Abstract
High-grade astrocytomas vary in incidence and mortality across populations, with Hispanic and Latino groups largely underrepresented in genomic epidemiology studies. This study characterizes the presence of known mutations of high-grade astrocytomas in a Latin American cohort through targeted genomic analysis of 70 Chilean patients. Molecular markers, including IDH, TERTp, H3, TP53, PTEN, EGFR, and CDKN2A, were assessed alongside survival analyses. Our results mostly aligned with international cohorts, confirming the importance of established molecular markers in glioblastoma. Novel damaging TP53 and PTEN mutations were identified, expanding the genetic spectrum of known mutations for these genes, while a lower-than-expected NF1 mutation frequency was observed (p < 0.01). These findings highlight the importance of examining underrepresented populations, providing insights into the molecular characteristics of high-grade astrocytomas in Latin America. Our findings contribute to understanding the diversity of genomic features across astrocytoma populations, setting a foundation for future international comparative studies.
Similar content being viewed by others
Introduction
High-grade astrocytomas (HGA) are a subset of aggressive and lethal brain cancers that constitute a significant challenge in oncology. A comprehensive understanding of the molecular mechanisms that drive HGA is crucial for the development of effective diagnostic and therapeutic approaches. Although extensive research has been conducted on HGA across diverse populations worldwide1,2,3,4,5, a notable knowledge gap exists concerning the incidence of mutations associated with HGA in Latin American populations. For example, in the largest cohort of patients with HGA and genomic data currently available, the TCGA pan-cancer atlas glioblastoma database, only approximately 2% are individuals of Hispanic or Latino ethnicity1.
Latin American ancestry refers to a highly admixed genetic background, as previously demonstrated in the Chilean population and other South American countries6. Genomic ancestry studies have shown an average genetic composition of approximately 40% Native American, 55% European, and 2% African ancestry7,8,9while studies on uniparental inheritance have reported up to 88% Native American mitochondrial DNA in populations from different Chilean cities10,11a pattern also observed in other regions of South America7.
With the increasing integration of molecular criteria into the widely used WHO classification of tumors of the central nervous system (CNS), the importance of accurate and reliable molecular techniques has increased in clinical practice12. Molecular diagnostics using next-generation sequencing (NGS)-based methods have become the gold standard for diagnostic workups12. The current criteria for diffuse high-grade astrocytoma classification include disease-defining alterations such as isocitrate dehydrogenase 1/2 (IDH) mutations and histone 3 (H3) gene family (H3F3A, H3C2) mutations, as well as grade-defining alterations such as telomerase reverse transcriptase promoter (TERTp) mutations, epidermal growth factor receptor (EGFR) gene amplification and the combined gain of entire chromosome 7 and loss of entire chromosome 10 in the case of IDH-wildtype tumors (glioblastoma), or cyclin-dependent kinase inhibitor 2 A/B (CDKN2A/B) deletion in the case of IDH-mutant tumors (astrocytoma, IDH-mutant)13. Molecular classification profoundly influences the prognosis and survival of glioblastoma patients, directly impacting their treatment, follow-up, and care14,15,16,17.
Here, we investigated the mutational status of known molecular drivers of HGA in a Latin American cohort, focusing on Chile as a nation characterized by its diverse ethnic composition, including Indigenous, European, and African influences18. By comparing the mutation frequencies in our cohort with those reported in international studies, we seek to determine the relevance of existing knowledge and treatment approaches for Latin American patients. This study aimed to provide insights into the genetic drivers of HGA in a South American population and underscore the broader importance of studying ethnic-specific genetic variations and their association with clinical outcomes.
Methods
Patients
We conducted an observational retrospective cohort study that included 70 patients aged 18 years and above with a confirmed diagnosis of glioblastoma (IDH-wildtype or IDH-mutant) or anaplastic astrocytoma, classified according to the WHO CNS tumor classification system in place at the time of each patient’s diagnosis. Patients who underwent total or partial resection surgery for a newly diagnosed high-grade astrocytoma at the Asenjo Neurosurgery Institute between January 2014 and January 2020 were included in this study. Patients with a previous diagnosis of diffuse glioma, incomplete follow-up, a positive fluorescence in situ hibridization for 1p/19q codeletion and/or insufficient tumor tissue samples for molecular studies were excluded. Additionally, five patients who underwent anterior temporal lobectomy for epilepsy treatment were included in the analysis as non-tumor tissue controls. A specific informed consent was obtained from all patients and controls. This study was approved by the Ethics Committee of the Metropolitan East Health Service in October 2017, and it was conducted in accordance with the principles of the Declaration of Helsinki.
Panel design
We utilized the AmpliSeq™ for Illumina tool to create a customized amplicon panel covering specific genomic regions of interest of several neuro-oncology-related genes. We used databases such as cBioPortal, NCBI/Clinvar, and the Catalogue of Somatic Mutations in Cancer (COSMIC) to identify relevant genomic coordinates. Amplicons were then designed to cover these mutations (see Supplementary Table 1). A coverage depth of greater than 200 reads per position of interest was planned to detect allelic frequencies as low as 5%, with a threshold of ≥ 10 altered reads, following literature recommendations19. For detailed methods, see Supplementary methods.
Results
Clinical variables
The study cohort included 70 patients diagnosed with HGA. All patients reported a “Latin American” ancestry, and no patients reported a purely indigenous, European or other ancestries. The most relevant clinical variables, including access to different treatments and outcomes for all patients, are presented in Table 1.
Frequency of the most common genomic alterations in HGA
We sequenced key glioma-related genes currently used in molecular classification (IDH1, IDH2, H3F3A, H3C2, EGFR, CDKN2A, ATRX, SMARCAL1, BRAF, NF1, TP53, TERTp and PTEN). Overall, 60 unique variants were identified in 66 of the 70 patients (Fig. 1and supplementary Table 2). Of the 70 patients, 67 showed genetic alterations when considering CNVs. The sequencing depths for each variant are shown in supplementary Table 3 for all the samples. Our findings and comparisons with those of other populations for specific genes are detailed in the following sections.
Oncoplot displaying genetic alterations in 14 genes included in our sequencing panel. Nex-generation sequencing results in 70 patients with high-grade astrocytoma are shown. Groups with different clinical outcomes, histological grade and tumor location for each patient are displayed below. All the multi-hit alterations in TERT promoter gene included the canonical mutations 113 and 135. Multi-hit 2 or more variants were identified, Complex event A copy number variation and another genomic alteration occurred simultaneously, Amp Gene amplification, Del Gene deletion, IDH Isocitrate Dehydrogenase, TERTp Telomerase Reverse Transcriptase Promoter, TMB Tumor mutational burden.
Mutations included in the WHO classification of CNS tumors
IDH is among the most relevant genetic markers currently in use for the classification of astrocytic tumors, and when mutated, it defines the recently termed IDH-mutant astrocytoma, which can be categorized as grade 2, 3, or 4 depending on additional histological and/or molecular criteria. This differentiation distinguishes IDH-mutated tumors from glioblastomas, which require IDH-wild-type confirmation. In our patient cohort, we found that ~ 13% of patients had IDH1 mutations (9 out of 70): 8 had the most frequent R132H mutation, whereas 1 had the least common R132S mutation. The clinical characteristics of these patients were similar to those reported in other cohorts20,21significantly affecting younger subjects than those carrying IDH-wildtype alleles (49 years old (range, 30–76) vs. 59 years old (range, 24–75)). When the subgroup of tumors histologically graded as grade 4 was assessed, the frequency of IDH1 mutations was 8.5% (6 out of 67), which was comparable to other cohorts5. As expected, due to their lower frequency20no IDH2 mutations were identified in our cohort. Consistent with previous reports21none of the patients with IDH1 mutations exhibited concomitant TERTp mutations.
Immunohistochemistry (IHC) for IDH1-R132H was routinely performed in patients diagnosed after 2017 (n = 38), showing a direct correlation with the sequencing results, with all six cases that had a positive R132 IHC showing a corresponding IDH1 mutation. Interestingly, although the mean age of the nine patients carrying IDH1 mutations was 49 years, two of these patients were older than 55 years, which is the most common cut-off for recommending sequencing in cases of negative R132H IHC13. The patient who carried the R132S mutation was older than 55 years and could not be accurately diagnosed using IHC, highlighting the relevance of using genomic techniques to evaluate IDH1 mutations in astrocytoma patients.
Less common mutations that define specific entities according to the current diagnostic criteria are those in the H3 gene family, including H3F3A and HIST1H3B genes22. In our cohort, we identified only one patient with an H3 mutation who developed a thalamic tumor and carried the most frequent H3F3A mutation (K28M, rs1057519903), consistent with a specific entity, a diffuse midline glioma, H3 K27-altered. No H3F3A G35 or HIST1H3B were observed in our patients. Consistent with international cohorts23,24our findings indicate that adult patients with tumors located in different supratentorial regions have a low frequency of H3 gene mutations.
In addition to the entity-defining markers, we evaluated the frequency of three genomic alterations that are currently considered in the grading of diffuse astrocytomas: TERTp mutations, EGFR amplification (molecular criteria for grade 4 glioblastoma), and CDKN2A deletion (molecular criterion for a grade 4 IDH-mutant astrocytoma). TERTp mutations typically occur in 70–80% of primary glioblastoma cases25. These mutations are closely associated with increased TERT expression, resulting in the preservation of telomeres, which play a pivotal role in promoting gliomagenesis5. We evaluated two canonical TERTp hotspots where mutations normally occur25. We found that 47 out of 70 patients (67.1%) carried one of these mutations, with 37 patients presented TERTp mutations at position g.chr5:1295113G > A and 10 patients with the less frequent g.chr5:1295135G > A mutation. These results are consistent with those of previous studies on European, North American, and Asian populations26,27,28. EGFR amplification is a recognized molecular hallmark of HGA. In our cohort, we found that 28 patients (40%) exhibited a significant increase in EGFR gene copy numbers. EGFR amplification was present only in IDH-wildtype tumors and was more frequent in TERTp-mutated cases (55.3% vs. 14.3%; p < 0.01). In contrast, CDKN2A deletion has been increasingly recognized as a prognostic marker in IDH-mutant astrocytomas29. CDKN2A partial or total deletions were present in 30 patients (42.9%). Only one patient with CDKN2A deletion also harbored an IDH1 mutation. The frequency distribution and co-existence of TERTp and IDH mutations reported here are consistent with the results obtained in other brain cancer cohorts30.
Regarding grading changes based on molecular markers, all patients with IDH-wildtype tumors carrying TERTp mutations and/or EGFR amplification, as well as those with IDH-mutant tumors carrying CDKN2A deletions, were already classified as grade 4 based on histological features. Therefore, no grading adjustments were made based on these findings.
Additional mutations related to HGA progression
TP53 and PTEN genomic alterations are among the most prevalent mutations in gliomas, and multiple targeted therapies focusing on these genes have been evaluated over the last few years31,32. We sequenced the genomic regions containing all the previously described mutations in TP53 and PTEN in glioblastomas and found a similar mutation rate for both genes compared to other cohorts33,34. Specifically, 25 out of 70 patients (allelic frequency (AF): 35.7%) harbored a TP53 mutation, three of which had not been previously reported in astrocytoma patients (g.chr17:7669615delT (AF: 45% SIFT indel: neutral effect), g.chr17:7674926 C > T (rs587778719; AF: 10.6%; SIFT: tolerated; Polyphen-2: benign), and g.chr17:7674966delC (AF: 84.4%; SIFT indel: damaging). On the other hand, PTEN alterations were found in 29 out of 70 patients (41.4%), with partial or total deletions present in 18 patients, while single nucleotide variants were present in 13 patients. Out Of 11 unique PTEN variants identified, two corresponded to novel variants in astrocytoma patients (g.chr10:87925553 A > G (AF: 43.6%; SIFT: deleterious; Polyphen-2: probably damaging) and g.chr10:87933224_87933225insG (AF: 43.7% SIFT indel: damaging). Thus, novel PTEN (frameshift deletion g.chr17:7674966delC) and TP53 variants (missense mutation g.chr10:87925553 A > G and frameshift insertion g.chr10:87933224_87933225insG) with predicted damaging effects were identified in our study, expanding the known mutations in these genes in patients with astrocytomas. However, each of these variants was only present in one patient, and four out of five carried an allelic frequency that suggests a somatic origin, therefore, it is unlikely that they constitute a specific hallmark of the Latin American population.
ATRX and SMARCAL1 mutations are linked to telomerase-independent telomere maintenance in HGA35,36. While ATRX mutations are typically present in approximately 20% of HGA36our study identified a lower-than-expected frequency of 10% (7 out of 70) (p < 0.01), and none of the commonly reported genetic alterations were present in our cohort. However, numerous infrequent ATRX variants not covered by our sequencing design have been described. We screened the most common SMARCAL1 cancer-associated mutations in our DNA sequencing panel, including Arg645Ser (R645S), Phe793del (del793), and Gly945fs*1 (945 fs) mutations, which have been previously described in up to 20% of TERTp-wt/IDH-wt tumors37. No SMARCAL1 mutations were detected in our cohort.
Regarding NF1, previous cohorts of glioblastoma (TCGA pan-cancer atlas glioblastoma database, MSKCC dataset) have reported approximately a 13% mutation rate for this gene38which is significantly higher than we observed in our cohort (p < 0.01). Interestingly, although our panel design covered most of the commonly reported variants, none of these variants were present in our cohort, suggesting that NF1 mutations are less frequent compared to other populations and/or are located in different genomic regions. Instead, we identified one variant of uncertain significance (rs2151559381) present in one patient, which has not been previously reported in patients with astrocytoma.
Finally, we examined the BRAF gene for the classic V600E mutation, which is traditionally associated with epithelioid features of glioblastoma39. Despite adequate coverage, sequencing revealed no BRAF mutations in our cohort.
Survival analyses—association with relevant genes and clinical data
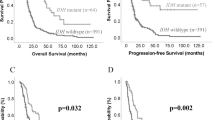
Next, we compared the overall survival curves between patient groups defined by the presence of IDH mutations, TERTp mutations, or a combination of both, as the prognostic significance of these markers has been well documented26,27. In our study, IDH mutations were associated with significantly longer overall survival than IDH wildtype patients (32.8 vs. 7.2 months; p < 0.01; Fig. 2A), while patients with TERTp mutations had lower overall survival than those without TERTp mutations (6.4 vs. 16.3 months; p < 0.01; Fig. 2B). Our analysis revealed that EGFR, TP53, PTEN, and CDKN2A were not associated with a significant difference in overall survival (Suppl. Figure 1), which is consistent with previous reports40. Moreover, comparing survival curves based on TERTp mutations in IDH wildtype patients resulted in sustained survival differences (10.5 vs. 6.4 months; mantel-cox and logrank test for trend p < 0.01; Fig. 2C), highlighting the additive role of these mutations in predicting patient survival outcomes.
Stratified overall survival curves. Stratified Kaplan-Meier curves and distribution of mutation status are presented for (A) IDH mutations, (B) TERTp mutations and (C) a combined classification using IDH and TERTp mutations. Log-rank test was performed to compare curves, and double asterisks indicate p-values < 0.01. IDH Isocitrate Dehydrogenase, TERTp telomerase reverse transcriptase promoter.
Regression analyses were conducted to evaluate the prognostic value of different clinical and molecular determinants in IDH wildtype tumors. We included all variables with univariate p-values < 0.25 in a multivariate analysis. The coefficients, confidence intervals, and significance values for univariate and multivariate analyses of each prognostic factor are shown in Table 2.
Our final multivariable regression model identified three factors capable of explaining overall survival in our HGA patients, all of them related to achieving optimal therapeutic goals: total versus subtotal resection, as well as radiotherapy and/or chemotherapy access. It is noteworthy that TERTp mutations exhibited significant interactions with other prognostic factors, such as age, postoperative Karnofsky Performance Status (KPS), and the utilization of radio- and chemotherapy (p < 0.01). These interactions could explain the lack of significance when analyzed together with these variables. For instance, patients who did not receive chemotherapy—often due to poorer clinical status or advanced age—were more likely to harbor TERTp mutations, limiting our ability to isolate the prognostic impact of this genetic alteration. Although this association is occasionally observed in clinical practice, it was likely exacerbated in our cohort due to limited access to chemotherapy. On the other hand, we did not include patients with IDH-mutant tumors in the regression analyses, to prevent any interaction effects that may influence the impact of other variables on survival.
Discussion
Numerous studies have elucidated ethnic variations in the molecular markers associated with various cancer types in Latin American populations41,42. However, Hispanic and Latino populations have largely been underrepresented in HGA research43. While some reports have indicated disparities in both the incidence and prognosis of HGA among individuals of Latin American and non-Latino backgrounds41,44no comprehensive comparisons have been made regarding the molecular features of astrocytomas between Latin American and other populations.
Here, we conducted the first comprehensive molecular profiling of a Latin American HGA cohort to establish a basis for potential international comparisons. Our study on HGA in a Chilean population revealed findings comparable to those of other cohorts of North American, European, and Asian origin, suggesting a universal relevance of key molecular markers such as IDH, TERTp, TP53, and PTEN mutations as well as EGFR amplification and CDKN2A deletions5,20,21,25,30. Novel TP53 and PTEN variants were identified and deemed likely pathogenic, based on in silico predictions. Mutations in the ATRX and SMARCAL1 genes related to Alternative Lengthening of Telomeres (ALT) were observed less frequently than in other studies36,37. Nevertheless, the disparity in our results might be attributed, at least partially, to the design of our panel and partial coverage of some of the most commonly mutated regions within these genes. In addition, lower than expected mutation rates were found in NF1.
Given the recent updates in central nervous system tumor classification, the capacity to clinically identify mutations will become increasingly important in the upcoming years. For instance, in our series, IDH sequencing allowed for the re-categorization of a patient with negative IDH R132H IHC and an R172K mutation identified by NGS, moving from a glioblastoma to a grade 4 IDH-mutant astrocytoma. In addition, the relatively older age of patients with IDH mutations in this study suggests that the clinical use of DNA sequencing could be more relevant than that currently considered in patients over 55 years of age in our population. Similarly, our results re-categorized a patient with a diffuse midline glioma, H3 K27-altered, which is a pediatric-type tumor with worse prognosis than glioblastoma22,24.
Although the cost of sequencing has dramatically decreased over the last 15 years, the implementation and overall costs of the complete sequencing process remain very high for use in most developing countries, making a simplified targeted sequencing strategy, such as that used in this study, an attractive alternative. In this study, we optimized the conditions to reduce the risk of false negatives (DNA extraction from highly cellular tumor tissues, designing sequencing panels with high coverage depth, and setting low read number limits for known variants). However, standardized clinical implementation also requires the use of positive controls to establish the sensitivity and specificity of the sequencing strategy in a local context45.
Study limitations
We acknowledge some limitations of the current study: (i) limited access to chemotherapy and radiotherapy has been a frequent occurrence in Latin American health systems during the last decade, and it generates a heterogeneous population to study the prognostic effect of genetic markers. (ii) the extent of resection was not systematically measured using immediate postoperative magnetic resonance imaging. (iii) O(6)-methylguanine-DNA methyltransferase (MGMT) promoter methylation was not determined, hence we could not evaluate its predictive value for temozolomide response46 in our cohort. (iv) Determination of gene copy number changes using amplicon sequencing as opposed to whole-genome sequencing has inherent limitations, mainly associated with coverage heterogeneity in different regions of interest19. These limitations were partially mitigated by bioinformatic processing of the data (see Methods). (v) Ancestry was determined by geography and self-report only, raising the possibility of an inadvertent mixture of genomic ancestries.
Our findings contribute to the global understanding of astrocytoma genetics and underscore the importance of inclusive research for advancing personalized medicine. Given that our study utilizes a targeted-sequencing approach, future research should expand upon these findings using more comprehensive methods and control data. This will help to further elucidate the genetic landscape of high-grade astrocytomas (HGA) in this population.
Conclusion
Our HGA cohort from Latin America exhibited an incidence of IDH, TERTp, PTEN, and TP53 mutations comparable to those reported in other studies with different ethnic origins. Conversely, NF1, ATRX, and SMARCAL1 mutations had a lower frequency than expected. The correlation between IDH and TERTp mutations and patient survival indicated a similar association, as reported in previous studies in populations of non-Latino American, European, and Asian origin. This study contributes to the advancement of future international comparative studies, potential strategies for treating HGA and population-specific health strategies. Our findings highlight the multifaceted nature of HGA, underscoring its complex molecular origins and the distinctive genetic diversity that exists among various populations. We emphasize the importance of ensuring equitable access to advanced molecular diagnostics, shedding light on the need for and approaches to the widespread adoption of state-of-the-art technologies in healthcare systems.
Data availability
All raw sequencing data can be found on the Gene Expression Omnibus (GEO) website: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE252870.
References
Weinstein, J. N. et al. The Cancer genome atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013).
Barthel, F. P. et al. Longitudinal molecular trajectories of diffuse glioma in adults. Nature 576, 112–120 (2019).
Vaubel, R. A. et al. Genomic and phenotypic characterization of a broad panel of Patient-Derived xenografts reflects the diversity of glioblastoma. Clin. Cancer Res. 26, 1094–1104 (2020).
Koo, H. et al. Ethnic delineation of primary glioblastoma genome. Cancer Med. 9, 7352–7359 (2020).
Ceccarelli, M. et al. Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell 164, 550–563 (2016).
Chacón-Duque, J. C. et al. Latin Americans show wide-spread Converso ancestry and imprint of local Native ancestry on physical appearance. Nature Communications 2018 9:1 9, 1–13 (2018).
Ruiz-Linares, A. et al. Admixture in Latin america: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet 10, (2014).
Eyheramendy, S., Martinez, F. I., Manevy, F., Vial, C. & Repetto, G. M. Genetic structure characterization of Chileans reflects historical immigration patterns. Nature Communications 2015 6:1 6, 1–10 (2015).
Verdugo, R. A. et al. Development of a small panel of SNPs to infer ancestry in Chileans that distinguishes Aymara and Mapuche components. Biol Res 53, (2020).
Rocco, P. Composición genética de La Población chilena. Distribución de Polimorfismos de DNA mitocondrial En grupos originarios y En La Población Mixta de Santiago. Rev. Med. Chil. 130, 125–131 (2002).
Vieira-Machado, C. D. et al. Uniparental ancestry markers in Chilean populations. Genet. Mol. Biol. 39, 573–579 (2016).
Sahm, F. et al. Molecular diagnostic tools for the world health organization (WHO) 2021 classification of gliomas, glioneuronal and neuronal tumors; an EANO guideline. Neuro Oncol. 25, 1731–1749 (2023).
Louis, D. N. et al. The 2021 WHO classification of tumors of the central nervous system: a summary. Neuro Oncol. 23, 1231–1251 (2021).
Horbinski, C., Berger, T., Packer, R. J. & Wen, P. Y. Clinical implications of the 2021 edition of the WHO classification of central nervous system tumours. Nature Reviews Neurology 2022 18:9 18, 515–529 (2022).
Berger, T. R., Wen, P. Y., Lang-Orsini, M. & Chukwueke, U. N. World health organization 2021 classification of central nervous system tumors and implications for therapy for Adult-Type gliomas: A review. JAMA Oncol. 8, 1493–1501 (2022).
Kurokawa, R. et al. Major changes in 2021 world health organization classification of central Nerus system tumors. Radiographics 42, 1474–1493 (2022).
Gritsch, S., Batchelor, T. T. & Gonzalez Castro, L. N. Diagnostic, therapeutic, and prognostic implications of the 2021 world health organization classification of tumors of the central nervous system. Cancer 128, 47–58 (2022).
Eyheramendy, S., Martinez, F. I., Manevy, F., Vial, C. & Repetto, G. M. Genetic structure characterization of Chileans reflects historical immigration patterns. Nat Commun 6, (2015).
Petrackova, A. et al. Standardization of sequencing coverage depth in NGS: recommendation for detection of clonal and subclonal mutations in Cancer diagnostics. Front. Oncol. 9, 851 (2019).
Yan, H. et al. IDH1 and IDH2 mutations in gliomas. N Engl. J. Med. 360, 765 (2009).
Killela, P. J. et al. Mutations in IDH1, IDH2, and in the TERT promoter define clinically distinct subgroups of adult malignant gliomas. Oncotarget 5, 1515 (2014).
Castel, D. et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic Pontine gliomas with different prognosis and phenotypes. Acta Neuropathol. 130, 815–827 (2015).
Schwartzentruber, J. et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 482, 226–231 (2012).
Sturm, D. et al. Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 22, 425–437 (2012).
Pekmezci, M. et al. Adult infiltrating gliomas with WHO 2016 integrated diagnosis: additional prognostic roles of ATRX and TERT. Acta Neuropathol. 133, 1001–1016 (2017).
Simon, M. et al. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro Oncol. 17, 45–52 (2015).
Labussière, M. et al. TERT promoter mutations in gliomas, genetic associations and clinico-pathological correlations. Br. J. Cancer. 111, 2024–2032 (2014).
Nguyen, H. N. et al. Human TERT promoter mutation enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol. 19, 394–404 (2017).
Appay, R. et al. CDKN2A homozygous deletion is a strong adverse prognosis factor in diffuse malignant IDH-mutant gliomas. Neuro Oncol. 21, 1519 (2019).
Brennan, C. W. et al. Somatic Genomic Landsc. Glioblastoma Cell 155, 462–477 (2013).
Hoxhaj, G. & Manning, B. D. The PI3K–AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer. 20, 74–88 (2019).
Hernández Borrero, L. J. & El-Deiry, W. S. Tumor suppressor p53: biology, signaling pathways, and therapeutic targeting. Biochim. Et Biophys. Acta (BBA) - Reviews Cancer. 1876, 188556 (2021).
Gao, J. et al. Integrative analysis of complex Cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 (2013).
Cerami, E. et al. The cBio Cancer genomics portal: an open platform for exploring multidimensional Cancer genomics data. Cancer Discov. 2, 401 (2012).
Liu, H. et al. Cancer-associated SMARCAL1 loss-of-function mutations promote alternative lengthening of telomeres and tumorigenesis in telomerase-negative glioblastoma cells. Neuro Oncol. 25, 1563–1575 (2023).
Haase, S. et al. Mutant ATRX: Uncovering a new therapeutic target for glioma. Expert Opin. Ther. Targets. 22, 599 (2018).
Diplas, B. H. et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat Commun 9, (2018).
Jonsson, P. et al. Genomic correlates of disease progression and treatment response in prospectively characterized gliomas. Clin. Cancer Res. 25, 5537–5547 (2019).
McNulty, S. N. et al. BRAF mutations May identify a clinically distinct subset of glioblastoma. Sci. Rep. 11, 1–10 (2021).
Kraus, J. A. et al. Molecular analysis of the PTEN, TP53 and CDKN2A tumor suppressor genes in long-term survivors of glioblastoma multiforme. J. Neurooncol. 48, 89–94 (2000).
Patel, N. P., Lyon, K. A. & Huang, J. H. The effect of race on the prognosis of the glioblastoma patient: a brief review. Neurol. Res. 41, 967–971 (2019).
Raez, L. E., Cardona, A. F., Arrieta, O. & Lopes, G. Lung Cancer disparities in hispanics: molecular diagnosis and use of immunotherapy. JCO Glob Oncol. 784–788. https://doi.org/10.1200/go.20.00004 (2020).
Guerrero, S. et al. Analysis of racial/ethnic representation in select basic and applied Cancer research studies. Sci. Rep. 8, 1–8 (2018).
Walsh, K. M. et al. Influence of county-level geographic/ancestral origin on glioma incidence and outcomes in US Hispanics. Neuro Oncol. 25, 398–406 (2023).
Strom, S. P. Current practices and guidelines for clinical next-generation sequencing oncology testing. Cancer Biol. Med. 13, 3 (2016).
Brandner, S. et al. MGMT promoter methylation testing to predict overall survival in people with glioblastoma treated with temozolomide: a comprehensive meta-analysis based on a Cochrane systematic review. Neuro Oncol. 23, 1457–1469 (2021).
Acknowledgements
This study is dedicated to Virgina Flores Kehr (CH family member), who died of glioblastoma during the study. We thank Claudia Sepulveda and Susana Manriquez for their laboratory management and administrative accounting. We also thank Pamela Moller for her technical support in immunohistochemistry and sample management.
Funding
Agencia Nacional de Investigación y Desarrollo (FONDECYT 1220573 to CH, FONDAP 15150012 to CH, FONDEF ID1ID22I10120 to CH, FONDEF NAM22I0057 to CH, FONDECYT 11180825 to HU, postdoctoral FONDECYT 3210294 to PP); Department of Neurological Sciences of the University of Chile; U.S. Air Force Office of Scientific Research (FA9550-21-1-0096 to CH); Department of Defense (W81XWH2110960 to CH), Millennium Institute (P09-015-F to CH); and Swiss Consolidation Grant -The Leading House for the Latin American Region (to CH).
Author information
Authors and Affiliations
Contributions
Project conceptualization: RF, JM, and CH; Methodology: RF, JM and CH; Formal analysis: RF, MS, GC and PP; Investigation: RF, HU, PP, BF, CS, DR, RM, and CT.Software: MS; Data curation: RF, HU, PP; Resources: CS and CH; Writing – Original draft: RF, HU and PP; Writing – Review and editing: all authors; Supervision and project administration: JM and CH; Funding acquisition: RF, DR, JM and CH.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fernández-Gajardo, R., Urra, H., Sáez, M. et al. Characterization of the mutational status of glioblastoma and high-grade astrocytomas in a Latin American cohort. Sci Rep 15, 34485 (2025). https://doi.org/10.1038/s41598-025-06129-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-06129-z