Abstract
MicroRNAs (miRNAs) have emerged as key regulators in the pathogenesis of amyotrophic lateral sclerosis (ALS). Despite growing evidence that miRNAs exhibit altered expression profiles in ALS, their utility as a biomarker remains limited. To address this, we conducted a meta-analysis using the Robust Rank Aggregation package, incorporating 20 differential miRNA profiling studies. Among these, miR-146a-5p emerged as the most dysregulated and significant miRNA in ALS (P = 0.0000142, P-adj = 0.0096), particularly in extracellular vesicle-derived studies. To evaluate its diagnostic accuracy, we validated miR-146a-5p expression in serum-derived exosomes and observed a significant increase in sporadic ALS (sALS) patients (n = 22) as compared to healthy controls (n = 18). Moreover, higher levels of miR-146a-5p were strongly associated with longer survival (≥ 5 years) (P = 0.0135), with a positive correlation between miR-146a-5p expression and survival duration (P = 0.0313) in sALS patients. Further, gene set enrichment analysis of miR-146a-5p target genes highlighted critical involvement of the Immune system and NF-kappa B signaling pathways in ALS pathophysiology. These findings highlight miR-146a-5p as a potential biomarker for ALS.
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating/debilitating motor neuron disease (MND) that appears as a combination of upper motor neuron (UMN) and lower motor neuron (LMN) dysfunction, primarily affecting the bulbar, cervical, and/or lumbar regions followed by respiratory involvement1. Muscle atrophy and weakness are the hallmark symptoms of the disease. ALS has a prevalence of approximately 4 per 100,000 population and an incidence of 1 per 100,000 person2,3. Majority of the individuals with ALS have a survival duration of 2–5 years, while only a few live more than 10 years after diagnosis4,5,6. Current diagnosis of ALS is based on revised El Escorial criteria, clinical assessment, electrophysiological examination or genetic screening. However, there are no reliable clinical biomarkers for understanding the complexity of symptoms and variability in disease progression. Consequently, there is a critical need for robust biomarkers for early identification and enable timely intervention.
MicroRNAs (miRNAs), a class of short (~ 22 nucleotides) non-coding RNA molecules, have emerged as a promising biomarker for several diseases, including neurodegenerative disorders7. miRNAs regulate gene expression by binding to the 3’ untranslated regions (UTR) of mRNA, resulting in mRNA degradation and translational repression. Each miRNA can regulate hundreds of mRNAs, broadly influencing diverse cellular functions, including intercellular signaling8. One of the most promising roles of miRNAs lies in their potential as biomarkers. In diseased cells, miRNAs are often dysregulated, making them valuable indicators of various pathological state9. Among these, circulating miRNAs are particularly appealing as biomarkers as they can be detected in biological fluids such as plasma, serum, cerebrospinal fluid, and saliva10,11. Their remarkable stability is attributed to their encapsulation within extracellular vesicles (EVs), predominantly exosomes, which shield them from degradation by endogenous ribonucleases12. Exosomes, formed during endocytosis and multivesicular body (MVB) maturation, not only protect miRNAs, but also facilitate their transport between cells, playing a vital role in cell–cell communication and disease progression13.
In patients with ALS, miR-206 has emerged as a promising biomarker candidate as it is closely linked to muscle damage and plays a role in ALS-related neurodegeneration and impaired neurogenesis through the down-regulation of brain-derived neurotrophic factor (BDNF)14. However, despite its potential, miR-206 has not been integrated into clinical diagnostics, leaving a significant gap in using miRNA-based biomarkers for early detection and disease monitoring. Two common approaches are used to study miRNAs as biomarkers; (1) profiling hundreds of miRNAs using high-throughput techniques such as microarray or next-generation sequencing (NGS) followed by qRT-PCR validation, or (2) focusing on a few known miRNAs for targeted analysis with qRT-PCR for higher sensitivity15. In recent years, numerous researchers have identified differentially expressed miRNAs in ALS patients. However, the overlap between different studies is relatively small. This inconsistency arises from variations in experimental techniques, types of biological samples, and sample size. To overcome the aforementioned limitations in conducting a meta-analysis, we utilized robust rank aggregation (RRA) method16. Through the analysis of miRNA expression profiling studies, we identified hsa-miR-146a-5p as a key marker. To validate this finding, we examined its expression in an Indian subset of sporadic ALS (sALS) patients, contributing to the growing evidence of its biological relevance.
Materials and methods
Search strategy
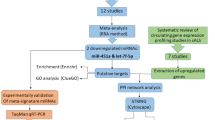
We executed a systematic search across multiple databases including PubMed, Science Direct and Cochrane Library to identify miRNA expression profiling studies related to ALS. Additionally, articles were searched manually from Google Scholar to ensure comprehensiveness of our search. The Table 1 lists the keywords used for the selection. Last search was performed in June, 2024, following Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines 202017.
Study selection
To access potentially relevant studies, we used Rayyan (https://www.rayyan.ai/), a free AI-powered web tool for systematic literature review. After removing duplicates both automatically and manually, two investigators (AR and SK) independently performed the article selection using Rayyan. Studies were included in the meta-analysis if they satisfied the following criteria: (i) miRNA expression profiling studies in ALS patients; (ii) original human case–control studies that compared miRNA expression according to respective cut-off criteria such as fold change or P-values; (iii) used technologies such as microarray, next generation sequencing (NGS) or qRT-PCR for reporting the miRNA expression. Following studies were removed from our meta-analysis: (i) Studies on cell lines or animal models; (ii) review of literature; (iii) studies with preselected and candidate miRNAs.
Data extraction and risk of bias assessment
We systematically collected and documented relevant information from the full text and supplementary data of each study, including (1) first author, (2) year of publication, (3) country of study, (4) sample size, (5) specimen type, (6) miRNA expression assay methods, (7) total number of up-regulated and down-regulated miRNAs, and (8) criteria used to determine dysregulated miRNAs. Firstly, an appropriately formatted analysis input file was created in MS Excel. The file consisted of ranked lists of miRNAs organized into columns, with the most dysregulated miRNAs positioned at the top based on their fold change or P-value as reported in the original studies. Each column had corresponding study names or identifiers as headings. To mitigate heterogeneity in miRNA expression among different sample types, we carried out a refined analysis, including studies that specifically focused on the differential expression profiling of miRNAs in extracellular vesicles (EVs). A similar input file was generated for these studies. To ensure consistency of the data, standard names of all miRNA were retrieved using miRBase version 22.1 (www.mirbase.org).
Quality assessment was conducted according to the Minimum Information About a Microarray Experiment (MIAME) guideline 2.018, Minimum Information for Publication of Quantitative Real-time PCR Experiments (MIQE) guideline19 and Minimum Information About a high-throughput SEQuencing Experiment (MINSEQE) guideline (https://www.fged.org/projects/minseqe) for evaluating microarray, PCR, and RNA-Seq studies, respectively.
Meta-analysis
Meta analysis was conducted using Robust Rank Aggregation method (RRA), available as a free package in R software (http://cran.R-project.org/). The RRA method assigns a P-value as a significance score to each miRNA in the ranked list assigning a new ranking to these miRNAs. This score indicates the extent to which the miRNA is ranked more favourably compared to a null model that assumes a random ordering. To reduce the risk of false positives, a Bonferroni correction was applied to the P-values. The miRNAs having adjusted P-value less than 0.01 were considered significant16.
Identification of target genes of top-ranked miRNA
The target genes of differentially expressed miRNAs were predicted using a MultiMiR package available on R software (http://multimir.org) which serves as an extensive repository of 14 databases encompassing both predicted and experimentally validated miRNAs-target genes interactions, along with their connections to various diseases and drugs. For predicted miRNA-target gene interaction, multiMir uses 8 external databases which includes DIANA-microT, ElMMo, MicroCosm, miRanda, miRDB, PITA, and Targetscan with the criterion of primary score listed in the top 35%. Furthermore, this package retrieves validated targets from 3 databases; miRTarBase, Tar Base, and miRecords. Only genes predicted by at least three of the databases and validated by at least two of the databases were considered for further study. For each miRNA, gene targets predicted by at least three of the eight databases were assigned a value of “1,” while those predicted by two or fewer were assigned a value of “0”. For validated gene targets, those appearing in at least two databases were assigned a value of “1”, while those appearing in two or fewer databases were assigned a value of “0”.
Enrichment analysis of top-ranked miRNA target genes
We conducted gene set enrichment analysis (GSEA) using an online web-based interface, Enrichr (https://maayanlab.cloud/Enrichr/) to elucidate the pathways and biological processes associated with the predicted and validated gene targets for top-ranked miRNA. Pathway analysis was done using Enrichr databases such as Reactome 2022, BioPlanet 2019, KEGG 2021 Human20, GO Biological Process 2023, GO Cellular Component 2023, GO Molecular Function 202321. The Enrichr tool employs Fisher’s exact test to calculate P-values and applies the Benjamini–Hochberg method for the adjustment of P-values, correcting for multiple hypothesis testing. Pathways and biological processes with adjusted P-value less than 0.05 were considered significantly enriched.
Validation of top-ranked miRNA in serum-derived exosomes of ALS patients
Serum samples were obtained from 22 sporadic ALS (sALS) patients and 18 age- and sex-matched healthy controls (HCs). Assessment included (1) detailed history and clinical neurological examination; (2) motor nerve conduction of median and ulnar nerves in both upper limbs and peroneal and posterior tibial nerves in both lower limbs; (3) sensory nerve conduction of median, ulnar, and sural nerves; (4) electromyography of distal and proximal muscles of all four limbs, bulbar and paraspinal muscles; and (5) magnetic resonance imaging of brain and spinal cord to exclude other neurological disorders. Based on the El Escorial-revised criteria22, patients diagnosed as ‘Clinically Definite ALS’ and ‘Clinically Probable ALS’ were included. ‘Clinically probable ALS-Laboratory supported’, ‘Clinically possible ALS’, and ‘Clinically Suspected ALS’ were excluded. Written informed consent was obtained from each participant. The study was approved by the Institutional Ethics Committee of Sir Ganga Ram Hospital (EC/05/20/1714) and was conducted in accordance with the Declaration of Helsinki.
Exosomal miRNAs were extracted from 1 ml of serum using exoRNeasy Midi kit (77144, Qiagen) according to the manufacturer’s instructions. 100 ng of isolated miRNAs were reverse-transcribed using miRCURY LNA RT Kit (339340, Qiagen). 0.5 µL of UniSp6 spike-in was used as a spike-in reverse-transcription control. Quantitative real-time PCR was performed via AriaMx Real-Time PCR System (Agilent) using miRCURY LNA SYBR Green PCR Kit (339345, Qiagen). Briefly, a master mix containing the 2 × miRCURY SYBR Green and RNase-free water was prepared. The cDNA was diluted at a ratio of 1:10 of a total volume. Finally, 3 μl of cDNA was used for qRT-PCR using miRCURY miRNA-specific primers. Data was analysed using the 2-ΔCt method and miR-103a-3p was used as a normalization control23. The sequence for miR-146a-5p and miR-103a-3p were 5’-UGAGAACUGAAUUCCAUGGGUU-3’ and 5’-AGCAGCAUUGUACAGGGCUAUGA-3’ respectively. Cyclic conditions used for qRT-PCR of miRNAs involved an initial denaturation step at 95 °C for 3 min, followed by 40 cycles of denaturation at 95 °C for 10 s and annealing at 60 °C for 30 s.
Statistical analysis
Two-tailed, unpaired, and non-parametric Mann–Whitney U-test was employed to compare the expression level of miRNAs in serum-derived exosomes of ALS patients with healthy controls. The relationship between miRNA expression and patient survival was analysed via Spearman’s rank correlation test. Further, to evaluate the diagnostic utility of differentially expressed miRNAs, Receiver Operating Characteristic (ROC) analysis was performed. Data was analysed with GraphPad Prism Version 10.4.0. P-values < 0.05 were considered statistically significant. Data in the graph are expressed as mean ± SEM.
Results
From an initial pool of 296 miRNA expression profiling studies identified through database search, 20 studies met the inclusion criteria and were selected for analysis. These studies were published between 2012 and 2023. Platforms like microarray, NGS, and PCR were used in the selected studies (Supplementary Table S1). The PRISMA flowchart for inclusion of studies in this meta-analysis is shown in Fig. 1. Of 20 studies, four studies assessed miRNAs from peripheral blood24,25,26,27, three from plasma28,29,30, three from serum31,32,33, two from muscle34,35, seven from EVs14,36,37,38,39,40,41, and one from CSF42. A total of 580 patients and 502 healthy controls were included in meta-analysis. After removing duplicates, 248 unique miRNAs were identified across the 20 datasets.
Risk of bias assessment
Using MIAME, MIQE and MINSEQE guidelines, it was identified that 85% of the included studies did not report raw data on hybridisation, which was classified as high risk. However, only 30% and 10% of studies did not provide sufficient information about annotation of array design and experiment design, respectively, which are prerequisite for replicability and reproducibility (Supplementary Fig. S1).
Differentially expressed miRNAs in ALS
In 20 miRNA expression profiling studies, 248 differentially expressed miRNAs were reported in ALS vs healthy control samples. Out of these, 41 miRNAs were observed in more than one independent study. After screening these miRNAs for directionality, 6 miRNAs were reported to be up-regulated and 6 miRNAs were down-regulated. Additionally, 29 miRNAs were reported to be dysregulated in both directions. A list of consistently and inconsistently dysregulated miRNAs is provided in Supplementary Table S2.
Top-ranked miRNA in ALS
In the RRA analysis of all studies, miR-146a-5p emerged as the top-ranked miRNA but it failed to achieve statistical significance after applying Bonferroni correction (P-adj = 1) (Supplementary Table S3). In addition to miR-146a-5p, three other miRNAs viz. hsa-miR-199a-5p, hsa-miR-10b-5p, and hsa-miR-151a-5p were identified with robust rank scores below 0.05. Given that most selected studies focused on EV-derived miRNAs (n = 7), we refined our analysis to these studies. The refined meta-analysis identified miR-146a-5p as the most significant miRNA, with a robust rank score of 1.42E−05 and an adjusted P-value of 0.0096 (Table 2). Our meta-analysis findings highlight the crucial involvement of miR-146-5p in sALS patients and support its biomarker potential.
Validation of miR-146a-5p in sALS patients
To validate the candidature of miR-146a-5p as a potential biomarker, we examined its expression levels in sALS patients (n = 22) and healthy controls (n = 18). Among 22 sALS patients, 16 patients were males and 6 patients were females with male to female ratio of 2.6:1. Mean age of the patients was 56.09 ± 12.34 years while mean age of disease onset was 59.05 ± 11.59 years. Sixteen patients presented with spinal-onset, while 6 patients exhibited bulbar-onset of disease. Median survival duration (95% CI) was 57 months (48.2–119.9). Demographic features of the patients are described in Supplementary Table S4.
We observed that the expression of miR-146a-5p was significantly up-regulated in sALS patients as compared to HCs (P = 0.0012, Fig. 2a). ROC analysis yielded an Area Under the Curve (AUC) of 0.7917, with a diagnostic sensitivity of 100% and a specificity of 50% indicating that miR-146a-5p could differentiate ALS patients from HCs (P = 0.0017, Fig. 2b). Interestingly, miR-146a-5p levels were significantly elevated in patients with a long survival duration (≥ 5 years) (P = 0.0135, Fig. 3a) and a significant positive correlation was found between miR-146a-5p expression and survival of ALS patients (r = 0.5106, P = 0.0152, simple linear regression: Y = 819.0*X—127.6, Fig. 3b). Further, miR-146a-5p was increased in females as compared to males and in patients presenting with spinal-onset compared to those with bulbar-onset, although these differences were not statistically significant (P = 0.0671 and 0.1824, Fig. 4a, b respectively). Additionally, miR-146a-5p levels were elevated in patients < 50 years of age of onset (P = 0.1491, Fig. 4c). A potentially negative correlation was identified between miR-146a-5p expression and the age of onset in ALS patients (r = -−0.3854, P = 0.0765, simple linear regression: Y = -82.01*X + 77.29, Fig. 4d).
Validation of miR-146a-5p expression in sALS patients and healthy controls. (a) Relative expression of miR-146a-5p in serum-derived exosomes from sALS patients (N = 22) and healthy controls (N = 18), analysed by qRT-PCR. Data was normalized to endogenous levels of miR-103a-3p and plotted as a relative expression [2^-ΔCt]. P-values were calculated using the Mann–Whitney U-test. (b) The Receiver-operating characteristic (ROC) curve analysis for serum-derived exosomal miR-146a-5p in discriminating ALS patients from controls, with Area Under the Curve (AUC) = 0.7917.
Analysis of miR-146a-5p expression in ALS patients based on survival duration. (a) In the sub-group analysis, miR-146a-5p was significantly up-regulated in patients with longer survival duration (N = 11) as compared to patients with short survival (N = 11). (b) The miR-146a-5p expression and survival duration in ALS patients showed positive-correlation (r = 0.5106, P = 0.0152, simple linear regression: Y = 819.0*X—127.6).
Analysis of miR-146a-5p expression in ALS patients based on demographic and clinical features. (a) Higher miR-146a-5p expression in females (N = 6) compared to males (N = 16) (P = 0.0671). (b) Increased expression in patients with limb-onset (N = 16) compared to bulbar-onset (N = 6) (P = 0.1824). (c) Higher expression in patients aged < 50 years (N = 9) compared to those aged > 50 years (N = 13) (P = 0.1491). (d) Potential negative correlation between miR-146a-5p expression and age of onset (r = -0.3854, P = 0.0765), modeled by simple linear regression (Y = −82.01*X + 77.29). P-values < 0.05 were considered significant.
Target prediction and pathway enrichment analysis of miR-146a-5p
The identification of potential target genes of miR-146a-5p was performed using the multimiR R package, which predicted 5370 targets. Among these, 1055 genes identified by at least three databases were considered for further analysis. These targets were derived from: DIANA-TarBase (1405), ElMMo (1905), miRcoSM (240), miRanda (1140), miRDB (160), PITA (3117), and TargetScan (1437). Pathway enrichment analysis revealed significant involvement in the immune system, actin filament organization, autophagy, and axonogenesis. Other significant pathways include signaling by Toll-like receptor, NF-kappa B and neurotrophin, down-regulation of ERBB4, and vitamin D metabolism (Supplementary Table S5). In addition, 4704 validated target genes were identified. Among these, 38 were identified by Mirecords, 4578 by Tarbase, and 203 by MirTarbase. According to the criteria defined in the methodology, 108 genes were considered for pathway enrichment analysis. Again, the immune system and NF-kappa B signaling pathway emerged as the most significantly enriched biological pathways. Additional pathways associated with the putative targets of miR-146a-5p included the regulation of the apoptotic process, protein ubiquitination, DNA damage response, Senescence and autophagy, response to vitamin D and signaling by NGF, MAPK, TGF-beta, RAGE, Notch, PIP3-AKT, as well as the BDNF signaling pathway (Supplementary Table S6). The association between validated as well as predicted target genes and their respective biological processes, GO terms and KEGG pathways are depicted in the GO Chord plot (Fig. 5a, b).
Chord plot generated using gene library sets (Gene Ontology, BioPlanet, Reactome and Kyoto Encyclopedia of Genes and Genomes) from Enrichr21for (a) validated and (b) predicted target genes of miR-146a-5p. Pathways and terms with an adjusted P-value < 0.05 were considered significant.
Discussion
Early diagnosis of ALS is crucial for potential therapeutic intervention and clinical management of the disease; however, this still remains a challenge due to the complexity of symptoms, variability in progression, and lack of definitive clinical biomarkers. Recently, miRNAs have been shown to regulate neuronal survival, autophagy, mitochondrial dysfunction, synapse integrity, oxidative damage, and neuroinflammation43,44. There is an increasing body of evidence supporting the crucial roles of microRNAs in ALS and its progression45. In the present study, we conducted a meta-analysis and miR-146a-5p was identified as a significantly dysregulated miRNA in the circulating EVs.
As per this meta-analysis, inconsistent dysregulation of miR-146a-5p was reported across different studies in ALS, for example, Waller et al. (2018) found down-regulation of miR-146a-5p in serum exosomes whereas Banack et al. (2020, 2022) and Liu et al. (2023) observed up-regulation in plasma-derived EVs38,39,42,46. Apart from these, several independent studies which did not fulfill the inclusion criteria also highlight the importance of miR-146a-5p in ALS. For instance, elevated miR-146a-5p expression was observed in the spinal cord and muscle of sALS patients, with evidence of negatively regulating Neurofilament light chain (NfL) levels, a key gene for maintaining neuronal morphology in the spinal cord47,48. Furthermore, consistent up-regulation was observed in the sciatic nerve and spinal cord of symptomatic SOD1-G93A transgenic mice49,50 while its down-regulation was reported in cortical astrocytes51 and serum of transgenic mice model and ALS patients49.
In the current study, significant up-regulation of miR-146a-5p was observed in serum-derived exosomes of sALS patients. Interestingly, miR-146a-5p levels were associated with prolonged survival, suggesting its potential as a biomarker, particularly for identifying patients with slower disease progression. Similar to our findings, a recent study by Banack et al. (2024) reported significant up-regulation of miR-146a-5p in neural-enriched plasma EVs from ALS patients, in comparison to patients with Parkinson’s disease, Primary lateral sclerosis as well as healthy controls52. Variability in miR-146a-5p expression likely reflects genetic and phenotypic heterogeneity associated with ALS.
Hsa‐miR‐146a-5p is located at chromosome 5q33.3. Previously known as miR-146a, this miRNA plays a crucial role in regulating inflammation and immune responses. miR-146a-5p has been studied extensively for its role in various cancers and neurodegenerative disorders. Further, miR-146a-5p has been found to be enriched in microglia53, where it communicates with neurons via exosomal transfer, influencing neurogenesis and glia-neuron communication54. Giagnorio et al. (2023) further highlighted the multifaceted role of miR-146a-5p in maintaining axon architecture and function49.
Functional enrichment analysis of miR-146a-5p identified the ‘Immune system’ and ‘NF-κB signaling pathway’ as the most significantly enriched biological pathways. Activation of NF-κB pathway in ALS has been shown to contribute to neuroinflammation and neuronal degeneration. Persistent activation of this pathway results in the production of pro-inflammatory cytokines, such as TNF-α, IL-1β, and IL-6, in glial cells and motor neurons. This chronic pro-inflammatory state not only accelerates motor neuron death but also leads to sustained microglial and astrocyte activation, further exacerbating neuroinflammation and contributing to neurodegeneration55,56,57. Previously, activation of inflammation and immune response was observed in the peripheral blood of juvenile ALS patient58.
The miR-146a has also been shown to target SMAD4, which plays a crucial role in the TGF-β signaling pathway59. In ALS, elevated astrocytic TGF-β levels activate PI3K/AKT/mTOR pathway leading to impaired autophagy60 and decreased motor neuron survival61,62. Further, it has been suggested that TGF-β mediated microglial activation contributes to neuroinflammation, neuromuscular junction dismantling, and subsequently muscle atrophy63. Gene Ontology enrichment revealed that miR-146a-5p is also implicated in ‘Regulation of protein ubiquitination (GO:0031396)’ and ‘Regulation of apoptotic process (GO:0042981)’. Notably, miR-146a-5p targets SQSTM1/p62, a critical adaptor protein in autophagy and proteostasis. Battaglia et al. (2019) demonstrated that up-regulation of miR-146a-5p may contribute to neurodegeneration in Alzheimer’s disease (AD) by inhibition of p62 activity, impairing autophagic flux64. Further, overexpression of p62 has been shown to mislocalize nuclear TDP-43 into cytoplasmic aggregates and promote the accumulation of ubiquitin-positive inclusions ultimately exacerbating neuronal death65,66. Furthermore, increased p62 aggregates in the spinal cord of ALS patients have been associated with shorter survival, which highlights a significant negative relationship between p62 accumulation, neuronal loss, and disease duration67.
While miR-146a-5p is well-known for its role in regulating inflammation and immune responses, miR-146a-3p has also been shown to contribute to innate immune regulation68and modulate key immune signaling genes such as IRAK1 and TRAF669. In sALS patients, miR-146a-3p is significantly upregulated in the spinal cord and directly targets the 3′ UTR of NfL mRNA47. This passenger strand has been shown to be significantly down regulated in plasma of symptomatic fALS and asymptomatic TARDBP mutation carriers70. Even, Banack et al. (2020) reported mir-146a-3p to be non-significant in plasma derived EVs of ALS patients38. Since miR-146a-3p was not identified in our meta-analysis, it was not further analysed. However, there can be commonality between the pathways regulated by the two 5p/3p strands of miR-146a. Pathway enrichment analysis of the validated target genes of miR-146a-5p/3p revealed shared regulated pathways including immune system, signaling by NGF, TGF-beta, PIP3-AKT, NF-kappaB, FoxO and Neurotrophin which are implicated in ALS (Supplementary Fig. S2).
Apart from ALS, previous studies have also shown the dysregulation of miR-146a-5p in the peripheral blood of patients with Alzheimer’s disease (AD) and Parkinson’s disease (PD)71,72. Up-regulation of miR-146a-5p has been linked to increased oxidative stress and amyloid-beta (Aβ) deposition in AD mice71,73, contributing to neurodegeneration. Similarly, in a PD mice model, miR-146a-5p was shown to regulate the Parkin gene, leading to mitochondrial fragmentation74. Additionally, miR-146a-5p has been associated with apoptosis and inflammatory response in status epilepticus (SE) rats75. Further, miR-146a-5p has also been demonstrated to attenuate glia-mediated neuroinflammation and neuropathic pain via targeting the TRAF6-mediated JNK/CCL2 signaling pathway, both in vivo and in vitro76. These findings suggest that miR-146a-5p plays a significant role in various neurodegenerative diseases, primarily through its regulation of inflammation, oxidative stress, and autophagy.
Limitations
This meta-analysis aimed to elucidate the diagnostic efficacy of miR-146a-5p in ALS. While the findings provide valuable insights, several limitations must be addressed. The studies included in the meta-analysis varied significantly in terms of their geographic origin, patient demographics, sample sizes, RNA isolation methods, and miRNA profiling techniques. These differences introduce variability that may affect the comparability and robustness of the findings. Many studies did not report clinical characteristics such as age at onset, site of onset, presence of genetic mutations, or survival duration. This limitation restricts the ability to assess how miR-146a-5p expression correlates with disease characteristics. Further, the absence of data on miR-146a-5p levels across different disease stages limits its potential use for patient stratification. Identifying miRNAs linked to specific disease stages could significantly enhance their utility in tailoring treatments or stratifying patients in clinical trials. Although miR-146a-5p shows promise as a biomarker, its clinical utility remains uncertain due to the lack of large-scale, multi-center validation studies.
Conclusions
Our study highlights miR-146a-5p as a promising biomarker for ALS. By targeting key inflammatory and autophagy-related pathways, the miR-146a-5p plays a multifaceted role in ALS pathophysiology. We demonstrated that miR-146a-5p up-regulation is associated with prolonged survival, highlighting its potential relevance in disease progression. However, these findings warrant further investigation in larger cohorts to validate its clinical utility.
Data availability
The data that support the findings of this study are available on request from the corresponding author.
References
Brown, R. H. & Al-Chalabi, A. Amyotrophic lateral sclerosis. N. Engl. J. Med. 377, 162–172 (2017).
Gourie-Devi, M. Neuroepidemiologic study in semiurban and rural areas in South India: pattern of neurological disorders including motor neuron disease. Mot. Neuron Dis. 11–21 (1987).
Talbott, E. O., Malek, A. M. & Lacomis, D. Chapter 13 - The epidemiology of amyotrophic lateral sclerosis. in Handbook of Clinical Neurology (eds. Aminoff, M. J., Boller, F. & Swaab, D. F.) vol. 138 225–238 (Elsevier, 2016).
Testa, D., Lovati, R., Ferrarini, M., Salmoiraghi, F. & Filippini, G. Survival of 793 patients with amyotrophic lateral sclerosis diagnosed over a 28-year period. Amyotroph. Lateral Scler. Mot. Neuron Disord. Off. Publ. World Fed. Neurol. Res. Group Mot. Neuron Dis. 5, 208–212 (2004).
Nalini, A., Thennarasu, K., Gourie-Devi, M., Shenoy, S. & Kulshreshtha, D. Clinical characteristics and survival pattern of 1153 patients with amyotrophic lateral sclerosis: experience over 30 years from India. J. Neurol. Sci. 272, 60–70 (2008).
Pupillo, E., Messina, P., Logroscino, G., Beghi, E., & SLALOM Group. Long-term survival in amyotrophic lateral sclerosis: a population-based study. Ann. Neurol. 75, 287–297 (2014).
Kuo, M.-C., Liu, S.C.-H., Hsu, Y.-F. & Wu, R.-M. The role of noncoding RNAs in Parkinson’s disease: biomarkers and associations with pathogenic pathways. J. Biomed. Sci. 28, 78 (2021).
Chevillet, J. R., Lee, I., Briggs, H. A., He, Y. & Wang, K. Issues and prospects of microRNA-based biomarkers in blood and other body fluids. Molecules 19, 6080 (2014).
Huang, W. MicroRNAs: Biomarkers, Diagnostics, and Therapeutics. in Bioinformatics in MicroRNA Research (eds. Huang, J. et al.) 57–67 (Springer, New York, NY, 2017). https://doi.org/10.1007/978-1-4939-7046-9_4.
Mitchell, P. S. et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. 105, 10513–10518 (2008).
Gilad, S. et al. Serum microRNAs are promising novel biomarkers. PLoS ONE 3, e3148 (2008).
Valadi, H. et al. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 9, 654–659 (2007).
Zhang, J. et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinform. 13, 17–24 (2015).
Liu, H. et al. Systematic review and meta-analysis on microRNAs in amyotrophic lateral sclerosis. Brain Res. Bull. 194, 82–89 (2023).
Motameny, S., Wolters, S., Nürnberg, P. & Schumacher, B. Next generation sequencing of miRNAs-strategies. Resour. Methods. Genes 1, 70–84 (2010).
Kolde, R., Laur, S., Adler, P. & Vilo, J. Robust rank aggregation for gene list integration and meta-analysis. Bioinformatics 28, 573–580 (2012).
Page, M. J. et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372, n71 (2021).
Brazma, A. et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 29, 365–371 (2001).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Kanehisa, M., Furumichi, M., Sato, Y., Matsuura, Y. & Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 53, D672–D677 (2025).
Xie, Z. et al. Gene set knowledge discovery with enrichr. Curr. Protoc. 1, e90 (2021).
Brooks, B. R., Miller, R. G., Swash, M., Munsat, T. L., & World Federation of Neurology Research Group on Motor Neuron Diseases. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. Mot. Neuron Disord. Off. Publ. World Fed. Neurol. Res. Group Mot. Neuron Dis. 1, 293–299 (2000).
Hoye, M. L. et al. MicroRNA profiling reveals marker of motor neuron disease in ALS models. J. Neurosci. 37, 5574–5586 (2017).
De Felice, B. et al. Wide-ranging analysis of MicroRNA profiles in sporadic amyotrophic lateral sclerosis using next-generation sequencing. Front. Genet. 9, 310 (2018).
De Felice, B. et al. A miRNA signature in leukocytes from sporadic amyotrophic lateral sclerosis. Gene 508, 35–40 (2012).
Chen, Y. et al. Aberration of miRNAs expression in leukocytes from sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 9, 69 (2016).
Liguori, M. et al. Dysregulation of MicroRNAs and target genes networks in peripheral blood of patients with sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 11 (2018).
Takahashi, I. et al. Identification of plasma microRNAs as a biomarker of sporadic amyotrophic lateral sclerosis. Mol. Brain 8, 67 (2015).
Kmetzsch, V. et al. Plasma microRNA signature in presymptomatic and symptomatic subjects with C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 92, 485–493 (2021).
Gomes, B. C. et al. Differential expression of miRNAs in amyotrophic lateral sclerosis patients. Mol. Neurobiol. 60, 7104–7117 (2023).
Freischmidt, A. et al. Serum microRNAs in patients with genetic amyotrophic lateral sclerosis and pre-manifest mutation carriers. Brain J. Neurol. 137, 2938–2950 (2014).
Waller, R. et al. Serum miRNAs miR-206, 143–3p and 374b–5p as potential biomarkers for amyotrophic lateral sclerosis (ALS). Neurobiol. Aging 55, 123–131 (2017).
Dobrowolny, G. et al. A longitudinal study defined circulating microRNAs as reliable biomarkers for disease prognosis and progression in ALS human patients. Cell Death Discov. 7, 4 (2021).
de Andrade, H. M. T. et al. MicroRNAs-424 and 206 are potential prognostic markers in spinal onset amyotrophic lateral sclerosis. J. Neurol. Sci. 368, 19–24 (2016).
Aksu-Menges, E. et al. Two distinct skeletal muscle microRNA signatures revealing the complex mechanism of sporadic ALS. Acta Neurol. Belg. 122, 1499–1509 (2022).
Saucier, D. et al. Identification of a circulating miRNA signature in extracellular vesicles collected from amyotrophic lateral sclerosis patients. Brain Res. 1708, 100–108 (2019).
Katsu, M. et al. MicroRNA expression profiles of neuron-derived extracellular vesicles in plasma from patients with amyotrophic lateral sclerosis. Neurosci. Lett. 708, 134176 (2019).
Banack, S. A., Dunlop, R. A. & Cox, P. A. An miRNA fingerprint using neural-enriched extracellular vesicles from blood plasma: towards a biomarker for amyotrophic lateral sclerosis/motor neuron disease. Open Biol. 10, 200116 (2020).
Banack, S. A., Dunlop, R. A., Stommel, E. W., Mehta, P. & Cox, P. A. miRNA extracted from extracellular vesicles is a robust biomarker of amyotrophic lateral sclerosis. J. Neurol. Sci. 442, 120396 (2022).
Pregnolato, F. et al. Exosome microRNAs in amyotrophic lateral sclerosis: A pilot study. Biomolecules 11, 1220 (2021).
Lo, T. et al. Extracellular vesicles in serum and central nervous system tissues contain microRNA signatures in sporadic amyotrophic lateral sclerosis. Front. Mol. Neurosci. 14 (2021).
Waller, R. et al. Small RNA sequencing of sporadic amyotrophic lateral sclerosis cerebrospinal fluid reveals differentially expressed miRNAs related to neural and glial activity. Front. Neurosci. 11 (2018).
Amini, J., Bibak, B., Afshar, A. R. & Sahebkar, A. Evaluation role of miR-124 in neurodegenerative diseases: Literature review and in silico analysis. 2021.10.17.464692 Preprint at https://doi.org/10.1101/2021.10.17.464692 (2021).
Catanesi, M. et al. MicroRNAs dysregulation and mitochondrial dysfunction in neurodegenerative diseases. Int. J. Mol. Sci. 21, 5986 (2020).
Ricci, C., Marzocchi, C. & Battistini, S. MicroRNAs as biomarkers in amyotrophic lateral sclerosis. Cells 7, 219 (2018).
Liu, Y. et al. MicroRNA-23a-3p is upregulated in plasma exosomes of bulbar-onset ALS patients and targets ERBB4. Neuroscience 524, 65–78 (2023).
Campos-Melo, D., Droppelmann, C. A., He, Z., Volkening, K. & Strong, M. J. Altered microRNA expression profile in amyotrophic lateral sclerosis: a role in the regulation of NFL mRNA levels. Mol. Brain 6, 26 (2013).
Pegoraro, V., Merico, A. & Angelini, C. Micro-RNAs in ALS muscle: Differences in gender, age at onset and disease duration. J. Neurol. Sci. 380, 58–63 (2017).
Giagnorio, E. et al. MiR-146a in ALS: Contribution to early peripheral nerve degeneration and relevance as disease biomarker. Int. J. Mol. Sci. 24, 4610 (2023).
Cunha, C. et al. Downregulated glia interplay and increased miRNA-155 as promising markers to track ALS at an early stage. Mol. Neurobiol. https://doi.org/10.1007/s12035-017-0631-2 (2017).
Gomes, C. et al. Cortical neurotoxic astrocytes with early ALS pathology and miR-146a deficit replicate gliosis markers of symptomatic SOD1G93A mouse model. Mol. Neurobiol. 56, 2137–2158 (2019).
Banack, S. A. et al. A microRNA diagnostic biomarker for amyotrophic lateral sclerosis. Brain Commun. 6, fcae268 (2024).
Jovičić, A. et al. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J. Neurosci. 33, 5127–5137 (2013).
Fan, C. et al. Microglia secrete miR-146a-5p-containing exosomes to regulate neurogenesis in depression. Mol. Ther. 30, 1300–1314 (2022).
Källstig, E., McCabe, B. D. & Schneider, B. L. The links between ALS and NF-κB. Int. J. Mol. Sci. 22, 3875 (2021).
Frakes, A. E. et al. Microglia induce motor neuron death via the classical NF-κB pathway in amyotrophic lateral sclerosis. Neuron 81, 1009–1023 (2014).
Swarup, V. et al. Deregulation of TDP-43 in amyotrophic lateral sclerosis triggers nuclear factor κB–mediated pathogenic pathways. J. Exp. Med. 208, 2429–2447 (2011).
Verma, S. et al. Multiomics approach reveal novel insights in FUS driven juvenile amyotrophic lateral sclerosis: A family quartet analysis. Ann. Neurosci. 09727531231194399 (2023) https://doi.org/10.1177/09727531231194399.
Sun, Y. et al. miR-146a-5p acts as a negative regulator of TGF-β signaling in skeletal muscle after acute contusion. Acta Biochim. Biophys. Sin. 49, 628–634 (2017).
Tripathi, P. et al. Reactive astrocytes promote ALS-like degeneration and intracellular protein aggregation in human motor neurons by disrupting autophagy through TGF-β1. Stem Cell Rep. 9, 667–680 (2017).
Peters, S. et al. The TGF-β system as a potential pathogenic player in disease modulation of amyotrophic lateral sclerosis. Front. Neurol. 8 (2017).
Endo, F. et al. Astrocyte-derived TGF-β1 accelerates disease progression in ALS mice by interfering with the neuroprotective functions of microglia and T cells. Cell Rep. 11, 592–604 (2015).
Galbiati, M. et al. Multiple roles of transforming growth factor beta in amyotrophic lateral sclerosis. Int. J. Mol. Sci. 21, 4291 (2020).
Battaglia, C. et al. Candidate genes and MiRNAs linked to the inverse relationship between cancer and Alzheimer’s disease: Insights from data mining and enrichment analysis. Front. Genet. 10, 846 (2019).
Foster, A. D. et al. p62 overexpression induces TDP-43 cytoplasmic mislocalisation, aggregation and cleavage and neuronal death. Sci. Rep. 11, 11474 (2021).
Mizuno, Y. et al. Immunoreactivities of p62, an ubiqutin-binding protein, in the spinal anterior horn cells of patients with amyotrophic lateral sclerosis. J. Neurol. Sci. 249, 13–18 (2006).
Pinkerton, M. et al. Survival in sporadic ALS is associated with lower p62 burden in the spinal cord. J. Neuropathol. Exp. Neurol. 82, 769–773 (2023).
Bogunia-Kubik, K. et al. Significance of polymorphism and expression of miR-146a and NFkB1 genetic variants in patients with rheumatoid arthritis. Arch. Immunol. Ther. Exp. (Warsz.) 64, 131–136 (2016).
Gronau, L. et al. Dual role of microRNA-146a in experimental inflammation in human pulmonary epithelial and immune cells and expression in inflammatory lung diseases. Int. J. Mol. Sci. 25, 7686 (2024).
Ruffo, P., Catalano, S., La Bella, V. & Conforti, F. L. Deregulation of plasma microRNA expression in a TARDBP-ALS family. Biomolecules 13, 706 (2023).
Lei, B. et al. NF-κB-induced upregulation of miR-146a-5p promoted hippocampal neuronal oxidative stress and pyroptosis via TIGAR in a model of Alzheimer’s disease. Front. Cell. Neurosci. 15 (2021).
Shu, Y., Qian, J. & Wang, C. Aberrant expression of microRNA-132-3p and microRNA-146a-5p in Parkinson’s disease patients. Open Life Sci. 15, 647–653 (2020).
Zhan-qiang, H. et al. miR-146a aggravates cognitive impairment and Alzheimer disease-like pathology by triggering oxidative stress through MAPK signaling. Neurologia 38, 486–494 (2023).
Jauhari, A., Singh, T., Mishra, S., Shankar, J. & Yadav, S. Coordinated action of miR-146a and Parkin gene regulate rotenone-induced neurodegeneration. Toxicol. Sci. Off. J. Soc. Toxicol. 176, 433–445 (2020).
Luo, Q. et al. Involvement of microRNA-146a in the inflammatory response of S tatus epilepticus rats. CNS Neurol. Disord. Drug Targets 16, 686–693 (2017).
Lu, Y., Cao, D.-L., Jiang, B.-C., Yang, T. & Gao, Y.-J. MicroRNA-146a-5p attenuates neuropathic pain via suppressing TRAF6 signaling in the spinal cord. Brain. Behav. Immun. 49, 119–129 (2015).
Acknowledgements
This work was supported by the Research Development Program of Sir Ganga Ram Hospital. We extend our sincere gratitude to all the patients and their families for their contributions to this study. We also thank Mrs. Parul Chugh from the Department of Biotechnology and Research, Sir Ganga Ram Hospital for her invaluable support in statistical analysis.
Funding
AR was supported by JRF scholarship from Indian Council of Medical Research (2020–2641/CMB/ADHOC-BMS). SK was supported by SRF scholarship from the Indian Council of Medical Research (3/1/2/151/Neuro/2021-NCD-I).
Author information
Authors and Affiliations
Contributions
A.R. and S.K. conceptualized and designed the study, conducted the literature review, performed data collection and experiments, analyzed and interpreted the data, and drafted the manuscript, S.V. provided critical feedback on the study design and methodology, and performed comprehensive editing and revisions, U.D. and N.K.G. reviewed the manuscript critically for intellectual content, provided feedback, and contributed to final revisions and editing, M.G.D. Conducted clinical examinations, diagnosis and interpretation of clinical data, and manuscript writing and editing, V.T. Conceptualized and designed the study, edited and revised the final manuscript . All authors have reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent to participate
Written informed consent was obtained from all participants. The study was conducted according to the principles of the Helsinki Declaration of 1964, as revised in 2013.
Ethics declarations
The study was approved by the Institutional Ethics Committee of Sir Ganga Ram Hospital, New Delhi (EC/05/20/1714).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rawat, A., Khurana, S., Verma, S. et al. Insights from meta-analysis and experimental validation identify exosomal miR-146a-5p as a potential biomarker for sporadic amyotrophic lateral sclerosis. Sci Rep 15, 33846 (2025). https://doi.org/10.1038/s41598-025-04322-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-04322-8
Keywords
This article is cited by
-
miRNA Biomarkers Diagnose Amyotrophic Lateral Sclerosis in Circulating Blood
Molecular Neurobiology (2026)