Abstract
Despite extensive research highlighting the pivotal role of NOX4 in the development of various malignancies, no systematic pan-cancer analysis has been conducted to evaluate its comprehensive prognostic value and potential immunological functions. In this study, we explored the prognostic significance of NOX4 and its role in immune modulation across different cancer types. We performed a pan-cancer analysis of NOX4 expression and its prognostic implications using multiple databases, including TCGA, GEPIA, GTEx, UALCAN, TISCH, GDSC, PDB, TIMER, and cBioPortal. This analysis provided a detailed assessment of NOX4 expression profiles, clinical correlations, genetic variants, tumor microenvironment interactions, immune relevance, therapeutic potential, and related gene functions through various multi-omics approaches. To further investigate these findings, we collected clinical samples from selected cancer types and validated NOX4 expression through immunohistochemistry, H&E staining, and RT-PCR. Our results revealed that NOX4 was overexpressed in most tumors, although its expression was significantly reduced in certain renal cancers. Moreover, we observed distinct correlations between NOX4 expression and patient prognosis. Notably, NOX4 expression was significantly associated with tumor infiltration, indicating its potential as a target for immunotherapy. Additionally, NOX4 expression showed significant correlations with immune checkpoint proteins (ICP), tumor mutation burden (TMB), microsatellite instability (MSI), RNA stemness scores (RNAss), DNA stemness scores (DNAss), RNA methylation, and DNA methylation.
Similar content being viewed by others
Introduction
The World Health Organization (WHO) reports that cancer is the second leading cause of death globally, responsible for approximately 10 million deaths annually. Cancer accounts for roughly one-sixth of all deaths worldwide1. Beyond these statistics, each case represents the profound impact on individuals and families, who face significant emotional, financial, and healthcare challenges. The growth of large biological databases, advancements in high-throughput technologies (e.g., genomics, proteomics, and metabolomics)2, and the development of novel assays have enabled a wealth of studies that provide new insights into genome-wide analyses and the identification of biomarkers for early detection of pan-cancer. These biomarkers include gene mutations, RNA alterations, driver genes, and copy number variations. In recent years, the ability to trace tumor origins has improved, facilitating earlier detection and potentially aiding in the prediction of clinical outcomes3. Currently, a variety of cancer treatment options are available, including chemotherapy, surgery, radiotherapy, immunotherapy, and targeted therapy. Pan-cancer analysis can identify both shared and unique characteristics across different tumors, offering valuable insights for cancer prevention, therapeutic target development, and the discovery of novel biomarkers4.
NADPH oxidases, members of the NOX family, are electron transport enzymes that function through the inner cell membrane. The NOX family was first identified in the membranes of phagocytes. Currently, the NOX family consists of seven members (NOX1-5 and DUOX1-2)5. Dysregulation of NOX members has been widely observed in cancer models, with NOX4 being the most commonly implicated. NOX4 is a homolog of the phagocyte NADPH oxidase GP91 PHOX (NOX2), and its unique enzymatic activity generates reactive oxygen species (ROS), exhibiting a cell-specific expression pattern across various organs. NOX4 produces ROS in a temporally and spatially regulated manner in response to specific extracellular and intracellular signals, thereby modulating numerous physiological processes6. However, dysregulated NOX-derived ROS production has been linked to a variety of pathological conditions, including carcinogenesis7,8. NOX4 is frequently dysregulated in tumors, leading to continuous production of hydrogen peroxide (H2O2), suggesting a key role for NOX4 in maintaining basal physiological REDOX homeostasis. Hypoxia-induced transcription and/or translation of NOX4 mRNA can further enhance its activity9,10, resulting in increased ROS production. This ROS production stimulates the expression of inflammatory cytokines in many cancer types, thus influencing the tumor microenvironment. Differential levels of NOX4 mRNA and protein have been observed in tumors from various sources11,12,13.
Most studies on the role of NOX4 in tumors have focused on individual cancer types. However, there is a lack of comprehensive research examining the association between NOX4 and multiple cancer types. Given the complexity of tumorigenesis, it is crucial to explore the NOX4 gene’s role and assess its relevance to clinical prognosis and underlying molecular mechanisms. In this study, we analyzed NOX4 expression and its prognostic significance across various cancers using multiple databases, including TCGA, GTEx, UALCAN, TISCH, TIMER, GEPIA, and cBioPortal. Clinical specimens were also collected for validation. Furthermore, we examined the relationship between NOX4 expression and factors such as survival, genetic variations, immune response, and drug sensitivity. Gene functional enrichment analysis and the STRING tool were employed to investigate the potential mechanisms by which the NOX4 family contributes to tumor development.
Materials and methods
Figure 1 shows the workflow of this study.
Gene expression analysis
Gene expression analysis was performed using the “Gene_DE” module of TIMER2 (http://timer.cistrome.org/), where NOX4 was input to assess expression differences between tumor and adjacent normal tissues across various cancer types or specific tumor subtypes from the TCGA project. For certain tumor types with limited normal tissue samples or less common cancers (e.g., TCGA-GBM [Glioblastoma multiforme], TCGA-LAML [Acute myeloid leukemia]), the UCSC Xena platform (https://xenabrowser.net/) was used to access datasets comparing expression levels between these tumor tissues and corresponding normal tissues from the GTEx database. TCGA normal data were then integrated with the GTEx data. Expression data for the NOX4 gene were extracted and log-transformed using log2(x + 0.001) to adjust for variations, and tumor species with fewer than three samples were excluded from the analysis. This process resulted in expression data for 34 cancer types. Statistical analysis was carried out using R (version 4.0.5), with differences in NOX4 expression between normal and tumor samples evaluated. The unpaired Wilcoxon rank-sum test and signed-rank test were employed to assess statistical significance. The abbreviations and full names of all cancer types in the datasets are provided in Table 1.
For protein expression analysis, the UALCAN portal (http://ualcan.path.uab.edu/analysis-prot.html), an interactive online resource for cancer omics data, was used to examine NOX4 protein expression (NP_058627.1) in primary tumor and normal tissues from the Clinical Proteomic Tumor Analysis Consortium (CPTAC). Datasets from six cancer types were selected for analysis: BRCA, OV, COAD, KIRC, UCEC, and LUAD. TISCH (http://tisch.comp-genomics.org), a large online database focused on the tumor microenvironment (TME) using single-cell RNA sequencing data, was accessed to obtain single-cell RNA-seq profiles for KIRC (Kidney Renal Clear Cell Carcinoma).
Patients and clinical specimens
A study was conducted at People’s Hospital in Yangjiang, Guangdong, China, from 2021 to 2022, involving 17 patients with KIRC, 8 patients with GBM, 11 patients with CHOL, 22 patients with BRCA, 21 patients with COAD, 16 patients with ESCA, 20 patients with LUAD, 18 patients with LUSC, 17 patients with LIHC, 11 patients with READ, 23 patients with STAD, and 14 patients with PRAD. The study was approved by the Ethics Committee and the Institutional Review Board of People’s Hospital in Yangjiang and was conducted in accordance with ethical standards, including the Declaration of Helsinki and Chinese Community Guidelines. Written informed consent was obtained from all participants. A total of 416 cancer tissue specimens were collected, including 4 paraffin-embedded specimens (2 KIRC and 2 normal samples) and 412 fresh specimens confirmed by surgery and histopathological examination.
RT-PCR assays
RT-PCR was performed to detect the relative expression of NOX4 mRNA, following the manufacturer’s instructions. The reaction system consisted of 10 µL SYBR Green, 0.4 µL of upstream and downstream primers, 2 µL of cDNA, and deionized water to a final volume of 20 µL. The thermal cycling reaction was carried out on a real-time fluorescence PCR instrument under the following conditions: pre-denaturation at 95 °C for 30 min, followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s. The melting curve was generated by heating the samples from 60 °C to 95 °C, with a temperature increase of 0.5 °C every 5 s, followed by data collection during the melting process. Each sample was analyzed in triplicate, and experiments were repeated three times. The relative expression levels of target genes were calculated using internal reference gene normalization. Primers were synthesized by Shanghai Biotechnology Co., Ltd. The gene-specific primer sequences for NOX4 and the housekeeping gene GAPDH were as follows:
NOX4: Forward: 5’-GTG TCT AAG CAG AGC CTC AGC ATC-3’, Reverse: 5’-CGG AGG TAA GCC AAG AGT GTT CG-3’; GAPDH: Forward: 5’-CTC CTC CTG TTC GAC AGT CA-3’, Reverse: 5’-GCT CCG CCC AGA TAC CATT-3’.
Immunohistochemical analysis
The primary antibody for NOX4 was purchased from Santa Cruz, and the secondary antibody was obtained from Fuzhou Meixin. All procedures were performed according to the manufacturer’s instructions. Tissue sections were blocked with 10% goat serum, incubated with NOX4 primary antibody (1:200 dilution) for 2 h, and then incubated with the secondary antibody for 30 min. DAB was used for color development, followed by dehydration and sealing. All specimens were fixed in 10% neutral formalin, embedded in paraffin, and sectioned at a thickness of 4 μm on slides pretreated with 10% poly-L-lysine. Staining intensity was scored as follows: 0 for no staining, 1 for weak staining, 2 for clear staining of granules and brownish cytoplasm, and 3 for dark brown cytoplasm. The positive cell count was scored as: 0 for no positive cells, 1 for ≤ 20% positive cells, 2 for 21-50% positive cells, and 3 for > 50% positive cells. The overall expression score was determined by multiplying the staining intensity score by the cell count score. A product of 0–2 was considered negative, while a product of ≥ 3 was considered positive expression.
Survival prognosis analysis
Survival and clinical phenotype data were retrieved from all samples available in TCGA. Four survival metrics: overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) were used to assess the relationship between NOX4 expression and patient prognosis. Forest plots were employed to visualize these associations. The OS and disease-free survival (DFS) statistics for NOX4 expression were obtained for all cancer types in TCGA using the “Survival Map” module of GEPIA2 (http://gepia2.cancer-pku.cn/#survival). A cut-off value of 50% was applied to categorize samples into high and low expression groups based on expression thresholds. Log-rank tests were used for statistical significance, and survival maps were also generated using the “Survival Analysis” module of GEPIA2. Kaplan-Meier analysis was performed, with a significance threshold of P < 0.05. Violin plots illustrating NOX4 expression across different pathological stages (I, II, III, and IV) of all tumors in TCGA were generated using the “Pathological Stage Map” feature of GEPIA2. The expression data were log-transformed using log2[TPM (Transcripts Per Million) + 1] before visualization in box plots or violin plots. The relationship between tumor stage, clinical phenotype, and NOX4 expression was evaluated, with statistical significance set at P < 0.05.
Genetic alteration analysis
After logging into the cBioPortal (https://www.cbioportal.org/), we selected the “TCGA Pan-Cancer Atlas Studies” under the “Quick Select” section and entered “NOX4” to query its genetic alteration profiles. The alteration frequency, mutation types, and copy number alterations (CNA) across all TCGA tumor types were analyzed using the “Cancer Types Summary” module. The mutation sites of NOX4 were visualized in a schematic format of the protein structure or in 3D (three-dimensional) using the “Mutations” module.
Correlation analysis of NOX4 expression with TMB or MSI
The Sangerbox platform (http://sangerbox.com/Tool) was used to explore the potential correlation between NOX4 expression and microsatellite instability (MSI) or tumor mutational burden (TMB) in different tumor types from the TCGA cohort. TMB and MSI scores were obtained from TCGA. Correlation analysis between NOX4 expression and TMB or MSI was conducted using Spearman’s rank correlation method. The x-axis of the plot represents the correlation coefficient between NOX4 and TMB or MSI, while the y-axis represents different cancer types. The size of the dots indicates the magnitude of the correlation coefficient, and the color of the dots reflects the statistical significance of the P-value.
Correlation analysis of NOX4 expression with RNA methylation and DNA methylation
We obtained a comprehensive pan-cancer dataset, TCGA-TARGET-GTEx (PANCAN), from the UCSC database, which includes 19,131 samples and 60,499 identifiers. From this dataset, we specifically extracted NOX4 gene expression data and the expression data of 44 RNA modification marker genes, categorized into m1A (10 genes), m5C (13 genes), and m6A (21 genes). These genes were selected from various sample types, including primary solid tumors, primary tumors, blood-derived cancer-bone marrow, and blood-derived cancer-peripheral blood samples. Normal samples were excluded, and each expression value was log2-transformed (x + 0.001). Pearson correlation analysis was performed to assess the relationship between NOX4 expression and RNA modification marker genes. Additionally, we investigated DNA methylation levels of NOX4 across different cancer types compared to their respective adjacent normal tissues using the UALCAN database. Statistical significance was assessed using Student’s t-test, with a significance threshold of P < 0.05.
Correlation analysis of NOX4 expression with tumor stemness
In previous studies, DNA and RNA tumor stemness scores were calculated based on methylation signatures for each tumor. We extracted the stemness index and gene expression data from the TCGA Pan-Cancer dataset, downloaded from the UCSC database. Each expression value was log2-transformed (x + 0.001) for normalization. Cancer types with fewer than three samples were excluded from the analysis. Pearson correlation analysis was then performed to investigate the relationship between NOX4 expression and tumor stemness scores for each tumor type.
Tumor microenvironment analysis
Gene expression profiles for each tumor were extracted from the TCGA TARGET GTEx dataset. These profiles were mapped to GeneSymbols, and the ESTIMATE scores for each sample were calculated using the ESTIMATE R package (version 1.0.13, https://bioinformatics.mdanderson.org/public-software/estimate/). Immune infiltration levels were estimated using several algorithms, including TIMER, CIBERSORT, IPS, QUANTISEQ, XCELL, MCPCOUNTER, and EPIC. We extracted expression data for 155 immune pathway marker genes (41 chemokines, 18 receptors, 21 MHC molecules, 24 immunoinhibitors, and 46 immunostimulators) and 62 immune checkpoint pathway genes (24 inhibitory and 36 stimulatory genes) from the pan-cancer dataset obtained from the UCSC database. Pearson correlation analysis was performed to examine the relationship between NOX4 expression and these immune-related genes. P-values and partial correlation coefficients were calculated using the purity-adjusted Spearman’s rank correlation test. The results were visualized using heatmaps and scatter plots.
Drug sensitivity and molecular docking analysis
The Genomics of Drug Sensitivity in Cancer (GDSC) database, developed by the Sanger Institute, provides data on tumor cell sensitivity and response to various drugs. This information is crucial for identifying potential therapeutic targets, as variations in the cancer genome can influence the effectiveness of treatments, and different targets may respond to drugs in distinct ways. Drug sensitivity data for screening were obtained from the GDSC portal. Sensitivity was quantified by calculating the 50% inhibitory concentration (IC50) values using the R package “Prophet”. A lower IC50 value indicates higher drug susceptibility, while a higher IC50 value indicates lower susceptibility. The crystal structures of the proteins encoded by NOX4 were retrieved from the Protein Data Bank (PDB, www.rcsb.org/pdb/home/home.do). The 3D structures of the compounds were obtained from PubChem (https://www.ncbi.nlm.nih.gov/pccompound). The molecular docking process involved preparing the protein and ligand, constructing the docking grid, and performing the docking of the compounds. Molecular docking simulations were conducted using AutoDock VINA software, following the Schrodinger Glide docking protocol. The best docking poses were selected based on hydrogen bonding, electrostatic, and hydrophobic interactions in the ligand-receptor complexes. Binding energy values (kcal/mol) were used to assess the docking results. PyMOL software was employed to visualize hydrogen bond interactions, binding affinities, interacting amino acid residues, and the 3D structure of the ligand-receptor complexes.
PPI network construction and hub gene recognition
We constructed a protein-protein interaction (PPI) network using the STRING database (https://cn.string-db.org/) to explore the relationships among genes co-expressed with NOX4. The interaction score was set to 0.15, and the top 50 nodes, supported by “Experimental Support” and “Text Mining” evidence, were selected for further visual analysis. Genes that directly interacted with NOX4 were considered central hub genes, while genes that interacted with these central hub genes were classified as subordinate hub genes.
Enrichment analysis of NOX4 co-expression genes
The top 100 genes co-expressed with NOX4 were identified from the STRING database, based on their P-values and correlation coefficients (R). We used the “Gene_Corr” module of TIMER2 to generate heatmaps for the selected genes, including partial correlation (COR) values and P-values derived from the purity-adjusted Spearman’s rank correlation test. We integrated this data for Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis. Gene identifiers (“OFFICIAL_GENE_SYMBOL”) and species (“Homo sapiens”) were uploaded to the Database for Annotation, Visualization, and Integrated Discovery (DAVID, https://davidbioinformatics.nih.gov/ ) for functional annotation. Enriched pathways were visualized using the “TidyR” and “ggplot2” R packages. Gene Ontology (GO) enrichment analysis was conducted using the “clusterProfiler” R package. All analyses were performed using R software (version 4.0.5), and statistical significance was considered at P < 0.05.
Statistical analysis
In this study, all gene expression RNA-seq data were standardized using log2 transformation. The Wilcoxon rank-sum test was employed to evaluate differential expression of NOX4 between normal tissue samples and tumor tissue samples across 34 cancer types. Clinical prognosis analysis was performed using the Kaplan-Meier method, log-rank test, and Cox proportional hazards regression model. Correlation analysis between variables was conducted using either the Spearman or Pearson test, as appropriate. A significance level of P < 0.05 was considered statistically significant. All statistical analyses were performed using R software (version 4.0.5) for data processing.
Results
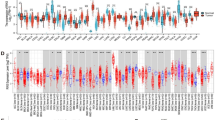
Expression of NOX4 in pan-cancer
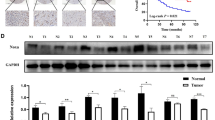
We conducted a comprehensive analysis of NOX4 expression across various tumor types using TCGA data. The results revealed that NOX4 expression was significantly higher in multiple cancers, including breast cancer (BRCA), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), head and neck squamous cell carcinoma (HNSC), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), pheochromocytoma (PCPG), prostate adenocarcinoma (PRAD), rectum adenocarcinoma (READ), stomach adenocarcinoma (STAD), and thyroid carcinoma (THCA), compared to adjacent normal tissues. However, in cervical squamous cell carcinoma (CESC), kidney renal clear cell carcinoma (KIRC), and kidney renal papillary cell carcinoma (KIRP), NOX4 expression was lower than in adjacent tissues (Fig. 2A). To further assess NOX4 expression, we compared tumor tissues with normal tissues from the GTEx dataset. We observed that NOX4 expression was elevated in 26 tumor types compared to their respective normal tissues. In contrast, expression was lower in six tumor types: KIRP, kidney papillary renal cell carcinoma (KIPAN), KIRC, kidney chromophobe (KICH), Wilms tumor (WT), and uterine carcinosarcoma (UCS) (Fig. 2B). RT-PCR analysis confirmed a significant upregulation of NOX4 mRNA in BRCA, ESCA, CHOL, COAD, STAD, LUSC, and GBM, while it was notably downregulated in KIRC (Fig. 2C). These findings were consistent with our database analysis. However, no significant differences were observed in READ, PRAD, and THCA (Figure S1). Analysis of the CPTAC dataset further confirmed a significant reduction in NOX4 protein expression in clear cell RCC tissues compared to normal tissues (Fig. 2D, P < 0.05). Unfortunately, data for other tumor types were unavailable, likely due to gaps in the database. Molecular subtyping can provide insights into the pathways and mechanisms deregulated in specific tumor subpopulations. Based on unsupervised clustering, tumors were categorized into eleven proteome-based pan-cancer subtypes. Each subtype (S1 to S11) corresponds to several tissue-specific tumor types14. Our results indicated that NOX4 protein expression was significantly higher in S4 tumors (associated with oxidative phosphorylation, epithelial functions, and the TCA cycle) and S8 tumors (related to the hemoglobin complex)15 (Fig. 2E). We investigated NOX4 expression in the tumor microenvironment (TME) of KIRC using the Single-Cell dataset from the TISCH database. The results revealed that NOX4 is expressed in malignant cells, endothelial cells, and epithelial cells within the TME of KIRC (Fig. 2F-H). Immunohistochemical analysis and H&E staining showed that NOX4 protein levels were low in KIRC tissues but highly expressed in normal tissues (Fig. 2I, J).
Expression levels of NOX4 in pan-cancer. (A) TIMER2.0 displays the expression of NOX4 across TCGA tumors and corresponding normal tissues. (B) The expression of NOX4 in tumor and normal tissues, as shown on the UALCAN website, which provides protein level data for NOX4 across various tumor types. (C) RT-PCR analysis of NOX4 expression in different tumor types, indicating potential variations in NOX4 regulation across cancers. (D) Expression of NOX4 protein in KIRC provided by the CPTAC website. (E) NOX4 expression through unsupervised cluster analysis for pan-cancer subtypes (S1 to S11) of different tumor types in CPTAC. (F) Single-cell map of KIRC from the GSE159115 dataset. (G) NOX4 expression in KIRC’s single-cell map. (H) Differential expression analysis of NOX4 in KIRC’s single-cell map, revealing significant upregulation in specific cell populations. (I) Hematoxylin and eosin staining of adjacent normal tissue and KIRC. (J) Immunohistochemical staining of NOX4 in KIRC and adjacent normal tissue. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
Prognostic value of NOX4 across tumors
To further investigate the relationship between differential NOX4 expression levels and the prognosis of cancer patients, we evaluated the correlation between NOX4 expression and overall survival (OS), disease-specific survival (DSS), disease-free interval (DFI), and progression-free interval (PFI) across various tumor types using the TCGA database. Univariate Cox regression analysis was performed for this purpose. As shown in Fig. 3A, NOX4 expression was significantly associated with the prognosis of several tumor types, including GBMLGG, LGG, LUAD, STES, STAD, COAD, COADREAD, MESO, PAAD, ACC, KIRP, KIPAN, KIRC, and SKCM. Among these, NOX4 had the most significant prognostic impact in GBMLGG (P = 3.2e-40, HR = 1.64, 95% CI [1.52, 1.77]), followed by LGG (P = 2.7e-7, HR = 1.37, 95% CI [1.22, 1.55]) and ACC (P = 5.7e-3, HR = 1.37, 95% CI [1.09, 1.71]). When examining the correlation between NOX4 expression and DSS in various tumors, most of the results were consistent with those observed for OS (Fig. 3B). The forest plot confirmed an association between high NOX4 expression and poor DFI in CESC, LUAD, PRAD, ESCA, STES, STAD, PAAD, HNSC, ACC, and CHOL. Conversely, in PCPG, low NOX4 expression was associated with poor DFI (Figure S2A). Further analysis of PFI, as shown in Figure S2B, revealed that high NOX4 expression in nine tumors (GBMLGG, LGG, BRCA, STES, PRAD, COAD, COADREAD, PAAD, and ACC) was linked to poor PFI, while low NOX4 expression in six tumors (KIRP, KIPAN, KIRC, SKCM-m, SKCM, and DLBC) was associated with poor PFI. To examine the prognostic implications of NOX4 expression in different tumor types, we categorized the TCGA dataset into high and low NOX4 expression groups. As shown in Fig. 3C, high NOX4 expression was significantly associated with poor OS in COAD (P = 0.013), LGG (P = 1.75e-5), LIHC (P = 0.033), MESO (P = 0.024), and LUAD (P = 0.037). In contrast, low NOX4 expression was significantly linked to poor OS prognosis in KIRC (P = 0.042) and SKCM (P = 0.00041). Analysis of disease-free survival (DFS) (Fig. 3D) indicated that high NOX4 expression was associated with poor prognosis in ACC (P = 0.021), COAD (P = 0.011), ESCA (P = 0.042), and PRAD (P = 0.0047). These findings suggest that NOX4 expression is a potential prognostic biomarker in various tumor types, with differential expression levels correlating with distinct patient outcomes.
Relationship between NOX4 expression and clinical phenotypes
Two key prognostic factors tumor infiltration depth (T category) and the number of positive lymph nodes (N category) have been well-established as significant indicators of cancer progression16,17. Therefore, we explored the relationship between NOX4 expression and clinical factors, including tumor pathological stages, across a range of cancer types. Using the TCGA dataset, we analyzed gene expression variations across different clinical stages in 33 tumor types. We performed unpaired Student’s t-tests and ANOVA to assess differences between samples. Significant variations in NOX4 expression were observed in 13 tumor types, including COAD (P = 1.0e-3), COADREAD (P = 3.0e-5), BRCA (P = 3.8e-3), ESCA (P = 9.0e-6), STES (P = 3.3e-9), KIRP (P = 0.03), KIPAN (P = 0.02), STAD (P = 1.0e-7), PRAD (P = 4.9e-14), THCA (P = 4.8e-4), and BLCA (P = 1.5e-7) at the T stage (Fig. 4A). We identified 10 tumors with significant variations in NOX4 expression across N stages, including COAD (P = 0.03), COADREAD (P = 2.2), BRCA (P = 3.3e-4), KIRP (P = 3.2e-3), KIPAN (P = 6.4e-5), PRAD (P = 3.3e-6), THCA (P = 4.1e-9), READ (P = 3.2e-3), KICH (P = 0.04), and CHOL (P = 0.02). NOX4 expression was consistently downregulated in KIRP and KIPAN, and progressively decreased in other tumors as the disease advanced from T1 to T4 (Fig. 4A) and from N0 to N3 (Fig. 4B). We further examined the correlation between NOX4 expression and tumor pathological stages using the “Pathological Stage Plot” module in GEPIA2. The analysis revealed that NOX4 expression was significantly correlated with pathological stages in several tumors, including BLCA, ESCA, COAD, TCGT, STAD, SKCM, and THCA (P < 0.05), but not in others (Fig. 4C). Interestingly, a significant difference in NOX4 expression was observed between stages I and II, suggesting a potential role of NOX4 in tumor progression at early stages.
Genetic alteration analysis of NOX4
Tumor mutation burden (TMB) is typically defined as the total number of nonsynonymous mutations occurring per 1 million bases in the coding region of the tumor genome, including single nucleotide variations (SNVs) and small insertions/deletions (INDELs). In this study, we analyzed the genetic alterations of NOX4 in specific tumor samples from the TCGA cohorts. We used cBioPortal to examine the mutational landscape and chromosomal abnormalities of NOX4 across various tumors in the TCGA dataset. As shown in Fig. 5A, the highest frequency of NOX4 mutations was observed in UCEC, with a mutation frequency exceeding 6%, representing the largest proportion among all mutation types. Notably, mutations in the NOX4 gene were found to be associated with alterations in several other genes, including DLBC, COAD, CHOL, LGG, and ACC. Furthermore, all cases exhibiting alterations in NOX4 also showed copy number amplification in AML. We further investigated the location and types of NOX4 mutations, as illustrated in Fig. 5B. Missense mutations were the most common mutation type, with the GS375E mutation being identified in four UCEC cases, one CHOL case, and two COAD cases. The 3D structure of the NOX4 protein, including the GS375E mutation site, can be viewed online (Fig. 5C). TMB is an important biomarker reflecting the overall mutational load in tumor cells. To assess the relationship between TMB and NOX4 expression, we performed a Spearman’s rank correlation analysis across various tumor types. The results revealed a positive correlation between NOX4 expression and TMB in LGG, COADREAD, KIRP, OV, and ACC (Fig. 6A). Additionally, we examined the correlation between NOX4 expression and microsatellite instability (MSI), a phenomenon that reflects changes in microsatellite size due to insertions or deletions in tumor cells compared to normal tissues. Our analysis showed that NOX4 expression was negatively correlated with MSI in GBMLGG, KIPAN, LUSC, and UCEC (Fig. 6B).
Correlation analysis of NOX4 expression with RNA and DNA methylation
RNA methylation refers to the chemical modification of RNA, particularly the methylation of adenine residues, catalyzed by methyltransferases. Common RNA modifications include 6-methyladenosine (m6A), 5-methylcytidine (m5C), and 1-methyladenosine (m1A). m6A methylation is a reversible process, involving methyltransferases (Writers), demethylases (Erasers), and methylated RNA-binding proteins (Readers). RNA methylation plays a crucial role in various biological processes, including immunity, tumorigenesis, metastasis, and stem cell renewal. We explored the correlation between NOX4 expression and RNA methylation-related genes in different tumor types (Fig. 7A). Our findings indicate a strong positive correlation between NOX4 expression and RNA methylation in several cancers, including adrenocortical carcinoma (ACC), pancreatic adenocarcinoma (PAAD), ovarian cancer (OV), and thyroid carcinoma (THCA). This suggests that NOX4 expression may influence tumorigenesis by regulating the expression patterns of RNA methylation genes in specific cancers.
(A) Pan-cancer correlation analysis of NOX4 expression with 44 RNA methylation genes. The correlation matrix illustrates the relationships between NOX4 expression and RNA methylation genes across tumors. Each cell in the matrix represents the correlation between NOX4 expression and a specific RNA methylation molecule (listed on the right) in a particular tumor type (listed at the bottom). The correlation coefficient and p-value are indicated by the color of each cell. (B) DNA methylation levels of NOX4 across tumors. *P < 0.05, **P < 0.01, ***P < 0.001.
In addition to RNA methylation, we also examined DNA methylation, which has been shown to impact the expression of genes associated with tumorigenesis18. Using the UALCAN database, we analyzed the methylation status of NOX4 and found that its promoter methylation levels were significantly lower in tumors such as TGCT, colon adenocarcinoma (COAD), and kidney renal clear cell carcinoma (KIRC), compared to control groups. In contrast, higher promoter methylation levels were observed in bladder cancer (BLCA), breast cancer (BRCA), prostate adenocarcinoma (PRAD), lung adenocarcinoma (LUAD), thyroid carcinoma (THCA), uterine corpus endometrial carcinoma (UCEC), stomach adenocarcinoma (STAD), cervical squamous cell carcinoma (CESC), and cholangiocarcinoma (CHOL) (Fig. 7B). No significant changes in NOX4 methylation levels were detected in other tumor types (Figure S3). Together, these findings suggest that the increased expression of NOX4 in tumors may be partly attributed to changes in its promoter methylation status.
Tumor stemness analysis
Cancer stem cells (CSCs) are a subset of cancer cells that share characteristics with normal stem cells, including the ability to give rise to all cell types within a tumor. These cells are thought to have the potential to initiate tumor formation and promote tumor growth, particularly as the tumor metastasizes to distant organs, resulting in the formation of secondary tumors. Compared to non-stem tumor cells, CSCs are associated with tumor progression, metastasis, drug resistance, and enhanced self-renewal capabilities. This is facilitated by self-protective mechanisms such as DNA damage repair, inhibition of apoptotic pathways, and the production of drug-resistant proteins. To quantify tumor stemness, we employed a machine learning algorithm called One-Class Linear Regression (OCLR)19, which integrates transcriptome and methylome data. This allowed us to calculate two stemness indexes: mRNAsi (reflecting gene expression profiles of stem cells) and MDNAsi (representing epigenetic characteristics of stem cells). Correlations between these indexes and NOX4 expression were calculated across various tumors. Our analysis (Fig. 8A) revealed a significant positive correlation between NOX4 expression and stemness indexes in several cancers. For example, in BRCA (R = 0.09, P = 0.006), KIRP (R = 0.17, P = 0.003), THYM (R = 0.19, P = 0.030), MESO (R = 0.32, P = 0.002), and UVM (R = 0.33, P = 0.002). A significant negative correlation was observed in one tumor, HNSC (R = -0.22, P = 2.45E-7). As shown in Fig. 8B, we found a strong correlation in 22 tumors, including a positive correlation in UVM.
Analysis of tumor immune microenvironment
A growing body of evidence highlights the critical role of the tumor microenvironment (TME) in tumorigenesis and progression20,21. Thus, understanding the relationship between the TME and NOX4 expression in pan-cancer is of utmost importance. In this study, we used the ESTIMATE algorithm to calculate immune scores for 34 tumor types from the TCGA, TARGET, and GTEx datasets and analyzed the correlation between NOX4 expression and these scores. Our findings showed that NOX4 expression was significantly positively correlated with the ESTIMATE score in 27 tumor types, including COAD, READ, BLCA, and COADREAD. However, in ALL-R, NOX4 expression was inversely correlated with the ESTIMATE score. The correlation coefficients for the top 9 tumors are presented in Fig. 9, while results for other tumors can be found in Figure S4. Given the clear relationship between NOX4 and immune response, we also examined the correlation between NOX4 expression and immune cell infiltration levels across pan-cancer. As shown in Fig. 10A, NOX4 expression was significantly associated with the abundance of various infiltrating immune cells in different tumors: B cells in 7 tumors, CD4 + T cells in 19 tumors, CD8 + T cells in 20 tumors, macrophages in 24 tumors, neutrophils in 27 tumors, and dendritic cells in 25 tumors. Notably, NOX4 expression was strongly correlated with immune cell abundance in COAD, LIHC, and COADREAD (Fig. 10B). Immune infiltration was assessed using multiple algorithms, including CIBERSORT, IPS, XCELL, QUANTISEQ, EPIC, and MCPCOUNTER, with results shown in Figure S5. We observed that NOX4 expression was positively correlated with M1 macrophages, M2 macrophages, Tregs, and CAFs in most tumors.
Immune checkpoints (ICP) play a pivotal role in regulating immune responses and determining the extent of immune activation22. Figure 11A illustrates the relationships between NOX4 expression and 60 immune checkpoint genes across different tumor types. Our analysis revealed a strong correlation between NOX4 expression and immune checkpoint genes in both inhibitory pathways (e.g., CD276, TGFB1) and stimulatory pathways (e.g., TNFSF4, CD28, VEGFB, CX3CL1, ILR4, IL2RA) in various tumors. Specifically, a close association was observed between NOX4 expression and immune checkpoints in ovarian (OV), liver (LIHC), prostate (PRAD), stomach (STES), and pancreatic (PAAD) cancers. These findings suggest that NOX4 may play a significant role in tumor immunity by modulating immune checkpoint expression across various tumor types. Furthermore, gene co-expression analysis was performed to explore the relationship between NOX4 expression and immune-related genes in different tumors. The analyzed genes included those involved in MHC presentation, immune activation and suppression, as well as chemokines and chemokine receptors. The heatmap of the results (Fig. 11B) showed that most immune-related genes were co-expressed with NOX4, with a majority showing positive correlations with NOX4 across various tumor types, except for DLBC, NB, and TGCT (P < 0.05).
Drug sensitivity and molecular docking analysis
Drug sensitivity has long been a key focus in personalized cancer chemotherapy. Investigating drug sensitivity in cancer is critical for tailoring treatments to individual patients and advancing precision medicine. However, due to significant inter-individual variability in drug response, relying solely on limited scientific resources is often ineffective. Therefore, identifying molecules linked to drug response is crucial for optimizing therapeutic strategies. The Genomics of Drug Sensitivity in Cancer (GDSC) is currently the largest public database containing drug susceptibility data and molecular markers in tumor cells. Based on multiple studies and integrated database information, the GDSC database re-analyzes drug sensitivity and response across three retrieval levels: cell, drug, and molecule. Our analysis revealed that NOX4 expression was negatively correlated with the sensitivity of tumor cells to Dabrafenib, TGX221, and CHIR-99,021, but positively correlated with the sensitivity to AICAR, Afatinib, Gefitinib, and EKB-569 (Fig. 12A). Bioinformatics analysis and molecular docking simulations further identified potential interactions between NOX4 and these drugs (Fig. 12B). The results indicated that NOX4 had the weakest binding affinity with Dabrafenib, with a binding energy of -8.3 kcal/mol. This interaction formed a conventional hydrogen bond with ASP7 at a distance of 2 Å. In contrast, the NOX4-CHIR-99,021 complex was stabilized by three hydrogen bonds with ALA299, ASP71, and TYR70, interacting at distances of 2.1 Å, 2.6 Å, and approximately 2.2 Å, respectively, with a binding energy of -8.0 kcal/mol. These findings suggest that Dabrafenib and CHIR-99,021 could be potential inhibitors of NOX4. However, we were unable to obtain meaningful molecular docking results for EKB-569 and Gefitinib. Detailed molecular docking results for the other drugs are provided in Table S1.
(A) Correlation between NOX4 expression and drug sensitivity (top 25 drugs) in pan-cancer, based on GDSC data. (B) NOX4 docking profile with a drug molecule, shown in the 3D structure of the ligand-receptor interaction in the left panel. The right panel displays a 2D representation of the interaction between the ligand and receptor within the binding pocket.
Functional enrichment analysis
To further explore the molecular mechanisms underlying the role of NOX4 in tumorigenesis, we focused on identifying NOX4-binding proteins and genes correlated with NOX4 expression for pathway enrichment analysis. Using the STRING tool, we identified 50 NOX4-binding proteins supported by experimental and co-expression evidence. The protein-protein interaction (PPI) network of these proteins, shown in Fig. 13A, includes 51 nodes and 403 edges, with a mean node degree of 15.8. Our analysis revealed that NOX4 was correlated with NOX1, NOX3, CYBB, DUOX1, DUOX2, and TLR4. Interestingly, all of these genes belong to the NOX family, except for TLR4. The corresponding heatmap (Fig. 13B) showed a strong correlation between NOX4 and these six genes across various tumor types. We then performed KEGG and GO enrichment analyses on the identified genes. The KEGG pathway analysis (Fig. 13C) suggested that the “AGE-RAGE signaling pathway,” “TNF signaling pathway,” and “pathways in cancer” might be involved in the role of NOX4 in tumor pathogenesis. The GO enrichment analysis indicated that many of these genes are associated with cellular processes related to RNA metabolism, including “response to oxidative stress,” “cellular response to oxidative stress,” “NADPH oxidase complex,” “endomembrane system,” “vesicle,” “oxidoreductase activity,” and “antioxidant activity” (Figs. 13D, E, and F).
NOX4-related gene enrichment analysis. (A) Using the STRING tool, we identified experimentally validated NOX4-binding proteins. (B) Heatmap of NOX4-binding proteins across different tumor types, including NOX1, NOX3, TLR4, CYBB, DUOX1, and DUOX2. (C) KEGG pathway analysis based on NOX4-binding and interacting genes. (D) Bubble plot of GO analysis (biological processes, BP). (E) Bubble plot of GO analysis (cellular components, CC). (F) Bubble plot of GO analysis (molecular functions, MF).
Discussion
In recent years, cancer has emerged as a significant public health challenge. To better understand the underlying molecular mechanisms of cancer and to develop more effective therapeutic strategies, numerous studies have conducted pan-cancer analyses to identify new prognostic and diagnostic biomarkers. NOX4, an important isoform of NADPH oxidase, plays a complex and crucial role in the initiation and progression of cancer. By generating low levels of reactive oxygen species (ROS), NOX4 regulates various biological processes such as cell proliferation, migration, and apoptosis23,24. Unlike other members of the NOX family, NOX4 exhibits constitutive activity, continuously producing ROS and acting as a second messenger in cell signaling pathways. It modulates key signaling pathways, including HIF2-α, p38 MAPK, TGF-β1/Smad2/3, Akt, and Caspase35. In tumor cells, NOX4 not only regulates cell growth and stress responses but also promotes metastasis and invasion through this mechanism. However, under certain external stimuli, NOX4 can generate high levels of ROS, activating downstream signaling pathways that further drive tumor progression25. Research has shown that NOX4 is highly expressed in various cancer types, particularly in tumors of the lung, ovary, and pancreas, where its expression in tumor tissues is typically higher than in adjacent normal tissues12,13,26. However, other studies suggest that NOX4 gene knockout enhances carcinogen-induced solid tumor formation27. In breast cancer, NOX4 has been shown to reduce the invasiveness of cancer cells by coordinating the PGC1α/Drp1 axis to regulate mitochondrial turnover28. Despite these findings, there remains a critical need for a comprehensive pan-cancer analysis of NOX4 to elucidate its broader relationship with cancer. In this study, we systematically examined the expression and function of NOX4 across 33 different cancer types using datasets from TCGA, GTEx, and CPTAC. Our analysis covered gene expression, prognostic significance, tumor heterogeneity, and immune cell infiltration within the tumor microenvironment.
In this study, we compared the expression of NOX4 in 34 different types of tumors and their corresponding normal tissues. The results showed that, with the exception of UCEC and CESC, NOX4 exhibited significant expression differences in most cancer types. Specifically, NOX4 was significantly overexpressed in 26 tumor types, while it was significantly underexpressed in 6 types (KIRP, KIPAN, KIRC, KICH, WT, and UCS). These findings are consistent with previous studies BLCA, BRCA, OV, PRAD, and GBM29. Notably, the expression of NOX4 in KIRC was significantly lower, as evidenced by mRNA expression data from TCGA, total NOX4 protein levels from CPTAC, and our own RT-PCR and immunohistochemistry analyses of KIRC tumor samples. Previous studies on renal cell carcinoma (RCC) have shown that NOX4 promotes tumor growth in renal cancer cells, with evidence suggesting that NOX4 plays a critical role in the onset and progression of RCC30. It does so by enhancing the expression and nuclear accumulation of HIF-2α, as well as influencing cell invasion and branching, thereby supporting the tumor phenotype in renal cancer cells30. Another study found that the nuclear localization of NOX4 in RCC tissues was significantly correlated with disease progression and mortality31. Higher nuclear NOX4 expression was strongly associated with increased disease progression and mortality, and elevated nuclear NOX4 levels were linked to resistance to targeted therapies and immunotherapies31. In RCC, NOX4 has been found to localize to the inner mitochondrial membrane or the outer mitochondrial membrane, and its co-localization with mitochondrial markers has been observed using MitoTracker Red staining and immunofluorescence techniques32. Although these findings seem inconsistent with our results, we hypothesize that this may be related to subcellular localization of NOX4 in RCC, tumor heterogeneity, and metabolic reprogramming32. Renal cancer cells often exhibit the Warburg effect, where they preferentially rely on aerobic glycolysis even in the presence of oxygen33. This metabolic shift provides rapid energy for the tumor and may reduce the cancer cells’ dependence on traditional oxidative phosphorylation pathways. Additionally, the low expression of NOX4 in KIRC might be related to tumor type. In breast cancer studies, NOX4 expression was found to be significantly overexpressed in Luminal-A MCF-7 cells (a model of estrogen receptor-positive breast cancer) compared to MDA-MB-231 cells (representing basal-like breast cancer) and normal mammary epithelial cells (MCF 10 A). In contrast, NOX4 expression was significantly downregulated in triple-negative breast cancer (TNBC) patients28. Prognostic analysis revealed that NOX4 expression levels are closely associated with prognosis in multiple cancers. Our analysis indicated that high NOX4 expression was associated with poorer overall survival (OS) and disease-free survival (DFS) in colon adenocarcinoma (COAD), low-grade glioma (LGG), and lung adenocarcinoma (LUAD). The prognostic impact of NOX4 expression has been confirmed in various cancers, including colorectal cancer, endometrial cancer, gastric cancer, retinoblastoma, and hepatocellular carcinoma12,34. NOX4 siRNA effectively inhibited tumor progression in xenograft models of hepatocellular carcinoma35. In head and neck cancer cells, epidermal growth factor receptor (EGFR) inhibitors induced autophagy via a NOX4-dependent mechanism36. These findings suggest that NOX4 expression may serve as a prognostic biomarker with varying clinical significance across different cancers. Additionally, we observed that NOX4 expression was associated with tumor stage in certain cancers, particularly in stages I and II of gastric cancer (STAD), where NOX4 expression was significantly elevated.
Tumor heterogeneity is a major challenge in cancer research and treatment. Within the same cancer type, there exists significant cell and molecular heterogeneity between molecular subtypes, patients, tumor lesions, and cells from specific lesions37. From a cellular perspective, tumor heterogeneity is characterized by the presence of subpopulations of cells with distinct morphological and phenotypic features. Although cancer cells originate from a single malignant cell, they accumulate mutations in their DNA as they proliferate. Over time, tumors evolve into a collection of highly aggressive clones with distinct abilities to survive, proliferate, and metastasize to distant organs. In this context, we explored various aspects of NOX4 in relation to pan-cancer, including mutations, methylation, and other factors. Our study revealed that the frequency of NOX4 mutations was highest in uterine corpus endometrial carcinoma (UCEC), with mutations in NOX4 detected in all cases of diffuse large B-cell lymphoma (DLBC), colon adenocarcinoma (COAD), cholangiocarcinoma (CHOL), lower-grade glioma (LGG), and adrenocortical carcinoma (ACC). Additionally, all cases with alterations in the NOX4 locus in acute myeloid leukemia (AML) were associated with copy number amplification. However, the prognostic significance of these mutations in these cancers remains unclear, and further investigation is needed to better understand their clinical implications. We also found that NOX4 expression was positively correlated with tumor mutational burden (TMB) in LGG, COAD, rectal adenocarcinoma (COADREAD), kidney renal papillary cell carcinoma (KIRP), ovarian cancer (OV), and ACC. Conversely, NOX4 expression was negatively correlated with microsatellite instability (MSI) in glioblastoma multiforme (GBM), LGG, KIPAN, lung squamous cell carcinoma (LUSC), and UCEC. Additionally, we examined the relationship between DNA promoter methylation, RNA methylation-modified genes, and NOX4 expression. The results consistently indicated that NOX4 expression was closely associated with tumor heterogeneity, thereby influencing tumor progression.
The tumor microenvironment, comprising tumor cells, inflammatory cells, and the surrounding stroma, includes both the immune microenvironment and the stromal microenvironment. It is a critical component of tumor tissue and plays a significant role in tumor progression, angiogenesis, and genomic instability. The immune microenvironment is often referred to as the “seventh hallmark” of cancer38. Therefore, we examined the role of NOX4 within the human tumor microenvironment. Our study found that NOX4 expression levels were significantly and positively correlated with immune scores in 27 types of tumors, including rectal adenocarcinoma (READ), colon adenocarcinoma (COAD), and bladder urothelial carcinoma (BLCA). Additionally, NOX4 expression was closely associated with the infiltration of immune cells, such as CD4 + T cells, dendritic cells, B cells, macrophages, CD8 + T cells, and neutrophils, particularly in COAD, liver hepatocellular carcinoma (LIHC), and COADREAD. These findings suggest that NOX4 interacts with immune cells, a relationship that warrants further investigation. In contrast, cancer-associated fibroblasts (CAFs) in the tumor stroma are known to regulate the infiltration and function of various immune cells39. Our study confirmed a strong positive correlation between NOX4 expression and CAFs in the TCGA dataset, aligning with previous research. This suggests that NOX4 may influence the tumor immune microenvironment by modulating the growth and activity of fibroblasts, which in turn affects tumor progression. In HCC, studies using NOX4-/- mice revealed a significant increase in M2-polarized macrophages, a reduction in liver fibrosis, and an enhancement of HCC development40. In NSCLC, NOX4 expression was closely associated with macrophage chemotaxis in NSCLC patients6. NOX4 stimulates the production of various cytokines, including CCL7, IL-8, CSF-1, and VEGF-C, through the generation of ROS, a process that is dependent on the PI3K/Akt signaling pathway6. In GBM cells, TGF-β1-induced metabolic reprogramming and epithelial-mesenchymal transition (EMT) are regulated by NOX4/ROS41. TGF-β1, a crucial cytokine released from the tumor microenvironment (TME), plays a key role in tumor initiation and malignant transformation. The metabolic alterations in tumor cells are a hallmark of cancer, providing ATP to fuel cellular processes and generating metabolic intermediates necessary for the synthesis of essential cellular components, which support cell proliferation, migration, and invasion41. These studies highlight the critical role of NOX4 in the invasive growth of tumor cells, particularly in the formation of invasive cellular protrusions and the migration and invasion activities of tumor cells.
Many therapeutic strategies are available for targeting immune cells in tumors, such as pericyte therapy with transgenic T cells and chimeric antigen receptor T (CART) cell therapy42. Additionally, the use of immune checkpoint inhibitors (ICIs) has emerged as a promising approach to enhance cancer immunity by blocking immune checkpoint receptors or ligands, thereby inhibiting tumor metastasis and recurrence, while also reducing off-target toxic effects43. In the context of chemoresistance, ICIs are particularly effective for tumors with high microsatellite instability (MSI) and high tumor mutational burden (TMB). Given these findings, NOX4 may serve as a potential target in combination with novel anti-cancer immunotherapeutic drugs or established immune checkpoint inhibitors to enhance cancer immunity and therapeutic response. Furthermore, several studies have suggested that NOX4 could be a promising target in antitumor immunotherapy. Combining chemotherapeutic agents with NOX4 depletion may offer a novel approach to overcoming resistance and improving treatment outcomes. Drug resistance remains a primary challenge in cancer chemotherapy, often limiting the effectiveness of treatment. Some studies have indicated that NOX4 is associated with chemoresistance. For instance, Karthigayan et al. found that NOX4 acts as a mitochondrial energy sensor, linking cancer metabolic reprogramming to drug resistance32. In ovarian cancer cells, NOX4 knockdown increased sensitivity to targeted therapy and radiotherapy by reducing the expression of HER3 and NF-κB p6544. Moreover, the combination of the NOX4 inhibitor GKT137831 with afatinib demonstrated a synergistic effect, reducing cell viability in ovarian cancer cells. In our study, we conducted drug sensitivity analysis and molecular docking assays, revealing that NOX4 expression was inversely correlated with the sensitivity to dabrafenib, TGX221, and CHIR-99,021, while positively correlated with the sensitivity to AICAR, afatinib, gefitinib, and EKB-569. These results suggest that NOX4 could be utilized as a novel drug target for anti-cancer immunotherapy, either alone or in combination with established immune checkpoint inhibitors, to enhance the immune response against tumors. Recent research has also highlighted a novel role for NOX4 in the regulation of cancer stemness, a key phenotypic hallmark that shares many characteristics with cell metastasis. Although the underlying mechanisms remain unclear, the regulation of cancer stemness by NOX4 may involve the modulation of stem cell-specific transcriptional networks45. However, further investigations are required to elucidate this potential relationship and its implications for cancer therapy.
Our multi-omics analysis has unveiled several potential molecular mechanisms through which NOX4 may contribute to cancer development and progression. The protein-protein interaction network and correlation analysis highlighted strong associations between NOX4 and other members of the NOX family (NOX1, NOX3, CYBB, DUOX1, DUOX2), as well as TLR4. This finding suggests intricate interactions within the NOX family, potentially playing a crucial role in cancer pathogenesis46. KEGG pathway analysis identified three key pathways potentially involved in NOX4-mediated tumorigenesis the AGE-RAGE signaling pathway, TNF signaling pathway, and various cancer-related pathways. The AGE-RAGE signaling pathway has been demonstrated to promote inflammation and oxidative stress, factors known to contribute to cancer development47. NOX4’s involvement in this pathway may exacerbate these pro-tumorigenic processes. Similarly, the TNF signaling pathway plays a critical role in inflammation and cell survival, and its dysregulation has been linked to multiple cancers48. The association of NOX4 with this pathway suggests it may modulate inflammatory responses and cell fate decisions within the tumor microenvironment. GO enrichment analysis provided further insights into NOX4’s potential mechanisms in cancer. Significant enrichment of terms related to oxidative stress response and NADPH oxidase complex activity aligns with NOX4’s established function as a major source of reactive oxygen species (ROS) in cells5. Elevated ROS levels can lead to DNA damage, genomic instability, and alterations in cell signaling pathways, all of which may promote cancer initiation and progression49. The enrichment of terms related to the endomembrane system and vesicles suggests that NOX4 may also play a role in intracellular trafficking and secretion processes. This could potentially influence the tumor microenvironment through the regulation of extracellular vesicle release or the secretion of pro-tumorigenic factors50. The association of NOX4 with RNA metabolism-related processes is a novel finding that warrants further investigation. This could indicate a previously unrecognized role for NOX4 in regulating gene expression at the post-transcriptional level, potentially affecting the expression of oncogenes or tumor suppressors51,52. NOX4 exhibits a strong correlation with TLR4, a key player in innate immunity that has been implicated in promoting tumor growth and immune evasion53. This association suggests that NOX4 may influence tumor-immune interactions, possibly through modulation of TLR4 signaling or by affecting the tumor inflammatory microenvironment.
Conclusions
In conclusion, our first pan-cancer analysis of NOX4 revealed that NOX4 expression was not significantly correlated with clinical prognosis, RNA and DNA methylation, tumor stemness, immune cell infiltration, tumor mutational burden, or microsatellite instability across a range of tumor types. These findings provide insights into the role of NOX4 in tumorigenesis from the perspective of clinical tumor samples. However, several limitations should be considered. The small sample size for certain tumor types, the quality of the clinical data, and the relatively limited body of research on NOX4 may introduce biases into our conclusions. Therefore, our next steps will involve obtaining additional tumor tissue samples and conducting further preclinical research to better understand the regulatory mechanisms of NOX4.
Limitations
Although our pan-cancer analysis provides a comprehensive evaluation of NOX4 expression and its prognostic and immunological correlations, several limitations remain. The majority of the data used in this study were sourced from publicly available databases, which rely on retrospective data and may be subject to inherent biases. These factors could affect the generalizability and accuracy of the study’s findings. Despite utilizing multiple databases and multi-omics approaches, our study lacks in vitro and in vivo functional validation across all cancer types, particularly regarding the specific mechanisms through which NOX4 impacts the tumor microenvironment and immune regulation. The heterogeneity among different cancer types, patient populations, and tissue samples may contribute to variations in NOX4 expression patterns, potentially limiting the consistency of its prognostic value across all malignancies. Immunohistochemistry and RT-PCR validation were performed on a limited number of selected cancer types, which may not fully represent the diversity of cancer scenarios. While we have identified correlations between NOX4 expression and immune checkpoint proteins, tumor mutation burden, and other biomarkers, the causal relationships and underlying molecular mechanisms require further investigation through functional studies and clinical trials to establish NOX4 as a viable prognostic and therapeutic target.
Data availability
All data are contained within the manuscript. The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71 (3), 209–249 (2021).
Hristova, V. A. & Chan, D. W. Cancer biomarker discovery and translation: proteomics and beyond. Expert Rev. Proteom. 16 (2), 93–103 (2019).
Zou, J. & Wang, E. Cancer biomarker discovery for precision medicine: new progress. Curr. Med. Chem. 26 (42), 7655–7671 (2019).
Priestley, P. et al. Pan-cancer whole-genome analyses of metastatic solid tumours. Nature 575 (7781), 210–216 (2019).
Vermot, A., Petit-Härtlein, I., Smith, S. M. E. & Fieschi, F. NADPH oxidases (NOX): an overview from discovery, molecular mechanisms to physiology and pathology. Antioxid. (Basel) 10, 6 (2021).
Zhang, J. et al. Tumoral NOX4 recruits M2 tumor-associated macrophages via ROS/PI3K signaling-dependent various cytokine production to promote NSCLC growth. Redox Biol. 22, 101116 (2019).
Roy, K. et al. NADPH oxidases and cancer. Clin. Sci. (Lond). 128 (12), 863–875 (2015).
Meitzler, J. L., Konaté, M. M. & Doroshow, J. H. Hydrogen peroxide-producing NADPH oxidases and the promotion of migratory phenotypes in cancer. Arch. Biochem. Biophys. 675, 108076 (2019).
Li, S. et al. NOX4 regulates ROS levels under normoxic and hypoxic conditions, triggers proliferation, and inhibits apoptosis in pulmonary artery adventitial fibroblasts. Antioxid. Redox Signal. 10 (10), 1687–1698 (2008).
Shi, Q. et al. Interplay between RNA-binding protein HuR and Nox4 as a novel therapeutic target in diabetic kidney disease. Mol. Metab. 36, 100968 (2020).
Azouzi, N. et al. NADPH oxidase NOX4 is a critical mediator of BRAF(V600E)-Induced downregulation of the sodium/iodide symporter in papillary thyroid carcinomas. Antioxid. Redox Signal. 26 (15), 864–877 (2017).
Lin, X. L. et al. Overexpression of NOX4 predicts poor prognosis and promotes tumor progression in human colorectal cancer. Oncotarget 8 (20), 33586–33600 (2017).
Meitzler, J. L. et al. Decoding NADPH oxidase 4 expression in human tumors. Redox Biol. 13, 182–195 (2017).
Chen, F., Chandrashekar, D. S., Varambally, S. & Creighton, C. J. Pan-cancer molecular subtypes revealed by mass-spectrometry-based proteomic characterization of more than 500 human cancers. Nat. Commun. 10 (1), 5679 (2019).
Zhang, Y., Chen, F., Chandrashekar, D. S., Varambally, S. & Creighton, C. J. Proteogenomic characterization of 2002 human cancers reveals pan-cancer molecular subtypes and associated pathways. Nat. Commun. 13 (1), 2669 (2022).
Reim, D. et al. Prognostic implications of the seventh edition of the international union against cancer classification for patients with gastric cancer: the Western experience of patients treated in a single-center European institution. J. Clin. Oncol. 31 (2), 263–271 (2013).
Sa, H. S. et al. Association of T and N categories of the American joint commission on cancer, 8th edition, with metastasis and survival in patients with orbital sarcoma. JAMA Ophthalmol. 138 (4), 374–381 (2020).
Lin, R. K. & Wang, Y. C. Dysregulated transcriptional and post-translational control of DNA methyltransferases in cancer. Cell. Biosci. 4, 46 (2014).
Chen, P., Hsu, W. H., Han, J., Xia, Y. & DePinho, R. A. Cancer stemness Meets immunity: from mechanism to therapy. Cell. Rep. 34 (1), 108597 (2021).
Peña-Romero, A. C. & Orenes-Piñero, E. Dual effect of immune cells within tumour microenvironment: Pro- and Anti-Tumour effects and their triggers. Cancers (Basel) 14, 7 (2022).
Jones, J. O., Moody, W. M. & Shields, J. D. Microenvironmental modulation of the developing tumour: an immune-stromal dialogue. Mol. Oncol. 15 (10), 2600–2633 (2021).
He, X. & Xu, C. Immune checkpoint signaling and cancer immunotherapy. Cell. Res. 30 (8), 660–669 (2020).
Moloney, J. N. & Cotter, T. G. ROS signalling in the biology of cancer. Semin Cell. Dev. Biol. 80, 50–64 (2018).
Wang, L. & Gong, W. NOX4 regulates gastric cancer cell invasion and proliferation by increasing ferroptosis sensitivity through regulating ROS. Int. Immunopharmacol. 132, 112052 (2024).
Wu, Y. et al. Activation of TLR4 is required for the synergistic induction of dual oxidase 2 and dual oxidase A2 by IFN-γ and lipopolysaccharide in human pancreatic cancer cell lines. J. Immunol. 190 (4), 1859–1872 (2013).
Hiraga, R., Kato, M., Miyagawa, S. & Kamata, T. Nox4-derived ROS signaling contributes to TGF-β-induced epithelial-mesenchymal transition in pancreatic cancer cells. Anticancer Res. 33 (10), 4431–4438 (2013).
Helfinger, V. et al. Genetic deletion of Nox4 enhances cancerogen-induced formation of solid tumors. Proc. Natl. Acad. Sci. U S A ;118, 11 (2021).
Bhadane, D., Kamble, D., Deval, M., Das, S. & Sitasawad, S. NOX4 alleviates breast cancer cell aggressiveness by co-ordinating mitochondrial turnover through PGC1α/Drp1 axis. Cell. Signal. 115, 111008 (2024).
Gong, S., Wang, S. & Shao, M. NADPH oxidase 4: a potential therapeutic target of malignancy. Front. Cell. Dev. Biol. 10, 884412 (2022).
Maranchie, J. K. & Zhan, Y. Nox4 is critical for hypoxia-inducible factor 2-alpha transcriptional activity in von Hippel-Lindau-deficient renal cell carcinoma. Cancer Res. 65 (20), 9190–9193 (2005).
Kaushik, D. et al. Nuclear NADPH oxidase-4 associated with disease progression in renal cell carcinoma. Transl Res. 223, 1–14 (2020).
Shanmugasundaram, K. et al. NOX4 functions as a mitochondrial energetic sensor coupling cancer metabolic reprogramming to drug resistance. Nat. Commun. 8 (1), 997 (2017).
Qureshi, A. S. & Ali, S. Review Warburg effect and renal cancer caused by errs in fumarate hydratase encoding gene. Pak J. Pharm. Sci. 32 (2), 743–749 (2019).
Chen, Y. H. et al. Nox4 overexpression as a poor prognostic factor in patients with oral tongue squamous cell carcinoma receiving surgical resection. J. Clin. Med. 7, 12 (2018).
Sancho, P. et al. The Inhibition of the epidermal growth factor (EGF) pathway enhances TGF-beta-induced apoptosis in rat hepatoma cells through inducing oxidative stress coincident with a change in the expression pattern of the NADPH oxidases (NOX) isoforms. Biochim. Biophys. Acta. 1793 (2), 253–263 (2009).
Sobhakumari, A. et al. NOX4 mediates cytoprotective autophagy induced by the EGFR inhibitor erlotinib in head and neck cancer cells. Toxicol. Appl. Pharmacol. 272 (3), 736–745 (2013).
McGranahan, N. & Swanton, C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell 168 (4), 613–628 (2017).
Gajewski, T. F., Schreiber, H. & Fu, Y. X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 14 (10), 1014–1022 (2013).
Mao, X. et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. Cancer. 20 (1), 131 (2021).
Kim, J. Y. et al. NADPH oxidase 4 deficiency promotes hepatocellular carcinoma arising from hepatic fibrosis by inducing M2-macrophages in the tumor microenvironment. Sci. Rep. 14 (1), 22358 (2024).
Su, X. et al. NOX4-Derived ROS mediates TGF-β1-Induced metabolic reprogramming during Epithelial-Mesenchymal transition through the PI3K/AKT/HIF-1α pathway in glioblastoma. Oxid. Med. Cell. Longev. 2021, 5549047 (2021).
Singh, A. K. & McGuirk, J. P. CAR T cells: continuation in a revolution of immunotherapy. Lancet Oncol. 21 (3), e168–e78 (2020).
Darvin, P., Toor, S. M., Sasidharan Nair, V. & Elkord, E. Immune checkpoint inhibitors: recent progress and potential biomarkers. Exp. Mol. Med. 50 (12), 1–11 (2018).
Liu, W. J. et al. NOX4 signaling mediates Cancer development and therapeutic resistance through HER3 in ovarian Cancer cells. Cells 10, 7 (2021).
Kang, X. et al. Nox2 and Nox4 regulate self-renewal of murine induced-pluripotent stem cells. IUBMB Life. 68 (12), 963–970 (2016).
Cho, S. Y. et al. Expression of NOX family genes and their clinical significance in colorectal Cancer. Dig. Dis. Sci. 63 (9), 2332–2340 (2018).
Muthyalaiah, Y. S., Jonnalagadda, B., John, C. M. & Arockiasamy, S. Impact of advanced glycation end products (AGEs) and its receptor (RAGE) on cancer metabolic signaling pathways and its progression. Glycoconj. J. 38 (6), 717–734 (2021).
Manohar, S. M. At the crossroads of TNF α signaling and Cancer. Curr. Mol. Pharmacol. 17 (1), e060923220758 (2024).
Nakamura, H. & Takada, K. Reactive oxygen species in cancer: current findings and future directions. Cancer Sci. 112 (10), 3945–3952 (2021).
Fiorilla, I. et al. Chronic inflammation, oxidative stress and metabolic plasticity: three players driving the Pro-Tumorigenic microenvironment in malignant mesothelioma. Cells 12, 16 (2023).
Szanto, I. NADPH oxidase 4 (NOX4) in cancer: linking redox signals to oncogenic metabolic adaptation. Int. J. Mol. Sci. 23, 5 (2022).
Crosas-Molist, E. et al. The NADPH oxidase NOX4 represses epithelial to amoeboid transition and efficient tumour dissemination. Oncogene 36 (21), 3002–3014 (2017).
Giallongo, C. et al. TLR4 signaling drives mesenchymal stromal cells commitment to promote tumor microenvironment transformation in multiple myeloma. Cell. Death Dis. 10 (10), 704 (2019).
Acknowledgements
We acknowledge the contributions from UCSC, TCGA, GTEx, GEPIA, TIMER, TISCH, cBioPortal for Cancer Genomics, GDSC, PDB, String and UALCAN databases.
Funding
The present study was supported by the National Natural Science Foundation of China (grant no. 82104629), the Natural Science Foundation of Guangdong Province (grant nos. 2021A1515010673 and 2023A1515011078), the Science and Technology Program of Yangjiang (grant nos. SF2021049, SF2022001, SF2023026 and SF2023027) and the Scientific Research Fund of Yangjiang People’s Hospital (grant nos. 2021003, G2021002 and G2021004).
Author information
Authors and Affiliations
Contributions
YS Liu, XS Zhao and XF Jiang mainly participated in literature search, study design, writing and critical revision,prepared Figs. 1, 2, 3, 4, 5, 6, 7, 8 and 9; Table 1. CY Wu and Q Wang mainly participated in data collection, data analysis and data interpretation, prepared Figs. 10, 11, 12 and 13. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Yangjiang People’s Hospital (20210021).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, Y., Wu, C., Wang, Q. et al. NOX4 as an emerging prognostic and immunological biomarker across pan-cancer analyses. Sci Rep 15, 18812 (2025). https://doi.org/10.1038/s41598-025-03499-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03499-2