Abstract
Endophytic actinomycetes of medicinal plants have recently been in focus for developing novel antimicrobial compounds to combat multidrug-resistant pathogens. In this study, we isolated and characterised endophytic actinomycetes of Panax sokpayensis rhizome traditionally used as medicine in Sikkim-Himalayan region and assessed their antimicrobial activity against multidrug-resistant (MDR) clinical isolates of Staphylococcus aureus. Saccharopolyspora dominated as the endophytic actinomycetes of P. sokpayensis rhizome. However, a novel actinomycete strain PSRA5T belongs to the genus Streptomyces, with the highest genome sequence similarity of 91.54% with its closest relative Streptomyces niveus NCIMB 11891 has shown an effective inhibition of six clinical isolates of MDR S. aureus during disc diffusion assay. Further comparative analysis of cellular fatty acids composition and phenotypic and biochemical characteristics of strain PSRA5T with its phylogenetically closely related strain of S. niveus, classified as representing a novel species of the genus Streptomyces, for which the name Streptomyces panacea sp. nov. is proposed here with type strain PSRA5T (= MCC5238T). The minimum inhibition concentration of ethyl acetate crude extract of PSRA5T culture supernatant against MDR S. aureus isolates was 5.5 to 13.5 µg/mL. Further correlation between biosynthetic gene clusters identified by genome search with LC-MS analysis-based chemical profiling of PSRA5T culture extract and antibacterial activity of the representative compounds detected several compounds of aminoglycosides and polyketides with antimicrobial activity against MDR S. aureus isolates. Scanning electron microscope imaging viewed an interesting change in the S. aureus cell morphology with flocs of thread-like biofilm formation in response to the PSRA5T culture crude extract treatment. We conclude that the antimicrobial compounds produced by S. panacea PSRA5T can be further purified, characterised, and evaluated for their mode of action and efficacy against MDR S. aureus for developing novel drugs.
Similar content being viewed by others
Introduction
The emergence of multidrug resistance in human pathogens is a global challenge1,2. Staphylococcus aureus is one of the most notorious multidrug-resistant (MDR) opportunistic bacterial pathogens, which is a major cause of nosocomial infection3. World Health Organisation (WHO) report that methicillin-resistant Staphylococcus aureus (MRSA) is the cause of 20% of nosocomial infections in most countries and even up to 80% in some countries4. Some MRSA strains have gradually increased their resistance to vancomycin, macrolides, trimethoprim-sulfamethoxazole (TMP-SMZ), streptogramins and lincosamides, thus compelling us to find new antibiotics against multidrug-resistant S. aureus5.
Actinomycetes, namely Streptomyces are the prolific producer of antibiotics and accounts for almost 75% of the world’s antibiotics6. Several compounds produced by Streptomyces, like polyketomycin, citreamicin, noseheptide, and marinopyrrole exhibit anti-MRSA activity7,8,9. Compounds like waldiomycin and walamycin that inhibit quorum sensing and streptogramin-B that disrupts biofilm formation are also produced by Streptomyces10. Most of the antibiotics mentioned above were discovered in the 1980s from Streptomyces isolated from soil and marine habitats. Some strains of MRSA have already developed resistance against the majority of these antibiotics5. The exploration of endophytes has recently been accelerated due to their capabilities towards the production of plethora of bioactive metabolites and antimicrobial agents11. The isolation of the endophytes that produce bioactive compounds is of great attention in the development of new molecules to fight against many pathogens especially with the emergence of antibiotic multi-resistant pathogens. Keeping in view of the potential of actinomycetes, it is appropriate and necessary to screen Streptomyces from unexplored unique habitats like endophytes of medicinal plants for developing novel antibiotics against multidrug-resistant MRSA.
Endophytic actinomycetes from several medicinal plants show antimicrobial activity against many pathogens12. Alkaloids, sesquiterpenes, lactones, organic acids and flavonoids are the range of compounds that have been derived from endophytic Streptomyces with antimicrobial properties13. Several antibiotic compounds like munumbicins, rhodostreptomycins, and alchivemycin were derived from endophytic Streptomyces of medicinal plants14,15,16. However, a recent review reported only three endophytic Streptomyces with anti-MRSA activity5. Actinomycetes occupy 20–40% of the culturable fraction of endophytes present in the rhizome of Panax17while their antimicrobial potential has not been explored, particularly against pathogens with multidrug resistance18. This study aims to assess the antimicrobial potential of endophytic actinomycetes of Panax sokpayensis, an endemic medicinal plant of Sikkim-Himalayas, against multidrug-resistant Staphylococcus aureus (MDRSA) by in-vitro assays, chemical profiling, and genome mining.
Materials and methods
Sample collection
The rhizome of P. sokpayensis was collected from Jorbotay, Gyalshing district, Sikkim, India (Latitude: 27°16′ 54.84″ N; Longitude: 88°5′ 8.12″ E; Altitude: 2297 m). The collected sample was placed in a sterilized plastic bag and brought to the laboratory in a cool box, and endophyte isolation was carried out within 24 h of collection. The collection of sample was done as per the guideline of the Sikkim State Government after availing research permit {letter no. 78/GOS/FEWMD/BD-R-2015/CCF(T&HQ)35 dated May 15, 2017}from the office of the Chief Conservator of Forest (T&HQ) cum CWLW, Department of Forest, Environment & Wildlife Management, Government of Sikkim, Deorali, Gangtok–737102, Sikkim. The plant material was identified by Ms. Dukchen Bhutia, Botanical Assistant at the Botanical Survey of India (BSI), Sikkim Himalayan Regional Centre (SHRC), Gangtok-737103, Sikkim, Ministry of Environment, Forest and Climate Change, Government of India and authentification letter issued by Dr. Rajib Gogoi, Scientist E & Head BSI, SHRC (vide letter no. SHRC/5/40/2021-Tech-273). The voucher specimen of the plant material has been deposited at BSI, SHRC, Gangtok, Sikkim, India and the accorded accession number is IBSD-SC-M11.
Isolation of endophytic actinomycetes
With little modifications, the methodology described by Mazumder et al.11 was followed to isolate endophytic actinomycte from the rhizome of P. sokpayensis. The rhizome sample was washed thoroughly under tap water to remove the soil and dust particles which was followed by rinsing with sterile distilled water and cut into small fragments (3–4 cm) under sterile conditions. The rhizome fragment was then subjected to surface sterilization in aseptic condition under laminar airflow. Surface sterilization is a decisive step to eliminate any epiphytic microorganism which was performed by adopting the standard protocol; the rhizome fragment was initially immersed in 70% (v/v) ethanol for 5 min, followed by treatment with sodium hypochlorite (6%) for 5 min and then with 70% (v/v) ethanol for 30 s. Finally, the sterilized rhizome fragment was washed with sterile distilled water. The sterilized rhizome was allowed to dry aseptically and then the outer tissues were removed with the help of sterile scalpel. The sterilized rhizome fragment was then cut into small pieces (0.5 × 0.5 cm) and placed in Petri dishes with Streptomyces agar (SA) medium (HiMedia Laboratories Pvt. Ltd, Mumbai, India) amended with cycloheximide (50 µg/mL) and nalidixic acid (50 µg/mL). The inoculated petri dishes were incubated at 28 ± 1 °C for 30 days and examined daily for the growth of new colonies. The tip of the colony emerging out of the plated rhizome pieces were picked and grown on SA medium. The endophytic actinomycete isolate was purified by the streak technique on SA medium and the pure culture was preserved as glycerol stock in cryovials at −80 °C. As an additional test of surface sterilization, aliquots (100 µl) of the final rinse water of rhizome fragment were also plated on SA medium. No growth on SA plate inoculated with final rinse water ensured the efficacy of the surface sterilization process and absence of any epiphytic contamination.
DNA extraction and 16 S rRNA gene sequencing
The genomic DNA of the endophytic actinomycetes culture was extracted using the phenol-chloroform method19. The 16S rDNA gene was amplified by using primer pair ACT283 F (5′-GGGTAGCCGGCCUGAGAGGG-3′) and 1360R (5′-CTGATCTGCGATTACTAGCGACTCC-3′). The PCR amplification was performed in thermal cycler (Bio-Rad, USA) (94 °C, 5 min; 10 cycles of denaturation at 94 °C (1 min), 65 °C (30 s), 72 °C (2 min) and 72 °C (5 min) followed by 20 cycles of denaturation at 92 °C (30 s), annealing at 65 °C (30 s), extension at 72 °C (2.5 min) and final extension at 72 °C (5 min). The PCR product was purified using a gel extraction kit (Promega) and subjected to sequencing by the Sanger method (Eurofins Genomics India Pvt Ltd., Bangalore, India). Taxonomic positioning was done by NCBI Blast-based sequence similarity with the closest relative. The evolutionary history was inferred by using the Maximum Likelihood method based on the Kimura 2-parameter model20 and evolutionary analyses were conducted in MEGA621.
MDR isolates and test microorganisms
Six clinical isolates of MDR S. aureus (resistant to oxacillin, trimethoprim/sulfamethoxazole, ciprofloxacin, and levofloxacin) were obtained from the Department of Microbiology, Sikkim Manipal Institute of Medical Sciences (SMIMS), Gangtok, Sikkim. The drug resistance and sensitive details of the MDR S. aureus strains are listed in supplementary Table S1. In addition, Escherichia coli MTCC739, Candida albicans MTCC227, Listeria monocytogenes MTCC839, and Mycobacterium phlei MTCC1724 were also used as test microorganisms. All the bacterial cultures were cultivated and maintained in freshly prepared Nutrient agar (NA) medium (Himedia Laboratories Pvt. Ltd, Mumbai, India) at 37 ± 1 °C and C. albicans in Potato Dextrose agar medium (Himedia Laboratories Pvt. Ltd, Mumbai, India) at 37 ± 1 °C.
Extract Preparation and antimicrobial activity determination
The antimicrobial activity of the endophytic actinomycete isolate was evaluated by growing the strain in CSPYME broth (Casein 0.1%, Corn starch 1%, di-Potassium hydrogen phosphate 0.5%, Peptone 0.1%, Yeast Extract 0.1%, Malt Extract 1%) (Himedia Laboratories Pvt. Ltd, Mumbai, India). A uniform suspension of the isolate PSRA5T (0.2 O. D measure at 600 nm) in Tween 20 (0.05%) was prepared by taking inoculum from fresh grown culture on SA plate at 28 ± 1 °C. Five ml of this suspension was inoculated into 100 ml of CSPYME broth contained in 250 mL Erlenmeyer flask and incubated at 28 ± 1 °C in a shaking incubator upheld at 140 rpm for 25 days. The broth culture was subjected to centrifugation at 15,000 x g (Sorvall Biofuge Primo R) at 4 °C for 20 min. The cell free supernatant was filter sterilized using a 0.2 μm disc filter (Merck Millipore) and tested for extracellular antimicrobial activity by adopting standard agar well diffusion method6 against the MDR S. aureus strains and other test microorganisms. Mueller Hinton agar (MHA) plates were seeded uniformly using disposable sterile L-spreader with 1.5 mL of overnight culture suspensions of MDR S. aureus strains and other test microorganisms containing 1.5 × 108 colony forming units (CFU/spores)/mL6. Sabouraud dextrose agar (SDA) plate was use for C. albicans. Agar wells (6 × 4 mm) were punctured on the MHA and SDA plates by scooping out the medium with a sterile cork borer. 100 µl of the sterilized supernatant of strain PSRA5T was then added to the wells separately. The MHA plates were incubated at 37 ± 1 °C for 24 h while SDA plate was incubated at 37 ± 1 °C for 48 h. Antimicrobial activity was determined by measuring the diameter zone of inhibition (DZI) in mm of the tested microorganisms around the well6. Each experiment was carried out in three replicates. Uninoculated CSPYME broth added to the wells serve as control.
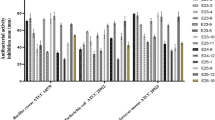
The crude bioactive secondary metabolite produced in the broth culture medium by the strain PSRA5T was extracted from the cell-free supernatant by shaking manually three times with equal volume of ethyl acetate (1:1) using a separating funnel. The solvent layer was collected in a round-bottomed flask and concentrated under vacuum using a rotary evaporator (Buchi R-114, Germany) maintained at 40 °C water bath. The antimicrobial activity of the crude metabolite was evaluated by the disc diffusion assay. Each strain of MDR S. aureus was inoculated on NA plates and incubated at 37 ± 1 °C for 24 h. Colonies from this fresh culture were used for the preparation of bacterial suspension. 1.5 mL of bacterial suspension (1.5 × 108 CFU/mL in sterile normal saline) was seeded on MHA plates by spreading uniformly using disposable sterile L-spreader as mentioned above. Sterile discs (6 mm) (Himedia Laboratories Pvt. Ltd, Mumbai, India) impregnated with 15 µg/mL of crude extract were then dispensed onto the surface of inoculated MHA plates and incubated at 37 ± 1 °C for 24 h. Disc saturated with ethyl acetate was used as control. Antimicrobial activity was determined by measuring the diameter zone of inhibition in mm around the disc. Each experiment was performed in triplicates. Minimum inhibitory concentration (MIC) was determined based on the methodology described by Singh et al.6. 1.5 mL of bacterial suspension (3 × 105 cfu/mL in sterile normal saline) of each test pathogen was added to 5 mL of MH broth in separate test tubes. Serial dilutions of the crude extract (ranging from 15 µg/mL to 1.0 µg/mL) were added in the respective tube at the same time. MIC of the crude extract against the test pathogen was evaluated after 48 h of incubation by removing 10 µl from each tube and spread inoculating on NA plates. Each experiment was performed in three replicates. Growth of the test pathogen were observed after 24 h of incubation at 37 ± 1 °C. MIC is considered as the lowest concentration needed to inhibit any visible growth. The representative compounds of Aminoglycosides (puromycin: TC198, spectinomycin, TC034, gentamicin, TC026, netilmicin, EM095, neomycin, TC301: Himedia Laboratories Pvt. Ltd, Mumbai, India), terpenoid-alkaloids (caffeine, xanthine, X0626: Sigma; hypoxanthine, PCT0206: Himedia Laboratories Pvt. Ltd, Mumbai, India) and polyketides (clindamycin, PCT1147, oxytetracycline, TC200, tetracycline, TC036: Himedia Laboratories Pvt. Ltd, Mumbai, India) with antimicrobial potential detected by the LC-MS in the crude extract of PSRA5T were analysed against six clinical isolates of MDR S. aureus by disc diffusion assay.
LC-MS profiling of the crude extract
The crude extract of strain PSRA5T was subjected to chemical profiling by LC-MS (C-CAMP facility, Bengaluru, India). The dried crude extract sample was reconstituted with 100% methanol. The reconstituted sample was centrifuged at 14,800 rpm for 10 min at 4 °C and passed through a 0.2 μm nylon membrane filter (Merck Millipore). The sample filtrate of 10 µl was injected (in duplicate) for analysis. Reserprine and Taurocholate-D8 were added as internal standards to check the method’s efficiency. The UHPLC (Ultimate 3000, Dionex) with C18 column (Phenomenex, Jupiter) and Q Exactive (Thermo Fisher Scientifc) were used for the mass spectroscopy. The filtered data of metabolites were annotated using mzCloud (Analysis on Compound Discoverer 3.2) and ChemSpider search from KEGG, HMDB, MassBank, E.coli Metabolome Database, and LipidMaps.
Genome sequencing and annotation
The genome sequence of PSRA5T was derived by using the Illumina HiSeq 2500 at the Genome Sequencing facility of the National Centre for Biological Sciences, Bengaluru, Karnataka, India. For the library preparation, Illumina Nextera XT DNA Library Preparation Kit (Catalog no-FC-131–1096) was used. The de novo assembly of raw sequence reads was done using Unicycler platform v0.4.8 and QUAST v5.0.2 was used to assess the quality on the PATRIC server22,23. Using a standalone Orthologous Average Nucleotide Identity Tool (OAT) with default parameters (https://www.ezbiocloud.net/tools/orthoani)24 pairwise Average Nucleotide Identity (ANI) values were calculated between the closest genomes of strain PSRA5T (S. camponoticapitis 2 H-TWYE14T and S. niveus NRRL 2466[T 25), and OrthoVenn web server was used to identify orthologous clusters and protein counts. The G + C content was calculated from the genomic sequence data. The antiSMASH server was used to identify, annotate and analyse potential secondary metabolite biosynthesis gene clusters in the genome of PSRA5[T 26.
Morphological, cultural, biochemical and physiological characterisation
The morphological characteristics of the strain PSRA5T were studied by adopting cover slip method wherein the culture was transferred to the base of cover slips buried in SA medium6 and incubated at 28 ± 1 °C for 14 days. The morphology of hyphae with spore chain was then observed under a phase contrast microscope (Nikon ECLIPSE Ci, Tokyo, Japan) and analysed as illustrated in Bergey’s manual27. Cultural characteristics were determined after 2 weeks of growth at 28 ± 1 °C using International Streptomyces Project 3 (ISP3), ISP5, ISP6, ISP7, and SA media28. The cellular fatty acids profiling of strain PSRA5T was carried out by FAME (Fatty Acid Methyl Ester) analysis29 based on the MIDI Microbial Identification System (MIS, Sherlock Version 6.1). The utilization of different carbohydrates (0.5%, w/v), hydrolysis of starch, esculin, urea, casein, reduction of nitrate, gelatin liquefaction, hydrogen sulfide (H2S) production and other biochemical tests were examined as described previously25. All the results were recorded after 2 weeks of incubation. Growth at different temperature ranges (15–45 ℃), pH ranges (pH 5–13), and NaCl tolerance (2–6%, w/v) were determined after incubation for 2 weeks in SA medium25.
Scanning electron microscopy (SEM)
The growth of MDR S. aureus culture in nutrient broth was stopped at 0.6 OD at 660 nm, and 100 µl was inoculated in a fresh nutrient broth. In one test tube, the crude extract of PSRA5T culture was reconstituted with ethyl acetate and filter sterilized, which served as the test. To the other, only ethyl acetate was added, which served as the control. The tubes were allowed to incubate at 37 ± 1 °C for 24 h. For fixing the sample for SEM, 0.5 ml of culture broth was centrifuged to a pellet in a sterile microcentrifuge tube, 250 µl of 2% glutaraldehyde was added to the cell pellet and kept at room temperature for 30 min. Further, the mixture was kept for 2 h at 4 °C. The mixture was then centrifuged, the supernatant was discarded, and one ml of phosphate buffer was then added and incubated at 4 °C for 15 min and then centrifuged. This process was repeated thrice for washing. To this, sequentially 30%, 50% and 70% ethanol was added to the pellet, centrifuged, and discarded. Finally, 100% ethanol was then added to the pellet and centrifuged and the resulting pellet was slowly suspended for some time so that no clumps were formed. Then on the stub, a circular coverslip was placed on the adhesive material (carbon tape) where a 20 µl drop of the suspended sample was taken over the cover slip. The sample was then prepared to dry by keeping it in the vacuum desiccator for 3–4 h. Once it was completely dry, the sample was sputter coated with gold for 30 mA and 82 s time duration. After that, scanning electron microscopy was performed in Zeiss-Merlin compact VP (Gemini), and the secondary and In-lens detector images were taken at electron energies between 4 keV and 5 keV.
Culture and sequence data deposition
The culture of strain PSRA5T was deposited to the Microbial Culture Collection (MCC), National Centre for Cell Science (NCCS), Pune, India, and the assigned accession number is MCC 5238T. The genome of strain PSRA5T was deposited to NCBI GenBank (BioProject PRJNA944806) and the assigned accession number is JARJHW000000000 having BioSample SAMN33762233.
Statistical analyses
Statistical analysis was done by calculating the means and standard deviations of the results obtained. Duncans multiple range test was performed to compare that the sample means were not significantly different from each other at a significant level of P > 0.00130.
Results
Endophytic actinomycetes of Panax sokpayensis showed antimicrobial activity against clinical isolates of MDR S. aureus
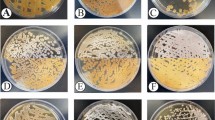
The endophytic actinomycetes of P. sokpayensis rhizome, dominated by Saccharopolyspora (Saccharopolyspora gloriosae and Saccharopolyspora dendranthemae) and Streptomyces spp. The taxonomic positions of 12 endophytic actinomycetes isolates of P. sokpayensis identified by 16S rDNA sequence similarity are shown in Fig. 1. Screening of these endophytic actinomycetes isolates against six clinical isolates of MDR S. aureus resistant to Oxacillin (1 1 µg), Trimethoprim/Sulfamethoxazole (1.25/23.75 µg), Ciprofloxacin (5 µg), and Levofloxacin (5 1 µg), showed an effective inhibition. The filter-sterilised ethyl acetate extract of strain PSRA5T culture supernatant showed an inhibition zone ranging from 19.1 to 22.5 mm against MDR S. aureus isolates during disc diffusion assay with no inhibition against other test pathogens (E. coli MTCC 739, L. monocytogenes MTCC 839, M. phlei MTCC 1724 and C. albicans MTCC 227). The colony and cell morphology of strain PSRA5T with potential antimicrobial activity against six clinical isolates of MDR S.aureus is shown in Fig. 2. The minimum inhibition concentration of ethyl acetate crude extract of strain PSRA5T culture supernatant was 5.5 to 13.5 µg/mL against MDR S. aureus isolates.
The phylogenetic tree based on 16S rDNA sequences constructed using the Maximum Likelihood method based on the Kimura 2-parameter model shows the taxonomic positions of 12 endophytic actinomycetes isolates of Panax sokpayensis. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
Description of Streptomyces panaceae sp. nov
The 16S rRNA gene sequence (1524 bases) similarity analysis of strain PSRA5T by NCBI-BLAST refseq_rna resulted in 98.07% similarity with Streptomyces niveus NRRL 2466T and 97.86% similarity with Streptomyces camponoticapitis 2 H-TWYE14T. The whole genome sequencing of PSRA5T by Illumina HiSeq 2500 and de novo assembly resulted in a genome size of 7,874,420 bp with a G + C content of 70.5%, 7478 coding sequences, 73 tRNAs and three rRNAs. The whole genome sequence-based ANI value comparison of strain PSRA5T with the closest species genomes showed 91.54% similarity with S. niveus NCIMB 11891 (NZCM002280) and 90.31% similarity with S. camponoticapitis DSM 100523T (NZ_BMMV01000083) (Fig. 3A), which were below the threshold ANI value of 95% that indicates that strain PSRA5T belong to a novel species. Moreover, comparative genomics by OrthoVenn showed a lack of 160 gene clusters (Fig. 3B) than the closest S. niveus NCIMB 11891 and the presence of 20 unique proteins in strain PSRA5T, indicating biologically much different that support its novelty. To further describe the novelty in the species, we analysed the composition of cellular fatty acids in the strain PSRA5T. The major cellular fatty acids of strain PSRA5T were iso-C16:0 (20.38%), anteisoA-C15:0 (18.58%), C16:00 (14.37%); anteiso-C17:0 (13.29%). anteisoA-C15:1 (8.68%), cis9-C16:1 (8.34%), anteisoC-C17:1 (2.13%), C15:00 (1.97%), iso-C15:0 (1.79%), isoH-C16:1 (1.39%), iso-C17:0 (1.20%), and cis9-C17:1 (1.12%). The comparative fatty acid profile of strain PSRA5T with the closest relative S. camponoticapitis strain 2 H-TWYE14T confirmed its novelty (Supplementary Table S2).
Besides the genotypic and cellular fatty acid profile, we could distinguish the strain PSRA5T from S. niveus JCM 4251T and S. camponoticapitis 2 H-TWYE14T based on several physiological, morphological and biochemical characteristics (Supplementary Table S3). Strain PSRA5T showed good growth on SA, ISP3, ISP5 and ISP7 while moderate growth on ISP6. The colour of the aerial mycelium was white, and greyish-brown soluble pigments were observed on the SA medium. The cellular morphology of strain PSRA5T showed spirals and a long chain, distinguishing it from S. niveus JCM 4251T and S. camponoticapitis 2 H-TWYE14T. The PSRA5T culture grew at a temperature range of 20–35 ℃ (optimum temperature being 28 ℃), at pH 5–11 (optimum pH being 7) and at NaCl concentrations of 2–5% (w/v). In contrast to S. camponoticapitis 2 H-TWYE14T, strain PSRA5T culture produced no pigments on ISP7 medium. The detail morphological, physiological, biochemical characteristics and carbohydrate utilization pattern of the strain PSRA5T is shown in supplementary Table S4. It is evident from the genotypic and phenotypic data that strain PSRA5T represents a novel species of the genus Streptomyces, for which the name Streptomyces panacea sp. nov. is proposed.
Streptomyces panacea (panəˈsiːə, named for its isolation from the rhizome of Panax with a potential remedy for antimicrobial resistance) is an aerobic, gram-positive bacteria that forms branched, non-fragmenting vegetative hyphae. Spores are non-motile with a smooth surface and are borne in chains on the aerial mycelium. Growth was observed on SA, ISP3, ISP5, ISP6, and ISP7 media. Greyish-brown soluble pigments were observed on SA medium not on ISP7. The strain tolerates up to 5% (w/v) NaCl and grows at temperatures between 20 and 35 ℃, with an optimum temperature of 28 ℃. Growth occurs at pH values between 5 and 11, the optimum being pH 7. Positive for production of urease, catalase, hydrolysis of starch, reduction of nitrate and utilisation of citrate. Among the tested carbohydtares, the strain PSRA5T utilizes galactose and glucose as carbon sources. The major fatty acids are iso-C16:00, anteiso-C15:00, C16:00, and anteiso-C17:00. The type strain is PSRA5T (= MCC5238T), isolated from the rhizome of Panax sokpayensis, collected from Jorbotay, Uttarey, Gyalshing district, Sikkim, India. The G + C content of the DNA of the type strain is 70.5%.
Genome analysis shows novelty in strain PSRA5T (A) UPGMA tree based on OrthoANI similarity values calculated from the phylogenetically closely related species of strain PSRA5T (B) Ven diagram shows the distribution of shared and unique gene clusters between phylogenetically closely related species of strain PSRA5T.
Genome mining identified gene clusters with antibacterial potential in Streptomyces panacea PSRA5T
The antiSMASH biosynthetic gene clusters (BGCs) similarity analysis identified 10 gene clusters in the genome of Streptomyces panacea PSRA5T with high similarity coding for potential antimicrobial compounds or their precursors of terpenes, T3PKS (Polyketide: Type III), non-ribosomal peptide synthetase (NRPS), lanthipeptide-class III and ectoine pathways (Table 1). We observed the presence of gene clusters for producing alkaloids-terpenoids like hopene, geosmin, raimonol and isorenieratene in the S. panacea PSRA5T. The presence of gene clusters for istamycin (an aminoglycoside antibiotic) with lesser gene cluster similarity was also observed in the antiSMASH results.
LC-MS chemical profiling of S. panacea PSRA5T culture extract detected antimicrobial metabolites
We correlated the potential antimicrobial compounds predicted (gene clusters and related secondary metabolite biosynthetic pathways) in the genome, by chemical profiling of S. panacea PSRA5T culture extract by mass spectrometry. LC-MS analysis of the ethyl acetate extract of S. panacea PSRA5T culture supernatant detected a total of 446 metabolites (after removal of the data with CV_QC > 20%) and assigned the identity of the possible compounds by tallying their mass on Compound Discoverer based on ChemSpider Database. Further literature search identified compounds with antimicrobial potential that mostly belong to the aminoglycosides, terpenoid-alkaloids, and polyketides types (Table 2). The detection of terpenoid-alkaloid isorenieratene (C40H48, m/z 528.3756), different types of polyketides like naringenin; and resorcinol, and ectoine (C6H10N2O2, m/z 142.0742) during the LC-MS analysis supported the genome mining based prediction results (Table 1). The presence of other antimicrobial compounds like benzoic and decanoic acid derivatives and linezolid and clindamycin in the crude extract of S.panacea PSRA5T supported the inhibition of clinical isolates of MDRSA. Further analysis of the antibacterial activity of the representative compounds detected by LC-MS in the culture extract of S. panacea PSRA5T resulted in the polyketides (clindamycin and tetracycline) and aminoglycosides (netilmicin and gentamicin) being the major antibacterial compounds against the MDR S.aureus clinical isolates. However, the terpenoid-alkaloids (caffeine and xanthine) did not show antibacterial activity against the MDR S. aureus isolates (Table 3).
SEM ultrastructure imaging visualised unusual cell deformation in the MDR S. aureus cells treated with S. panacea PSRA5T extract
The cells of MDR S. aureus isolates formed flocs of thread-like fibres and settled at the bottom of the test tube while treated with S. panacea PSRA5T culture crude extract (Fig. 4B), whereas the control ethyl acetate (used for the crude extract) treatment maintained the culture turbidity. Figure 4A shows the zone of inhibition exhibited by crude extract of S.panacea PSRA5T against MDR S. aureus P-435 in comparison to oxacillin which does not show zone of inhibition.We used SEM ultrastructure imaging to visualise the changes in the S. aureus cell morphology due to the antimicrobial activity of the S. panacea PSRA5T culture extract. Interestingly, SEM imaging visualised distinct changes in the morphology of S. panacea PSRA5T extract treated MDRSA strains, with unexplained clumping with thread-like structures similar to biofilm formation, whereas untreated control MDRSA cells maintain the spherical shape (Fig. 5). This observation confirmed a drastic physical change in the cell morphology of MDRSA cultures by the antimicrobial compound present in the crude extract of S. panacea PSRA5T.
Antimicrobial activity of S. panacea PSRA5T against MDR S. aureus strain. (A) Agar disc diffusion assay shows zone of inhibition exhibited by the crude extract of S. panacea PSRA5T against MDR S. aureus P-435, OX indicates oxacillin 1 µg/disc. (B) S. aureus P-435 culture treated with crude extract of S. panacea PSRA5T shows flocculation in broth culture, whereas untreated control maintains the turbidity.
Scanning electron micrographs show the bizarre changes in the cell morphology of MDR S. aureus P-435 after treatment with the crude extract of S. panacea PSRA5T. (A) Untreated control culture of MDR S. aureus P-435. (B) MDR S. aureus P-435 treated with ethyl acetate crude culture extract of S. panacea PSRA5T.
Discussion
Endophytic actinomycetes of medicinal plants have recently been a prime focus in recent days for developing novel antimicrobial compounds to combat the MDR S. aureus31. In our study, though Saccharopolyspora presents dominantly as endophytic actinomycetes in the rhizome of P. sokpayensis, a novel strain of S. panacea PSRA5T showed effective inhibition on the clinical isolates of MDR S. aureus. In addition, the 16S rRNA gene similarity threshold of Streptomyces sp. PSRA3 also indicates a possible novel species that supports rich biodiversity in the Himalayan region, particularly in the plant endophytes. The members of the genus Streptomyces are well known for antibiotic production and prominent resources for developing anti-MRSA drugs5. Uncovering the genomes of actinomycetes has promoted several researchers to explore biosynthetic gene clusters (BGC) and pathways responsible for antibiotic production32. S. panacea PSRA5T contains BGC that codes for antibiotic production pathways, including terpene, polyketide (T3PKS), non-ribosomal peptide synthetase (NRPS), lanthipeptide-class III and ectoine. In recent days, naturally occurring terpenoid-alkaloids of plant origin with antimicrobial activity have been proposed to combat MDR S. aureus33. Caffeine, evocarpine, trigonelline, arecoline, Norharman (β-carboline), xanthine, and cotinine are a few examples of plant terpenoid–alkaloids with antimicrobial effects against MDR S. aureus34,35,36,37,38,39,40. Our study detected terpene-type secondary metabolite biosynthetic pathways and gene clusters for hopene, geosmin, raimonol and isorenieratene production in S. panacea PSRA5T genome. The LC-MS analysis results of S. panacea PSRA5T culture extract supported the genome mining results with the detection of several terpenoid-alkaloid compounds with antimicrobial activity namely caffeine, evocarpine, trigonelline, arecoline, xanthine, norharman, xanthine, and cotinine34,35,36,37,38,39,40. However, the MDR S. aureus clinical isolates were resistant to the representative terpenoid-alkaloids (caffeine and xanthine) screened during this study.
The genus Streptomyces is one of the best producers of polyketides, which are the major source of antibiotics41,42. The presence of polyketides (Polyketide: Type III, T3PKS) type secondary metabolites biosynthetic pathways related gene clusters (naringenin and alkylrescorcinol) in S. panacea PSRA5T genome, along with LC-MS detection of several polyketides namely oxytetracycline, anhydrotetracycline, ieodomycin, anisomycin, lankacidin, ambruticin, and clindamycin with antimicrobial potential in the S. panacea PSRA5T culture extract (Table 2) supported its antibacterial activity42,43,44,45,46,47. However, the screening of representative polyketides detected by LC-MS against MDR S.aureus clinical isolates showed that isolates were sensitive to clindamycin and tetracycline, not oxytetracycline.
Aminoglycosides are the major group of antimicrobial compounds mostly used against infection by gram-negative pathogens48recently proposed as effective against MDR S. aureus. The major antimicrobial mechanism involved in the aminoglycoside is its ability to bind to the coding region of the 16S ribosomal RNA (rRNA) component of the 30S subunit of the bacterial ribosome. Also, aminoglycosides inhibit aminoglycoside-6′-N-acetyltransferase and 2″-O-phosphotransferase which are produced by MDR S. aureus49. We have observed the presence of gene clusters for istamycin production with lower similarity level in S. panacea PSRA5T genome and detected several aminoglycosoides (istamycin, spectinomycin, acmimycin, puromycin, bekanamycin, apramycin, dihydrodesoxystreptomycin, streptobiosamine, 2′′′-acetyl-6′′′-hydroxyneomycin, fortimicin, astromicin, gentamicin, n(2’)-acetyl gentamycin, netilmicin) during the LC-MS analysis of S. panacea PSRA5T culture extract, with antimicrobial activity reported by earlier researchers50,51,52,53,54,55,56,57,58,59,60,61. However, the screening of representative aminoglycosides detected by LC-MS against MDR S. aureus clinical isolates showed that the isolates were sensitive to netlilmicin, gentamicin, puromycin and spectinomycin, not to neomycin, supported the strong antibacterial activity visualised by S. panacea PSRA5T culture extract against clinical isolates of MDR S. aureus during this study. Our correlation studies relate the genome mining results with detected compounds during LC-MS analysis of S. panacea PSRA5T culture extract. Also, the results of activity screening of the detected representative pure compounds against the MDR S. aureus support the findings.
Non-ribosomal protein synthetases (NRPS) gene clusters have been associated with several drug discoveries62with an example of non-ribosomal peptide echinomycin with excellent anti-MRSA activity63. Detection of NRPS gene cluster with 100% similarity supported the antimicrobial activity visualised in this study.
Interestingly, the changes in the ultrastructure of MDR S. aureus in response to the crude extract of S. panacea PSRA5T revealed peculiar cell morphology changes. Prior research works on antimicrobial compounds have shown bacterial cell wall collapse, small surface depression or overall stop in growth rate64. In the case of daptomycin-treated bacterial cells, the authors report “bizarre” formations along the line of cell cleavage65. While in our study, the cells of clinical isolates of MDR S. aureus treated with ethyl acetate extract of S. panacea PSRA5T culture showed unusual cell deformations and progressed further to flocs formation with unexplained thread-like structural changes like a biofilm formation. This structure deformation could be due to the inherent resistant mechanism to neutralise the antimicrobial activity of S. panacea PSRA5T extract by biofilm formation66. The most known component of biofilm in S. aureus is the intercellular adhesion of bacterial cells by polysaccharide (poly-β(1–6) - N-acetylglucosamine (PNAG) formation67. This ability of cell deformation can be attributed to the variety of compounds (predicted by genome mining and detected by LC-MS analysis) with antimicrobial potential produced by S.panacea PSRA5T. More detailed studies are required to explain the mode of action, particularly bioactivity-guided purification and characterisation of molecules responsible for the bizarre effect visualised in MDR S. aureus culture by S.panacea PSRA5T culture extract.
Conclusion
To the best of our knowledge, this is the first report of endophytic actinomycetes of the medicinal plant, P. sokpyensis endemic to the Sikkim-Himalayan region, with the dominant presence of the Saccharopolyspora in its rhizome. We identified a novel endophytic actinomycete for which the name Streptomyces panacea sp. nov. is proposed here with type strain PSRA5T (= MCC5238), which shows the importance of studying the endophytes of medicinal plants in the Himalayan biodiversity hotspot. Due to its novelty in the genome, S. panacea PSRA5T could be a potential source of novel antimicrobial compounds. The genome mining, LC-MS analysis and antimicrobial activity of representative compounds showed the potential of S. panacea PSRA5T against MDR S. aureus. The MICs of the crude extract of S. panacea PSRA5T against the test pathogens indicates the effective activity that highlights its potential and could be a contender in the generation of new antimicrobial agents to fight against MDR S. aureus. The finding paves the way forward for further detailed investigation on purification, characteriastion of bioactive compound produce by S. panacea PSRA5T, mode of action, and efficacy in animal models for developing a novel drug. Such a detailed characterization and structure elucidation of the bioactive compound may lead to a new entity reported from this untapped location.
Data availability
The datasets generated and/or analysed during the current study are available in the respective database. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE [1] partner repository with the dataset identifier PXD053596. The Whole Genome (of Streptomyces sp. PSRA5) Shotgun project has been deposited at GenBank under the accession JARJHW000000000. The version described in this paper is version JARJHW010000000. The BioProject accession number is PRJNA944806.
References
Salvia, T. et al. Molecular characterization of extended-spectrum beta-lactamases and carbapenemases producing Enterobacteriaceae isolated from North Eastern region of India. J. Lab. Physicians. 16 (3), 245–252. https://doi.org/10.25259/jlp-2023-5-17-%281795%29 (2004).
Salvia, T., Dolma, K. G., Dhakal, O. P., Khandelwal, B. & Singh, L. S. Phenotypic detection of ESBL, ampc, MBL, and their co-occurrence among MDR Enterobacteriaceae isolates. J. Lab. Physicians. 14, 329–335. https://doi.org/10.1055/s-0042-1744239 (2022).
Livermore, D. M. Antibiotic resistance in Staphylococci. Int. J. Antimicrob. Agents. 16, 3–10. https://doi.org/10.1016/S0924-8579(00)00299-5 (2000).
Alvarez, A. et al. Methicillin-resistant Staphylococcus aureus in hospitals: latest trends and treatments based on bacteriophages. J. Clin. Microbiol. 57, 10–1128. https://doi.org/10.1128/JCM.01006-19 (2019).
Kemung, H. M. et al. Streptomyces as a prominent resource of future anti-MRSA drugs. Front. Microbiol. 9, 377137. https://doi.org/10.3389/fmicb.2018.02221 (2018).
Singh, L. S., Sharma, H. & Talukdar, N. C. Production of potent antimicrobial agent by actinomycete, Streptomyces sannanensis strain SU118 isolated from Phoomdi in Loktak lake of manipur, India. BMC Microbiol. 14, 278. https://doi.org/10.1186/s12866-014-0278-3 (2014).
Hughes, C. C., Prieto-Davo, A., Jensen, P. R. & Fenical, W. The marinopyrroles, antibiotics of an unprecedented structure class from a marine Streptomyces Sp. Org. Lett. 10, 629–631. https://doi.org/10.1021/OL702952N (2008).
Haste, N. M. et al. Activity of the thiopeptide antibiotic nosiheptide against contemporary strains of methicillin-resistant Staphylococcus aureus. J Antibiot (Tokyo). 65, 593–598. https://doi.org/10.1038/JA.2012.77 (2012).
Liu, L. L. et al. Four new antibacterial Xanthones from the marine-derived actinomycetes Streptomyces caelestis. Mar. Drugs. 10, 2571. https://doi.org/10.3390/MD10112571 (2012).
Suzuki, N. et al. A compound inhibits biofilm formation of Staphylococcus aureus from Streptomyces. Biol. Pharm. Bull. 38, 889–892. https://doi.org/10.1248/BPB.B15-00053 (2015).
Mazumder, S., Singh, L. S. & Bora, T. Endophytic fungi associated with medicinal plants of Gibbon wild life sanctuary, india, and their antagonistic properties. In: (eds Tayung, K., Barik, S. B. & Mohapatra, U. B.) Advances in Life Sciences. Studium; 213–224. (2012).
Hong, C. E., Jo, S. H., Jo, I. H. & Park, M. Diversity and antifungal activity of endophytic bacteria associated with Panax ginseng seedlings. Plant. Biotechnol. Rep. 12, 409–418. https://doi.org/10.1007/s11816-018-0504-9 (2018).
Wu, W. et al. Beneficial relationships between endophytic bacteria and medicinal plants. Front. Plant. Sci. 12, 646146. https://doi.org/10.3389/fpls.2021.646146 (2021).
Castillo, U. F. et al. Munumbicins, wide-spectrum antibiotics produced by Streptomyces NRRL 30562, endophytic on Kennedia nigriscans. Microbiol 148, 2675–2685. https://doi.org/10.1099/00221287-148-9-2675 (2002).
Kurosawa, K. et al. Rhodostreptomycins, antibiotics biosynthesized following horizontal gene transfer from Streptomyces Padanus to Rhodococcus fascians. J. Am. Chem. Soc. 130, 1126–1127. https://doi.org/10.1021/JA077821P (2008).
Onaka, H., Mori, Y., Igarashi, Y. & Furumai, T. Mycolic acid-containing bacteria induce natural-product biosynthesis in Streptomyces species. Appl. Environ. Microbiol. 77, 400–406. https://doi.org/10.1128/AEM.01337-10 (2011).
Rai, S. et al. Endophytic fungi of Panax sokpayensis produce bioactive ginsenoside compound K in flask fermentation. Sci. Rep. 14, 9318. https://doi.org/10.1038/s41598-024-56441-3 (2024).
H Goodwin, P. PH The endosphere Microbiome of ginseng. Plants 11, 415. https://doi.org/10.3390/plants11030415 (2022).
Barnett, R., Larson, G. A. & Phenol-Chloroform, Protocol for extracting DNA from ancient samples. In Ancient DNA: Methods and Protocols (eds Shapiro, B. & Hofreiter, M.) 13–19 (Humana, 2012). https://doi.org/10.1007/978-1-61779-516-9_2.
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120. https://doi.org/10.1007/BF01731581 (1980).
Tamura, K., Stecher, G., Peterson, D., Filipski, A. & Kumar, S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. https://doi.org/10.1093/molbev/mst197 (2013).
Davis, J. J. et al. The PATRIC bioinformatics resource center: expanding data and analysis capabilities. Nucleic Acids Res. 48 https://doi.org/10.1093/nar/gkz943 (2020). D606-D612.
Gurevich, A., Saveliev, V., Vyahhi, N. & Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29, 1072–1075. https://doi.org/10.1093/bioinformatics/btt086 (2013).
Lee, I., Kim, Y. O., Park, S. C., Chun, J. & OrthoANI An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. https://doi.org/10.1099/IJSEM.0.000760 (2016).
Li, Y. et al. Streptomyces Camponoticapitis sp. nov., an actinomycete isolated from the head of an ant (Camponotus japonicus Mayr). Int. J. Syst. Evol. Microbiol. 66, 3855–3859. https://doi.org/10.1099/ijsem.0.001276 (2016).
Blin, K. et al. AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. https://doi.org/10.1093/nar/gkab335 (2021).
Locci, R. Streptomycetes and related genera. In Bergey’s Manual of Systematic Bacteriology Vol. 4 (ed. Williams, S. T.) 2451–2469 (Williams and Wilkins Company, 1989).
Shirling, E. B. & Gottlieb, D. Methods for characterisation of Streptomyces species1. Int. J. Syst. Evol. Microbiol. 16, 313–340. https://doi.org/10.1099/00207713-16-3-313 (1966).
Sasser, M. Identification of Bacteria by gas chromatography of cellular fatty acids. Technical Note 101. MIDI, Inc. Newark, 1–7. (2001).
Gomez, K. A. & Gomez, A. A. Statistical Procedures for Agricultural Research. Second Edition, A Wiley-Interscience Publication207–215 (Wiley, 1984).
Mahdi, R. A., Bahrami, Y. & Kakaei, E. Identification and antibacterial evaluation of endophytic actinobacteria from Luffa cylindrica. Sci. Rep. 12, 18236. https://doi.org/10.1038/s41598-022-23073-4 (2022).
Cruz-Morales, P. et al. The genome sequence of Streptomyces lividans 66 reveals a novel tRNA-dependent peptide biosynthetic system within a metal-related genomic Island. Genome Biol. Evol. 5 (6), 1165–1175. https://doi.org/10.1093/gbe/evt082 (2013).
Pervaiz, A. et al. An emerging antibacterial modality against Methicillin-resistant Staphylococcus aureus. Curr. Pharm. Des. 22, 4420–4429. https://doi.org/10.2174/1381612822999160629115627 (2016).
Almeida, A. A. P., Farah, A., Silva, D. A. M., Nunan, E. A. & Gloria, M. B. A. Antibacterial activity of coffee extracts and selected coffee chemical compounds against Enterobacteria. J. Agric. Food Chem. 54, 8738–8743. https://doi.org/10.1021/jf0617317 (2006).
Casciaro, B. et al. Naturally-occurring alkaloids of plant origin as potential antimicrobials against antibiotic-resistant infections. Molecules 25, 3619. https://doi.org/10.3390/molecules25163619 (2020).
Zhou, J., Chan, L., Zhou, S. & Trigonelline A plant alkaloid with therapeutic potential for diabetes and central nervous system disease. Curr. Med. Chem. 19, 3523–3531. https://doi.org/10.2174/092986712801323171 (2012).
Jubair, N., Rajagopal, M., Chinnappan, S., Abdullah, N. B. & FatimaA. Review on the antibacterial mechanism of Plant-Derived compounds against Multidrug-Resistant Bacteria (MDR). Evid. Based Complement. Alternat Med. 2021 (663315). https://doi.org/10.1155/2021/3663315 (2021).
Zheng, L., Chen, H., Han, X., Lin, W. & Yan, X. Antimicrobial screening and active compound isolation from marine bacterium NJ6-3-1 associated with the sponge Hymeniacidon perleve. World J. Microbiol. Biotechnol. 21, 201–206. https://doi.org/10.1007/s11274-004-3318-6 (2005).
Ozturk, G. et al. J.M.L.N. The antimicrobial activity of bovine milk Xanthine oxidase. Int. Dairy. J. 102, 104581. https://doi.org/10.1016/j.idairyj.2019.104581 (2020).
Machova, M., Bajer, T., Silha, D., Ventura, K. & Bajerova, P. Volatiles composition and antimicrobial activities of areca nut extracts obtained by simultaneous distillation-extraction and headspace solid-phase Microextraction. Molecules 26 (24), 7422. https://doi.org/10.3390/molecules26247422 (2021).
Risdian, C., Mozef, T. & Wink, J. Biosynthesis of polyketides in Streptomyces. Microorganisms 7, 124. https://doi.org/10.3390/microorganisms7050124 (2019).
Robertsen, H. L. & Musiol-Kroll, E. M. Actinomycete-derived polyketides as a source of antibiotics and lead structures for the development of new antimicrobial drugs. Antibiot 8, 157. https://doi.org/10.3390/antibiotics8040157 (2019).
Pickens, L. B. & Tang, Y. Oxytetracycline biosynthesis. J. Biol. Chem. 285, 27509–27515. https://doi.org/10.1074/jbc.R110.130419 (2010).
Mondol, M. A. M. et al. Ieodomycins A-D, antimicrobial fatty acids from a marine Bacillus Sp. J. Nat. Prod. 74, 1606–1612. https://doi.org/10.1021/np200223r (2011).
Jimenez, A., Vazquez, D. & Hahn, F. E. Anisomycin and related antibiotics. In: Gottlieb D,Antibiotics Vol 5. Edited, Mechanism of action of antieukaryotic and antiviral compounds. 1–19. Springer-Verlag, Berlin Heidelberg New York (1979).
Arakawa, K., Sugino, F., Kodama, K., Ishii, T. & Kinashi, H. Cyclization mechanism for the synthesis of macrocyclic antibiotic Lankacidin in Streptomyces rochei. Chem. Biol. 12, 249–256. https://doi.org/10.1016/j.chembiol.2005.01.009 (2005).
Liu, P. & Jacobsen, E. N. Total synthesis of (+)-ambruticin. J. Am. Chem. Soc. 123, 10772–10773. https://doi.org/10.1021/ja016893s (2001).
Serio, A. W., Keepers, T., Andrews, L. & Krause, K. M. Aminoglycoside revival: review of a historically important class of antimicrobials undergoing rejuvenation. EcoSal Plus. 8 (1), 1–20. https://doi.org/10.1128/ecosalplus.esp-0002-2018 (2018).
Hirai, Y. et al. In vitro and in vivo antimicrobial activity of TS2037, a novel aminoglycoside antibiotic. J. Antibiot. 71, 363–371. https://doi.org/10.1038/s41429-017-0002-2 (2018).
Hotta, K., Yoshida, M., Hamada, M. & Okami, Y. Studies on new aminoglycoside antibiotics, Istamycins, from an actinomycete isolated from a marine environment III. Nutritional effects on Istamycin production and additional chemical and biological properties of Istamycins. J. Antibiot. 33, 1515–1520. https://doi.org/10.7164/antibiotics.33.1515 (1980).
Washington, J. A. & Yu, P. K. In vitro antibacterial activity of spectinomycin. Antimicrob. Agents Chemother. 2, 427–430. https://doi.org/10.1128/aac.2.6.427 (1972).
Awata, M., Hayashi, M., Muto, N. & Sakakibara, H. Acmimycin, a new aminoglycoside antibiotic. Agric. Biol. Chem. 50, 239–241. https://doi.org/10.1080/00021369.1986.10867367 (1986).
Aviner, R. The science of puromycin: from studies of ribosome function to applications in biotechnology. Comput Struct. Biotechnol J. 18, 1074–1083. https://doi.org/10.1016/j.csbj.2020.04.014 (2020).
Fosso, M. Y., Shrestha, S. K., Green, K. D. & Garneau-Tsodikova, S. Synthesis and bioactivities of Kanamycin B-derived cationic amphiphiles. J. Med. Chem. 58, 9124–9132. https://doi.org/10.1021/acs.jmedchem.5b01375 (2015).
Juhas, M. et al. In vitro activity of Apramycin against multidrug-, carbapenem and aminoglycoside-resistant Enterobacteriaceae and Acinetobacter baumannii. J. Antimicrob. Chemother. 74, 944–952. https://doi.org/10.1093/jac/dky546 (2019).
Obuchi, S. et al. Clinical studies with Dihydrodesoxystreptomycin preliminary report. J Antibiot. Ser. A. 11, 199–201. https://doi.org/10.11554/antibioticsa.11.5_199 (1958).
Jawetz, B. E., Gunnison, J. B. & Coleman, V. R. Observations on the mode of action of antibiotic synergism and antagonism. J. Gen. Microbiol. 10, 191–198. https://doi.org/10.1099/00221287-10-2-191 (1958).
Robertson, J. H., Baas, R., Yeager, R. L. & Tsuji, K. Antimicrobial activity of neomycin C against Staphylococcus epidermidis. Appl. Microbiol. 22, 1164–11655. https://doi.org/10.1128/am.22.6.1164-1165.1971 (1971).
Thornsberry, C. et al. Antibacterial activity of Fortimicin A compared with those of five other aminoglycosides, and factors affecting susceptibility tests. Antimicrob. Agents Chemother. 19, 122–129. https://doi.org/10.1128/aac.19.1.122 (1981).
Yoshizawa, S., Fourmy, D. & Puglisi, J. D. Structural origins of gentamicin antibiotic action. EMBO J. 17, 6437–6448. https://doi.org/10.1093/emboj/17.22.6437 (1988).
Miller, G. H., Arcieri, G., Weinstein, M. J. & Allan Waitz, J. Biological activity of netilmicin, a broad-spectrum semisynthetic aminoglycoside antibiotic. Antimicrob. Agents Chemother. 10, 827–836. https://doi.org/10.1128/aac.10.5.827 (1976).
Felnagle, E. A. et al. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol. Pharm. 5 (2), 191–211. https://doi.org/10.1021/mp700137g (2008).
Jiang, K., Chen, X., Zhang, W., Guo, Y. & Liu, G. Nonribosomal antibacterial peptides isolated from Streptomyces agglomeratus 5-1-3 in the Qinghai-Tibet plateau. Microb. Cell. Fact. 22, 5. https://doi.org/10.1186/s12934-023-02018-0 (2023).
Greenwood, D. & O’grady, F. Scanning Electron microscopy of Staphylococcus aureus exposed to some common anti-staphylococcal agents. J. Gen. Microbiol. 70, 263–270. https://doi.org/10.1099/00221287-70-2-263 (1972).
Wale, L. J., Shelton, A. P. & Greenwood, D. Scanning electron microscopy of Staphylococcus aureus and Enterococcus faecalis exposed to daptomycin. J. Med. Microbiol. 30, 45–49. https://doi.org/10.1099/00222615-30-1-45 (1989).
Idrees, M., Sawant, S., Karodia, N. & Rahman, A. Staphylococcus aureus biofilm: morphology, genetics, pathogenesis and treatment strategies. Int. J. Environ. Res. Public. Health. 18 (14), 7602. https://doi.org/10.3390/ijerph18147602 (2021).
Panda, S. K., Das, R., Lavigne, R. & Luyten, W. Indian medicinal plant extracts to control multidrug-resistant S. aureus, including in biofilms. S Afr. J. Bot. 128, 283–291. https://doi.org/10.1016/j.sajb.2019.11.019 (2022).
Acknowledgements
LSS thank the Department of Biotechnology (DBT), Government of India, New Delhi for supporting the work in terms of project grant [DBT/NER/Agri/24/2013]. The authors acknowledge the financial help & assistance provided to SR by Dr. Uma Ramakrishnan, National Centre for Biological Sciences (NCBS-TIFR), Bengaluru and Mrs. Tirtha Tamang during work at NCBS-TIFR Bengaluru. The assistance provided by Dr. Preethi and Anjali, Electron Microscopy facility, NCBS-TIFR; Mr. Awadhesh, NGS facility, NCBS-TIFR; Padma and Theja, C-CAMP facility Bengaluru and Dr. Shannon Olsson and Prof. Mahesh Sankaran (NCBS-TIFR) for working space. We acknowledged Ms. Anjali Rai and Mr. Bishal Tiwari for assisting in antimicrobial assay experiments using pure compounds. We thank the Department of Forests, Environment and Wildlife Management, Government of Sikkim for cooperation in sample collection.
Author information
Authors and Affiliations
Contributions
LSS conceived the idea and designed the experiments; SR, KL, and LSS conducted the experiments and collected the data. SR, KL, KJ, and LSS performed the analysis (analyzed the data). SR, LSS, and KJ wrote the paper (contributed to manuscript writing). LSS, TP, DS and KJ reviewd and edited the manuscript. LSS and DS supervised the work. All the authors discussed the results and contributed to the final manuscript. All authors have duly checked and approved the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rai, S., Singh, L.S., Liriina, K. et al. Novel endophytic actinomycetes species Streptomyces panacea of Panax sokpayensis produce antimicrobial compounds against multidrug resistant Staphylococcus aureus. Sci Rep 15, 19863 (2025). https://doi.org/10.1038/s41598-025-05333-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-05333-1