Abstract
A novel electrochemical biosensor with high amplification efficiency was explored for serum microRNA-21 (miRNA-21) detection. This biosensor consisted of the target recycling amplification (TRA) and non-linear hybridization chain reaction (NHCR). When the target miRNA-21 was presented, hairpin probe 1 (HP1) could be opened and hybridized with target miRNA-21, the formation of DNA/RNA complexes could hybridize with hairpin probe 2 (HP2) and released target miRNA-21, resulting in the target recycling amplification. Then, the opened HP2 acted as a trigger chain and bonded to substrate-A to trigger NHCR. After the occurrence of NHCR, large amounts of high molecular weight double-stranded DNA were produced. Because the two basic chains, substrate-A and substrate-B, of the synthesized NHCR product were labeled with biotin, the NHCR product carried a large amount of biotin. When streptavidin-alkaline phosphatase (ST-AP) was added, it could combine with the biotin and catalyze the hydrolysis of α-naphthyl phosphate (α-NP) to α-naphthol. Since α-naphthol was electrically active, an ultrasensitive electrochemical readout was obtained. Based on the highly efficient signal amplification, the established biosensor showed an outstanding sensitivity for target miRNA-21 detection. The limit was as low as 0.8 fM, and the linear range was 1 fM to 10 nM. The research also verified that the biosensor had good stability and repeatability. Moreover, the detection results of spiked miRNA-21 in serum by this biosensor were in good agreement with those in the buffer solution. Due to its excellent ability, this new biosensor might have great potential for application in the detection of biomolecules and clinical disease diagnosis.
Similar content being viewed by others
Introduction
MicroRNAs (miRNAs) play an important role in gene expression and regulation1. Aberrant expression of miRNAs is highly associated with the occurrence of numerous diseases2,3. In recent years, a variety of miRNAs have emerged as significant biomarkers for the clinical diagnosis and prognosis of diseases. Among these miRNAs, miRNA-21 is the focus of investigation. MiRNA-21 is a typical oncogenic miRNA overexpressed in various malignant tumors and promotes tumor development by inhibiting tumor suppressor genes and apoptosis-related genes4. High expression of miRNA-21 is associated with poor prognosis and chemotherapy resistance, and it can serve as a diagnostic marker or therapeutic target. Targeted inhibition of miRNA-21 is a potential anti-cancer strategy. Due to its stable presence in the blood, miRNA-21 can be used as a non-invasive biomarker for early cancer screening or disease monitoring. Due to the low abundance of miRNAs in the human serum, a high sensitivity of the assay is essential. However, the sensitivity of the existing detection methods is limited and insufficient to meet the clinical requirements for miRNAs detection. Hence, a sensitive test method for miRNA is urgently needed. At present, there are several methods for miRNA detection have been established, such as surface plasmon resonance5 northern blotting6 quantitative polymerase chain reaction7 mass spectrometry8 in situ hybridization9surface-enhanced Raman spectroscopy10 and electrochemical biosensors11,12. Among these detecting techniques, the electrochemical biosensors have gained wide attention because it has a relatively high sensitivity, less expensive to fabricate, simple to operate, and very few samples to be used13. To achieve high sensitivity, lots of classic isothermal amplification methods have been used to amplify the detection signal, such as hybridization chain reaction (HCR)14, target recycling amplification (TRA)15, strand displacement amplification (SDA)16, rolling circle amplification (RCA)17 and exonuclease III-assisted signal amplification (Exo III-assisted CA)8. Among them, HCR has been used in a variety of assays. However, the sensitivity of this method is not very satisfactory. Nonlinear hybridization chain reaction (NHCR) is a new isothermal amplification method in recent years9. Compared with classical HCR, the amplified product of NHCR is not linear double-stranded DNA, which can be branched during assembly, thus greatly increasing the molecular weight of the product. These large molecular weight nonlinear double-stranded DNA products significantly increase the detection signal, thus improving the sensitivity of the detection method.
Target recycling amplification (TRA) is also an effective signal amplification method, including duplex-specific nuclease-assisted target recycling20exonuclease I-assisted target recycling21 APE1 enzyme-assisted target recycling22 etc. These signal amplification methods could make a finite target material go through an infinite cycle, thereby achieving the effect of signal amplification. However, enzymatic-mediated cyclic amplification strategies could be influenced by enzyme activity and reaction environment. These characteristics limit the broad application of this method.
To solve this problem, we have developed a novel signal amplification strategy without an enzyme. In this study, as the target to be detected, miRNA-21 was also the trigger chain of TRA. The central part of the TRA was two specially designed probes, both of which existed in a stable hairpin structure in the absence of the target miRNA-21. In the presence of the target miRNA-21, hairpin probe 1 (HP1) was opened, and a DNA and RNA complex with miRNA-21 was formed, while ends that could bind to hairpin probe 2 (HP2) were exposed. When HP2 was bound to HP1, the target miRNA-21 could be replaced from the DNA/RNA complex, and a new HP1 would be opened by the target miRNA-21. In the process, target miRNA-21 could realize its own cycle while catalyzing the hybridization of HP1 and HP2. A small amount of target miRNA-21 could produce a large amount of HP1/HP2, thus achieving the effect of signal amplification. To detect low-abundance miRNA-21 in serum, NHCR and TRA were combined to achieve cascade signal amplification and further improve the sensitivity of detection. HP1/HP2 triggered NHCR, producing large amounts of nonlinear high-molecular weight double-stranded DNA. Double-stranded DNA was labeled with biotin, which could bind to the streptavidin-alkaline phosphatase (ST-AP). ST-AP catalyzed the hydrolysis of α-naphthyl phosphate (α-NP) to α-naphthol, producing an amplified electrochemical signal. Compared with other methods, the biosensor achieved signal amplification without the involvement of enzymes, which made it cheaper to detect and more stable. The ingeniously designed pair of hairpin structures minimized non-specific binding and significantly enhanced the specificity of the sensing system. TRA enabled limited targets to be fully reused and increased sensitivity. The addition of NHCR enabled the ultra-sensitive detection of miRNA-21. Overall, the double signal amplification effect made the detection method more sensitive, and the double trigger made the sensor more specific.
Experimental sections
Materials and methods
Reagent
Nucleotide sequences in Supplementary Table 1. Nucleotide sequences were obtained from Beijing Tsingke Biotech Co., Ltd. (Beijing, China). Kept HP1 and HP2 in a 95 °C water bath for 5 min, and then slowly cooled them to room temperature. Substrate-A was prepared by heating a mixture containing 1 µM of the L-strand and 1.5 µM of the LF-strand of substrate-A to 95 °C for five minutes, followed by gradual cooling to room temperature. Substrate-B was prepared with the corresponding L-strand and LF-strand by the same method. Streptavidin-alkaline phosphatase (ST-AP), bovine serum albumin (BSA), α-naphthyl phosphate (α-NP) and 6-mercapto-1-hexanol (MCH) were bought from Sigma (St. Louis, MO, USA). DNA PAGE electrophoresis Kit was purchased from Beyotime Biotech Co., Ltd. (Shanghai, China). Blank human serum was purchased from Huizhi Heyuan Biotechnology Co., Ltd. (Suzhou, China).
The hybridization buffer used in the experiment containing 10 mM Tris–HCl, 50 mM NaCl, 10 mM MgCl2 (pH 8.0). 0.01 M phosphate buffered solution (PBS) containing 0.01 M Na2HPO4, 0.01 M NaH2PO4, 0.01 M MgCl2 and 0.05% (w/v) Tween-20 (pH 7.4) was used as the washing buffer. Diethanolamine (DEA) buffer was the working buffer containing 0.1 M DEA, 1 M MgCl2 and 100 mM KCl (pH 9.6). All buffers were prepared by ultrapure water, which was obtained from Wuhan Kexin water purification system (18.3 MΩ). The experimental buffers were all prepared with 0.1% of DEPC.
Apparatus
In this experiment, the CHI660D electrochemical workstation (manufactured by Shanghai Chen Hua Instruments, Shanghai, China) served as the primary instrument for conducting electrochemical measurements. Classical threeelectrode system was used for detection: Ag/AgCl reference electrode, platinum wire auxiliary electrode and gold working electrode. The dimensions of the gold electrode were 3 mm in diameter and 5 mm in length (GaossUnion Technology Co., Ltd, Wuhan, China). PAGE was completed on a Bio-Rad vertical electrophoresis apparatus (Bio-Rad Laboratories, Inc, USA), and gel results were observed and processed on Tanon 3500 Gel imaging analysis System (Tanon Science & Technology Co., Ltd.). All measurements in this study were performed under ambient temperature conditions.
Preparation of the sensing electrode
0.05 mm alumina slurry was used to polish the gold disk electrode. When the surface of the electrode was polished, it was treated with ultrasound in ultra-pure water. Piranha solution was prepared by mixing 98% concentrated sulfuric acid and 30% hydrogen peroxide in a 3:1 volume. Piranha acid was dropped onto the surface of the gold electrode 3 times and rinsed with ultrapure water thoroughly. After that, 10 µL of 100 nM thiol-modified HP1 was dropped onto the surface of the gold disk electrode and then placed in the refrigerator at 4 °C. 12 h later, 10 µL of 1 mM MCH solution was dropped on the obtained electrode for 1 h to remove the nonspecific DNA adsorption. Then, washed the electrode surface with a washing buffer. After that, 1% BSA was added and allowed to remain on the electrode surface for half an hour. These treatments could avoid the nonspecific DNA and enzyme adsorbed on the electrode surface. The obtained electrode was rinsed with PBS and DEA buffer for the following operation.
Firstly, miRNA-21 and HP2 were mixed at a volume ratio of 1:4, obtaining the mixtures with a final concentration of 200 nM HP2 and different concentrations of miRNA-21. 10 µL of the mixture was added to the prepared electrode with the incubation for 1 h. At the same time, substrate-A and substrate-B were mixed with 2 µM corresponding auxiliaries separately and incubated for 30 min. Thus, solutions A and B were obtained. Excess label-free strands in the two solutions hybridized with the auxiliaries and formed byproducts. Then 30 µL of solution A and 60 µL of solution B were mixed, obtaining the reaction solution with the concentrations of 0.17 µM substrate-A, 0.33 µM auxiliary-A, 0.33 µM substrate-B, and 0.67 µM auxiliary-B. 10 µL of the obtained reaction solution was dripped on the electrode and incubated for 1 h. After washing with DEA, 10 µL of 1 µg mL−1 ST-AP was added on the surface of the gold electrode and incubated at 37 °C. Thirty minutes later, the output signal was detected using differential pulse voltammetry (DPV) in a DEA solution containing 1 mg mL⁻¹ of α-NP.
Results and discussion
Principle of miRNA-21 biosensor design
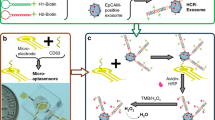
Figure 1 illustrates the principle of the biosensor developed for the detection of miRNA-21. First, the thiol-modified HP1 was assembled on the surface of the gold disk electrode through the Au-S bond. When target miRNA-21 hybridized with HP1 and made the stem-loop structure of HP1 open, the complementary sequence of HP2 could be exposed. When HP2 was presented, it bonded to the opened HP1 and displaced the target miRNA-21. The displaced miRNA-21 could open another HP1 and start a new cycle. At the same time, the exposed sequence of HP2 became a critical step for the successful triggering of NHCR. HP2 would hybridize with the end of substrate-A, and the partial biotin-labeled strand (L-strand) was displaced from the label-free strand (LF-strand). This made the first loop of the substrate-A open and exposed part of the LF-strand. LF-strand could be displaced from the L-strand when auxiliary-A hybridized with it. Through this step, byproduct-A was released, and the second loop of the substrate-A was opened. The exposed two loops could hybridize with two substrate-B at the same time. After auxiliary-B hybridized with substrate-B, byproduct-B was generated with two of the same single-stranded regions exposed. The exposed regions were used as the triggers to start a new cycle. The triggers could initiate more cycles, ultimately resulting in the non-linear hybridization chain reaction. When ST-AP was added, it could combine with the biotin in the dendritic DNA and catalyze the hydrolysis of α-NP to electroactive α-naphthol. The electrochemical signal was detected by differential pulse voltammetry (DPV).
Characterization of biosensor
In this experiment, electrochemical impedance spectroscopy (EIS) and square wave voltammetry (SWV) were useful methods to prove the layerbylayer assembly process on the working electrode. EIS and SWV measurements were performed in 0.4 M KCl containing 0.5 mM [Fe(CN)6]3-/4-. Figure 2 A showed the results of the EIS measurements. The semicircle diameters of the obtained curves represented the electrontransfer resistance (Ret) on the surface of the biosensor. The curve of the bare gold electrode was nearly a straight line because the electroactive ions could transport easily on the bare electrode surface (curve a). After HP1 was modified on the electrode, an obvious increase in Ret was noted because HP1 could impede the transport of electroactive ions toward the electrode surface (curve b). As blocking agents, MCH and BSA made a further increase of the Ret (curve c). When miRNA-21 and HP2 were added to the electrode surface, the TRA was completed, and the electrode surface produced many HP1 and HP2 hybridized double strands, resulting in a further increase in the resistance and enlargement of the semicircle diameters (curve d). When NHCR was finished, the dendritic products were loaded on the surface of the electrode, which gave fabrication of this biosensor. Large molecular weight DNA accumulated on the surface of the electrode, leading to an extreme increase in impedance (curve e). Figure 2B showed the verification process of the sensor by SWV. The peak current reflected the rate of REDOX reactions on the electrode surface. Curve a was the bare electrode without any modification, with low Ret. The probe freely approached the electrode, and SWV showed a high and sharp [Fe(CN)6]3-/4- peak. When HP1 modified the electrode surface, it slightly hindered the electron transfer on the electrode surface, resulting in a slight decrease in the peak current (curve b). When MCH and BSA blocked the electrode surface, it significantly hindered the probe diffusion, and the Ret increased sharply, resulting in a significant decrease in the peak current (curve c). When miRNA-21 and HP2 were introduced into the system to complete TRA, the Ret was greater, and the peak current was further reduced (curve d). After the NHCR assembly was completed, the electrode surface was covered with high-molecular-weight DNA, the Ret reached the maximum, and the peak current was the lowest (curve e). The characterization results of these two methods were consistent, which fully demonstrated that the biosensor has been successfully constructed.
Verification of signal amplification effect
In order to check the signal amplification effect of the sensor, DPV was used for verification. As shown in Supplementary Fig. 1, the detection signal was weak in the absence of target miRNA-21 (curve a) because a small amount of ST-AP was non-specifically adsorbed on the electrode surface. To validate the detection efficacy of TRA, HP2 was labeled with biotin. When biotin-labeled HP2 was added alone, it hybridized with HP1 opened by target miRNA-21. The biotin of HP2 bonded to ST-AP and AP catalyzed the α-NP to an electroactive product, yielding a relatively high detection signal (curve b). When the same amount of miRNA-21 was detected with the TRA and NHCR dual amplification system, a higher output signal was obtained (curve c). Experiments show that the sensor is successfully established and has a good signal amplification function.
Validation by native polyacrylamide gel electrophoresis
The designed detection system was validated by native polyacrylamide gel electrophoresis (Fig. 3). The nucleic acid in the detection system was prepared in the hybridization buffer at pH 8.0. Lane 1 is the 0.5 µM HP1, Lane 2 is the product of the hybridization of the target miRNA-21 and HP1 with a concentration of 0.5 µM. Lane 3 is the addition of 0.5 µM HP2. Separately, mixed L-strand-A (1 µM) and LF-strand-A (1.5 µM) at 95 °C for 5 min, slowly cooled to room temperature, and obtain Substrate -A (Lane 5). The same method was used to process L-strand-B and LF-strand-B to obtain Substrate-B (Lane 6). Substrate -A and Substrate -B were incubated at room temperature with their respective auxiliaries (2 µM) for 20 min to obtain solutions A and B. The solutions A and B were mixed in a 1:2 ratio, resulting in a final concentration of approximately 0.15 µM Substrate-A, 0.23 µM auxiliary-A, 0.3 µM Substrate-B, and 0.46 µM auxiliary-B in the reaction solution. Finally, HP2 was introduced as the trigger chain into the reaction solution after the mixture of Solutions A and B, and Lane 5 disappeared, with a low-migrating bright band appearing, indicating that Substrate-A was assembled into a larger molecular weight product (Lane 4). Lane 7 is the product of the hybridization of LF-strand-A and auxiliary-A, and Lane 8 is the product of the hybridization of LF-strand-B and auxiliary-B.
Optimization of experimental conditions
Optimization of experimental conditions was essential to improving the accuracy and reliability of results. The primary conditions influencing the TRA were optimized, specifically the concentrations of HP1 and HP2, as well as the incubating time of TRA. As illustrated in Supplementary Fig. 2 A, the peak current response varied with the concentration of HP1 in a 10 µL TRA mixture, ranging from 50 nM to 150 nM. The peak current response increased with rising HP1 concentration until it surpassed 100 nM. Accordingly, 100 nM was chosen in further experiments. Supplementary Fig. 2B showed that the peak current response was increased with the increase of HP2 concentration from 100 nM to 200 nM. Over 200 nM, the signal was reduced. Thus, 200 nM was considered the optimal condition. Supplementary Fig. 2 C described the effect of the incubation time of the TRA process on the electrochemical signal. The signal was increased from 20 min to 60 min, and there was no more increase after that. Therefore, 60 min was considered the optimum incubation time. Reaction time was a crucial factor affecting NHCR products. Supplementary Fig. 2D showed that with the extension of the reaction time of NHCR, DPV response enhanced. After the reaction time reached 60 min, the response remained stable. Consequently, a reaction time of 60 min was identified as optimal for the NHCR.
Sensitivity of the biosensor
After each reaction condition was optimized, the influence of the concentration of target miRNA-21 on the output signal was studied. As shown in Fig. 4 A, the higher the concentration of the target miRNA-21, the higher the peak value of the current. Figure 4B displayed that in the range of 1 fM to 10 nM, the peak current has a linear relationship with the logarithm of the target miRNA-21 concentration, and the correlation coefficient is 0.9973. The regression equation was given by I (µA) = 1.59 + 0.61 X, where I represented the peak current, and X represented the logarithm of the target miRNA-21 concentration (n = 3). Based on the definition of the detection limit, we calculated the detection limit to be 0.8 fM.
The sensitivity of the biosensor we developed was compared with those of other miRNA detection strategies in Table 1. The results demonstrate that this strategy has a comparable sensitivity with the other signal amplification.
Specificity, repeatability and stability of the assay
To identify the target miRNA-21 from the highly similar miRNAs is a great challenge because of the high homology. Single-base mismatch (SM), double-base mismatch (DM), and noncomplement sequence (NC) are commonly used specific verification tools. Meanwhile, miRNA-155 was selected as an interfering substance in serum samples to verify the specificity of the biosensor. As illustrated in Fig. 5, the current intensity of the SM, DM, NC, and miRNA-155 were comparable to that of the blank. At the same time, the current signal was significantly enhanced when miRNA-21 was added. Clearly, the biosensor we designed could distinguish the target miRNA-21 from the homologous miRNAs and showed good specificity.
The biosensor constructed in this experiment was used to determine miRNA-21 at two concentration levels of 10 nM and 10 pM for repeatability analysis. The target miRNA-21 with two concentrations were divided into 10 EP tubes and stored at −20℃. Then each concentration was tested five times in the optimal experimental condition (Supplementary Table 2). The relative standard deviations (RSD) of the detection results at 10 nM was 3.6% and 10 pM was 3.1%, showing the good reproducibility.
Stability was a critical property that influenced biosensor applications. Electrodes modified with the probe were stored at 4 °C for two weeks to evaluate the stability of the biosensor. The 10 nM miRNA-21 was detected by the sensors every two days. On the 14th day, the current response was approximately 93.4% of the initial current response, indicating the superior stability of this proposed method.
MiRNA-21 analysis in human serum samples
The recovery experiment is an effective method to verify the effectiveness of biosensors in analyzing and detecting targets in real samples29,30. To ensure the standardization of the validation process, blank human serum purchased from a biotechnology company were used for the experiment. Different concentrations of miRNA-21 were added to human serum samples and tested by the established biosensor. As shown in Table 2, the recovery rate obtained was 97.3–102%, and the relative standard deviation was 2.62–3.61%. The result of this recovery experiment is satisfactory, indicating that this sensor has the potential to detect miRNA-21 in real samples.
Conclusions
In this study, an enzyme-free electrochemical biosensor combining TRA and NHCR for miRNA-21 detection was successfully established. The successful completion of the TRA was the first step in sensor development. TRA allowed the limited target miRNA to be reused, resulting in the production of large amounts of double-stranded DNA that could trigger NHCR. After completion of NHCR, large amounts of biotin-labeled high molecular weight DNA were produced. Finally, biotin bound to ST-AP and catalyzed α-NP to produce an amplified electrochemical signal for miRNA-21 detection. The biosensor demonstrated ultra-high sensitivity with a detection limit as low as 0.8 fM, attributable to the implementation of dual signal amplification. In addition, taking advantage of the specificity recognition performance of the hairpin probe, this electrochemical biosensor exhibited satisfactory specificity. This biosensor also produced satisfactory results for miRNA-21 detection in human serum samples. Hence, with the outstanding analytical performance, the biosensor we developed would play an important role in the early diagnosis of diseases in the future.
Data availability
Data is provided within the manuscript and supplementary information files.
References
Iqrar, S. et al. Set of MiRNAs involved in sulfur uptake and the assimilation pathway of Indian mustard (B. juncea) in response to sulfur treatments. ACS Omega. 7, 13228–13242. https://doi.org/10.1021/acsomega.2c00676 (2022).
Ahmadi, S. M. et al. Recent advances in novel MiRNA mediated approaches for targeting breast cancer. J. Drug Target. 31, 777–793. https://doi.org/10.1080/1061186X.2023.2240979 (2023).
Koi, Y. et al. Assessment of the expression of microRNAs–221–3p, –146a–5p, –16–5p and BCL2 in oncocytic carcinoma of the breast: A case report. Oncol. Lett. 26, 535. https://doi.org/10.3892/ol.2023.14123 (2023).
Seo, S. B. et al. Nucleic acid amplification Circuit-Based hydrogel (NACH) assay for One-Step detection of metastatic gastric Cancer-Derived Exosomal MiRNA. Adv. Sci. (Weinh). 11, e2407621. https://doi.org/10.1002/advs.202407621 (2024).
Chang, Y. F. et al. Amplification-free detection of cytomegalovirus MiRNA using a Modification-free surface plasmon resonance biosensor. Anal. Chem. 93, 8002–8009. https://doi.org/10.1021/acs.analchem.1c01093 (2021).
Esmaeilzadeh, A. A. et al. Recent advances on the electrochemical and optical biosensing strategies for monitoring microRNA-21: a review. Anal. Methods. 14, 4449–4459. https://doi.org/10.1039/d2ay01384c (2022).
Yanai, K. et al. Quantitative Real-time polymerase chain reaction evaluation of MicroRNA expression in kidney and serum of mice with Age-dependent renal impairment. J. Vis. Exp. https://doi.org/10.3791/63258 (2022).
Kuang, Y., Cao, J., Xu, F. & Chen, Y. Duplex-Specific Nuclease-Mediated amplification strategy for mass spectrometry quantification of MiRNA-200c in breast Cancer stem cells. Anal. Chem. 91, 8820–8826. https://doi.org/10.1021/acs.analchem.8b04468 (2019).
Boen, J. R. A., Pintelon, I., Gevaert, A. B., Segers, V. F. M. & van Craenenbroeck, E. M. Fluorescent in situ hybridization for MiRNA combined with staining of proteins. Curr. Protocols. 3, e880. https://doi.org/10.1002/cpz1.880 (2023).
Li, D. et al. Label-Free detection of MiRNA using Surface-Enhanced Raman spectroscopy. Anal. Chem. 92, 12769–12773. https://doi.org/10.1021/acs.analchem.0c03335 (2020).
Su, Z. et al. An electrochemical determination strategy for miRNA based on bimetallic nanozyme and toehold-mediated DNA replacement procedure. Mikrochimica Acta 190, 149, doi: 10.1007/s00604-023-05720-3 (2023).
Wang, Q. et al. Ultrasensitive electrochemical detection of MiRNA based on polymerization signal amplification. Talanta 235, 122744. https://doi.org/10.1016/j.talanta.2021.122744 (2021).
Hou, T. L., Zhu, L., Zhang, X. L., Chai, Y. Q. & Yuan, R. Multiregion linear DNA Walker-Mediated ultrasensitive electrochemical biosensor for MiRNA detection. Anal. Chem. 94, 10524–10530. https://doi.org/10.1021/acs.analchem.2c02004 (2022).
Ma, F. et al. Hybridization chain reaction assisted multicolor immunosensor for sensitively detection of human chorionic gonadotropin. Talanta 270, 125578. https://doi.org/10.1016/j.talanta.2023.125578 (2024).
Yang, Y. et al. Catalytic hairpin Assembly-Enhanced graphene transistor for ultrasensitive MiRNA detection. Anal. Chem. 95, 13281–13288. https://doi.org/10.1021/acs.analchem.3c02433 (2023).
Du, Y., Qi, Y., Kang, Q., Yang, X. & Xiang, H. A fluorescent sensor based on strand displacement amplification and primer exchange reaction coupling for label-free detection of MiRNA. Anal. Chim. Acta. 1279, 341780. https://doi.org/10.1016/j.aca.2023.341780 (2023).
Sun, S. et al. A sensing system constructed by combining a structure-switchable molecular beacon with nicking-enhanced rolling circle amplification for highly sensitive MiRNA detection. Analyst 147, 1937–1943. https://doi.org/10.1039/d1an02218k (2022).
Sun, Y. et al. The compact integration of multiple exonuclease III-Assisted Cyclic amplification units for High-Efficiency ratiometric electrochemiluminescence detection of MRSA. Anal. Chem. 96, 943–948. https://doi.org/10.1021/acs.analchem.3c05410 (2024).
Zhou, L. et al. A label-free electrochemical biosensor for MicroRNAs detection based on DNA nanomaterial by coupling with Y-shaped DNA structure and non-linear hybridization chain reaction. Biosens. Bioelectron. 126, 657–663. https://doi.org/10.1016/j.bios.2018.11.028 (2019).
Han, Y. et al. MicroRNA detection based on duplex-specific nuclease-assisted target recycling and gold nanoparticle/graphene oxide nanocomposite-mediated electrocatalytic amplification. Biosens. Bioelectron. 127, 188–193. https://doi.org/10.1016/j.bios.2018.12.027 (2019).
Wu, H., Wu, J., Liu, Y., Wang, H. & Zou, P. Silver nanoclusters-based fluorescent biosensing strategy for determination of mucin 1: combination of exonuclease I-assisted target recycling and graphene oxide-assisted hybridization chain reaction. Anal. Chim. Acta. 1129, 40–48. https://doi.org/10.1016/j.aca.2020.06.040 (2020).
Liu, Z. et al. A cascade signal-amplified fluorescent biosensor combining APE1 enzyme cleavage-assisted target cycling with rolling circle amplification. Analyst 149, 82–87. https://doi.org/10.1039/d3an01727c (2023).
Wang, W., Jayachandran, S., Li, M., Xu, S. & Luo, X. Hyaluronic acid functionalized nanostructured sensing interface for voltammetric determination of MicroRNA in biological media with ultra-high sensitivity and ultra-low fouling. Mikrochim Acta. 185, 156. https://doi.org/10.1007/s00604-018-2694-9 (2018).
Li, Q., Zeng, F., Lyu, N. & Liang, J. Highly sensitive and specific electrochemical biosensor for microRNA-21 detection by coupling catalytic hairpin assembly with rolling circle amplification. Analyst 143, 2304–2309. https://doi.org/10.1039/c8an00437d (2018).
Yan, Y., Zhao, H., Fang, Y., Ma, C. & Chen, J. Label-Free miRNA-21 analysis based on strand displacement and terminal Deoxynucleotidyl Transferase-Assisted amplification strategy. Biosens. (Basel). 12. https://doi.org/10.3390/bios12050328 (2022).
Xu, Y. et al. Tetrahedral DNA framework based CRISPR electrochemical biosensor for amplification-free MiRNA detection. Biosens. Bioelectron. 217, 114671. https://doi.org/10.1016/j.bios.2022.114671 (2022).
Shahsavar, K., Shokri, E. & Hosseini, M. Sensitive colorimetric detection of miRNA-155 via G-quadruplex DNAzyme decorated spherical nucleic acid. Mikrochim Acta. 189, 357. https://doi.org/10.1007/s00604-022-05455-7 (2022).
Chen, S., He, Y., Liu, L., Wang, J. & Yi, X. DNA walking system integrated with enzymatic cleavage reaction for sensitive surface plasmon resonance detection of MiRNA. Sci. Rep. 12, 16093. https://doi.org/10.1038/s41598-022-20453-8 (2022).
Chen, X. et al. Switch-conversional ratiometric fluorescence biosensor for MiRNA detection. Biosens. Bioelectron. 155, 112104. https://doi.org/10.1016/j.bios.2020.112104 (2020).
Miao, X. et al. Ultrasensitive electrochemical detection of miRNA-21 by using an iridium(III) complex as catalyst. Biosens. Bioelectron. 86, 454–458. https://doi.org/10.1016/j.bios.2016.07.001 (2016).
Funding
This work was financially supported by the Qingdao Key Medical and Health Discipline Project, Medicine and Health Scientific Research program of Qingdao in 2023 (2023-WJZD181), 2023 Shandong Medicine and Health Science and technology project (202311000432), 2023 Shandong Medicine and Health Science and technology project (202302050293), 2024 Shandong Medicine and Health Science and technology project (202403020702).
Author information
Authors and Affiliations
Contributions
Jing Guo: Conceptualization, Methodology, Investigation, Data curation, Writing-original draft. Shou-feng Zhao: Visualization, Conceptualization, Resources, Software, Formal analysis. Shuang Chen: Methodology, Data curation, Supervision, Writing-review & editing, Project administration. Li-qun Li and Wei-qing Song: Project administration, Formal analysis, Supervision, Funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, J., Zhao, Sf., Chen, S. et al. An enzyme-free electrochemical biosensor for sensitive and specific detection of microRNA-21 based on target recycling amplification and non-linear hybridization chain reaction. Sci Rep 15, 22272 (2025). https://doi.org/10.1038/s41598-025-06003-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-06003-y