (A–C) Cell cycle distribution was analyzed by flow cytometry. The percentage of cell number in each phase was calculated. (D.E) Proliferation of DRP2 knockdown and control cells was detected, and the percentage of Edu-positive nuclei was counted after 48 h of culture.Scale bar = 100 μm. (F) CCK8 analysis was used to detect the cell viability of knockdown and control DRP2 cells. (G,H) A549 and H1299 cells depleted of DRP2 (sh-NC as a negative control) were subjected to colony formation assays. Representative images were shown in (G). Colonies were quantified in (H). Each experiment was performed three times and the data were expressed as mean ± standard deviation (SD, n = 3). *P < 0.05, **P < 0.01, ***P < 0.001 versus “sh-NC” group.
To further validate these effects, we assessed cell viability and proliferation rates using the EdU incorporation assay (Figs. 5D, E), the CCK-8 assay (Fig. 5F) and the Colony formation assay (Figs. 5G, H). In line with our previous results, both A549 and H1299 cells with DRP2 knockdown displayed marked reductions in viability and proliferation.
Collectively, these findings underscore the inhibitory role of DRP2 knockdown on LUAD cell proliferation. Further exploration of the underlying mechanisms by which DRP2 promotes tumorigenesis will not only enhance our comprehension of LUAD pathogenesis but also pave the way for the identification of novel therapeutic targets aimed at intervening in this aggressive cancer subtype.
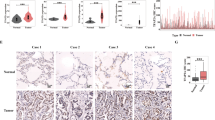
In vivo, DRP2 deficiency inhibits lung adenocarcinoma progression
To substantiate the functional significance of DRP2 in lung cancer progression, we conducted in vivo experiments utilizing animal models (Fig. 6A). Xenograft tumors were generated in immunocompromised nude mice by implanting A549 cells that had been stably transfected with either sh-NC or sh-DRP2-2 lentiviral constructs. Tumor growth was rigorously monitored and documented, culminating in the euthanasia of all mice and subsequent tumor weight assessment (Figs. 6B-D). Consistent with our hypothesis, the suppression of DRP2 significantly impeded the growth of lung cancer cells within the host environment. Immunohistochemical (IHC) analysis (Fig. 6E) further validated this finding, revealing a decrease in the expression of proliferation markers ki67 and PCNA following DRP2 knockdown.
In vivo, DRP2 deficiency inhibits lung adenocarcinoma progression. (A) Schematic diagram of an animal model of LUAD cells. (B–D) Tumor growth and weight were monitored in mice. (E) Immunohistochemical staining was performed to detect the expression of DRP2, PCNA and ki 67 in the xenografts of two groups of nude mice. (F) Left section: Photograph of a mouse lung with metastatic nodules after inoculation with sh-NC or sh-DRP2-2 A549 cells. Scale bar = 3 mm. Right section: Representative microscopic image of H&E staining from the established micrometastases in the lungs of the pair of mice mentioned in the left section. Scale bar = 1 mm. Red arrows indicate metastatic nodules and micrometastases in the lung. (G,H) Dot plots showed differences in metastatic nodules (G) and micrometastases (H) in lung tissue between DRP2 silenced group and control group (4 per group). Data were expressed as mean ± standard deviation (SD, n = 4). **P < 0.01, ***P < 0.001 versus “sh-NC” group.
To investigate the potential impact of DRP2 on metastasis, we established a caudal vein injection model. Intriguingly, despite the development of lung metastases in all mice, those in the DRP2 knockdown cohort exhibited notably smaller metastatic foci compared to the control group (Figs. 6F-H). Collectively, these results suggest that DRP2 may play a pivotal role in facilitating lung cancer progression in vivo, potentially through its modulation of EMT.
Discussion
DRP2, a component of the Dystrophin family, predominantly expresses in the brain and spinal cord, holding important cell biology and clinical significance in the nervous system18. Among the Dystrophin family, dystrophin is a 427-kDa cytoskeletal protein belonging to the β-spectrin/α-actin in family19. DRP2 participates in the formation of the dystrophin-glycoprotein complex (DGC), which bridges the intracellular cytoskeleton (e.g., actin) and the extracellular matrix (e.g., laminin) to maintain the mechanical stability of the cell membrane. This mechanism is critical for resisting mechanical stress, particularly in muscle and nerve cells20. Additionally, DRP2 interacts with signaling molecules (e.g., nitric oxide synthase and ion channels) through the DGC, regulating processes such as intracellular calcium homeostasis, mechanotransduction, and cellular responses including survival, differentiation, and inflammation21.Prior research by Wang et al. indicated that dystrophin exhibits a general tumor suppressive effect in certain myogenic human cancers, notably gastrointestinal stromal tumors (GIST), rhabdomyosarcomas, and leiomyosarcomas22. Conversely, DRP2 has been recognized as an up-regulated gene in breast cancer samples10, and Studies have also shown a link between DRP2 and lung cancer23 and brain cancer24. Nevertheless, the precise function of DRP2 in the development and advancement of lung cancer is still not fully understood and warrants further investigation.
In our study, we sought to clarify the elevated expression of DRP2 in LUAD and its impact on proliferation, metastasis, and invasive capabilities of LUAD cells following knockdown. Our findings revealed a significant increase in DRP2 expression in LUAD, which correlated with poor prognosis in patients and was identified as an independent poor prognostic factor. These results were validated using online datasets such as GEO and TCGA, as well as through analysis of LUAD tissues and cell lines.
Cancer metastasis is a multifaceted process involving the migration of cancer cells from the primary site to distant tissues for colonization25,26, contributing to the majority of cancer-related deaths27. In lung cancer patients, metastasis is the primary cause of mortality, and many patients with early-stage lung cancer relapse due to undetected distant metastases28. The EMT, marked by the disappearance of epithelial traits and the development of mesenchymal characteristics, plays a crucial role in initiating metastasis29. Our study employed TCGA, IHC and GSEA to identify an association between DRP2 and EMT. We further found that down-regulating DRP2 inhibits EMT, upregulates E-cadherin, and downregulates N-cadherin, Snail, and Vimentin. These findings suggest that DRP2 facilitates the invasion and migration of LUAD cells by modulating the EMT pathway. But The lack of suppression in TGF-β-induced EMT (Fig. S3A-B) demonstrates that DRP2 does not directly regulate the canonical TGF-β/Smad pathway, a cornerstone of EMT activation in lung adenocarcinoma30. This finding challenges the initial hypothesis that DRP2 might function as a universal EMT modulator. However, the observed baseline EMT marker changes (e.g., Snail and Vimentin downregulation in Fig. 4) suggest that DRP2 may influence EMT-like phenotypes through alternative mechanisms. One plausible explanation is that these changes arise indirectly from DRP2’s primary role in proliferation suppression. For instance, reduced cell density due to inhibited proliferation could alter cell-cell adhesion dynamics or mechanical stress signaling, thereby secondarily affecting EMT markers31. The robust suppression of Cyclin D1 (70% reduction, p < 0.001) upon DRP2 knockdown, compared to the modest effect on EMT markers (e.g., 30–40% Snail downregulation, p < 0.05), strongly suggests that DRP2’s primary function in LUAD lies in cell cycle regulation rather than direct EMT modulation (Figure S3C). This experimental hierarchy aligns with the stage-specific prognostic significance of DRP2 revealed by TCGA analysis (Fig. S2C-F), where its high expression predicts poor survival exclusively in early-stage (I-II) patients—a phase dominated by proliferative expansion rather than metastatic dissemination32. Such concordance between molecular mechanism and clinical relevance underscores DRP2 as a potential driver of proliferation-dependent tumorigenesis.
However, our study has certain limitations. Further confirmatory experiments are needed to clarify the downstream target of DRP2 in the pathogenesis of LUAD. Furthermore, obtaining more samples from hospitals would strengthen the findings derived from TCGA and facilitate the translation of our research into clinical practice.
Moreover, our future research agenda will emphasize the incorporation of an analysis on EMT and therapeutic resistance within our ongoing investigation of DRP2’s role in lung adenocarcinoma (LUAD) progression. This focus is prompted by insightful studies, including “Involvement of FoxQ1 in NSCLC through regulating EMT and increasing chemosensitivity"33 and “THBS2 + cancer-associated fibroblasts promote EMT leading to oxaliplatin resistance via COL8A1-mediated PI3K/AKT activation in colorectal cancer"34 which have illuminated the crucial interplay between EMT and therapeutic outcomes across various cancer types. We look forward to exploring this intricate relationship further in our subsequent endeavors.
Conclusion
In essence, our study highlights the upregulation of DRP2 as an oncogenic factor in LUAD cell lines and clinical samples. Importantly, the targeted silencing of DRP2 via shRNA technology exerts inhibitory effects on both cell proliferation and metastatic capabilities. These observations collectively point towards DRP2 as a promising novel therapeutic target for LUAD treatment. This discovery not only offers fresh insights into the underlying mechanisms of LUAD pathogenesis but also presents opportunities for enhancing clinical outcomes. Consequently, targeting DRP2 emerges as a potentially innovative therapeutic strategy aimed at curbing the aggressive progression of LUAD.
Data availability
The datasets used and/or analysed during the current study available from the corresponding author on reasonable request.
References
Nasim, F., Sabath, B. F. & Eapen, G. A. Lung cancer. Med. Clin. North. Am. 103, 463–473 (2019).
Gridelli, C. et al. Non-small-cell lung cancer. Nat. Rev. Dis. Primer. 1, 15009 (2015).
Shi, Y. & Shin, D. S. Dysregulation of SWI/SNF chromatin remodelers in NSCLC: its influence on Cancer therapies including immunotherapy. Biomolecules 13, 984 (2023).
Srivastava, S. et al. Emerging role in prognosis, heterogeneity, and therapeutics. Semin. Cancer Biol. 86, 233–246 (2022).
Alexandar, M., Kim, S. Y. & Cheng, H. Update 2020: management of non-small cell lung cancer. Lung 198, 897–907 (2020).
Roberts, R. G. et al. Characterization of DRP2, a novel human dystrophin homologue. Nat. Genet. 13, 223–226 (1996).
Sherman, D. L., Fabrizi, C., Gillespie, C. S. & Brophy, P. J. Specific disruption of a Schwann cell dystrophin-related protein complex in a demyelinating neuropathy. Neuron 30, 677–687 (2001).
Brennan, K. M. et al. Absence of dystrophin related Protein-2 disrupts Cajal bands in a patient with Charcot-Marie-Tooth disease. Neuromuscul. Disord NMD. 25, 786–793 (2015).
Roda, R. H., McCray, B. A., Klein, C. J. & Ahmet, H. Novel hemizygous nonsense mutation in DRP2 is associated with inherited neuropathy. Neurol. Genet. 4, e220 (2018).
Bao, Y. et al. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that May be associated with breast cancer. Cell. Mol. Biol. Lett. 24, 38 (2019).
Warde-Farley, D. et al. The genemania prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214–220 (2010).
Damian, S. et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019).
Brlek, P., Kafka, A., Bukovac, A. & Pećina-Šlaus, N. Integrative cBioPortal analysis revealed molecular mechanisms that regulate EGFR-PI3K-AKT-mTOR pathway in diffuse gliomas of the brain. Cancers 13, 3247 (2021).
Duma, N., Santana-Davila, R. & Molina, J. R. Non-small cell lung cancer: epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc. 94, 1623–1640 (2019).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677 (2025).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes Genomes.
Roberts, R. G. et al. Characterization of DRP2, a novel human dystrophin homologue. Nat. Genet. 13, (1996).
Koenig, M., Monaco, A. P. & Kunkel, L. M. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell 53, 219–228 (1988).
Deconinck, A. E. et al. Utrophin-dystrophin-deficient mice as a model for Duchenne muscular dystrophy. Cell 90, 717–727 (1997).
Kramarcy, N. R., Vidal, A., Froehner, S. C. & Sealock, R. Association of utrophin and multiple dystrophin short forms with the mammalian M(r) 58,000 dystrophin-associated protein (syntrophin). J. Biol. Chem. 269, 2870–2876 (1994).
Wang, Y. & Fletcher, J. A. Cell cycle and dystrophin dysregulation in GIST. Cell. Cycle Georget. Tex. 14, 2713–2714 (2015).
Devaraj, S. & Natarajan, J. miRNA-mRNA network detects hub mRNAs and cancer specific MiRNAs in lung cancer. Silico Biol. 11, 281–295 (2011).
Pooladi, M. et al. The study of ‘dihydropyrimidinase related proteins (DRPs)’ expression changes influence in malignant Astrocytoma brain tumor. Iran. J. Cancer Prev. 7, 130–136 (2014).
Lambert, A. W., Pattabiraman, D. R. & Weinberg, R. A. Emerging biological principles of metastasis Cell. 168, 670–691 (2017).
Zada, S. et al. Control of the epithelial-to-mesenchymal transition and cancer metastasis by autophagy-dependent SNAI1 degradation. Cells 8, 129 (2019).
Mehlen, P. & Puisieux, A. Metastasis: a question of life or death. Nat. Rev. Cancer. 6, 449–458 (2006).
Wy, Z. et al. A prediction rule for overall survival in non-small-cell lung cancer patients with a pathological tumor size less than 30 mm. Dis. Markers (2019).
Neito, M.A., Huang, H. Y., Jackson, R. A. & Theiry, J. P. EMT: 2016. Cell 166 (2016).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell. Biol. 15, 178–196 (2014).
Kagawa, Y. et al. Cell cycle-dependent Rho GTPase activity dynamically regulates cancer cell motility and invasion in vivo. PLoS ONE. 8, e83629 (2013).
Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46 (2022).
Feng, J. et al. Involvement of FoxQ1 in NSCLC through regulating EMT and increasing chemosensitivity. Oncotarget 5, 9689–9702 (2014).
Zhou, X. et al. THBS2 + cancer-associated fibroblasts promote EMT leading to oxaliplatin resistance via COL8A1-mediated PI3K/AKT activation in colorectal cancer.
Funding
This work was supported by grants from Soochow University Technology Program (H230093).
Author information
Authors and Affiliations
Contributions
Zhimeng Chen: Writing – original draft, Conceptualization.Hao Shi: Software, Methodology. Wenxuan Hu: Project administration.Jian Yang:Validation.Yuxuan Xing: Data Curation.Xin Lv: Visualization.Chenzhuo Wu: Software, Methodology.Cheng Ding: Writing – review & editing. Jun Zhao: Supervision, Writing – review & editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The authors assume responsibility for the entire scope of this research, including the assurance that any inquiries concerning the precision or authenticity of any component are thoroughly examined and addressed accordingly. This undertaking has been granted ethical clearance by the Ethics Committee of Soochow University. In this study, we strictly adhered to ethical guidelines for human research. Informed consent was obtained from all patients or their legal guardians for lung adenocarcinoma and adjacent non-cancerous tissue experiments. We informed them of the purpose, methods, risks, and their rights. All participants voluntarily signed the consent form. We promise to protect their privacy and ensure data confidentiality. We will not disclose identifiable information or use data for other purposes. We believe in ethical conduct and thank all participants for their trust and support. We confirm full compliance with ARRIVE guidelines (https://arriveguidelines.org) for animal research reporting, as required by the journal’s editorial policies (https://www.nature.com/srep/journal-policies/editorial-policies#experimental-subjects). All methods were performed in accordance with the relevant guidelines and regulations.The experimental protocols were approved by the Ethics Committee of Soochow University.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, Z., Shi, H., Hu, W. et al. DRP2 promotes EMT and serves as a potential therapeutic target for LUAD treatment. Sci Rep 15, 16590 (2025). https://doi.org/10.1038/s41598-025-01611-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-01611-0
Keywords
This article is cited by
-
Screening and identification of differentially expressed miRNA and mRNA for intervertebral disc degeneration on bioinformatics
European Journal of Medical Research (2025)