Abstract
Recent advances in state space models (SSMs) have demonstrated remarkable efficiency in modeling long-range dependencies, yet their application to 3D medical image segmentation remains underexplored. This paper introduces CDA-Mamba (Cross-Directional Attention Mamba), a novel hybrid architecture that combines the efficiency of SSMs with the strengths of convolutional and attention mechanisms to address the unique challenges of 3D medical image segmentation. CDA-Mamba features three key innovations: a Multi-Frequency Gated Convolution (MFGC) module to enhance spatial and frequency-domain feature integration, a Tri-Directional Mamba module to capture volumetric dependencies across orthogonal dimensions, and Selective Self-Attention integration in high-semantic layers to balance computational efficiency with global context modeling. Comprehensive experiments on the BraTS2023 brain tumor segmentation dataset highlight the competitive performance of CDA-Mamba, which achieves an average Dice score of 91.44. Moreover, evaluations on the AIIB2023 airway segmentation dataset further validate its effectiveness, with CDA-Mamba attaining the highest IoU of 88.72 and a DLR of 71.01. These results underscore its ability to balance accuracy and efficiency in 3D medical image segmentation.
Similar content being viewed by others
Introduction
Accurate and efficient 3D medical image segmentation is of paramount importance for a wide range of clinical applications, including patient diagnosis, treatment planning, and disease monitoring1,2. Precisely delineating anatomical structures and pathological regions within volumetric data improves patient outcomes and significantly reduces the workload for medical professionals. Conventional convolutional neural networks (CNNs) have demonstrated considerable promise in medical image analysis3,4,5. However, due to the inherent locality of convolutional operations, CNN-based methods are typically limited in effectively capturing long-range spatial dependencies and global contextual information. In practical segmentation tasks, this limitation manifests as difficulties in accurately segmenting large-scale anatomical structures or pathological regions that span multiple slices or exhibit complex spatial relationships, leading to suboptimal segmentation performance6,7.
Recently, inspired by the success of Transformers8 in modeling long-range dependencies, researchers have reframed volumetric (3D) medical image segmentation as a sequence-to-sequence prediction problem. The Transformer architecture9,10,11, –12,10,11,, leveraging self-attention mechanisms capable of capturing global relationships, has garnered significant attention in medical image segmentation. Models such as UNETR13 and SwinUNETR14, which follow the popular “U-shaped” encoder-decoder structure, demonstrate promising capabilities in capturing contextual information within 3D medical images. Nevertheless, the self-attention mechanism’s quadratic computational complexity with respect to sequence length poses substantial computational burdens, including excessive GPU memory usage and slow inference speed. These drawbacks severely hinder the practical deployment of Transformer-based methods in clinical scenarios, especially in resource-constrained environments such as edge devices or smaller healthcare facilities15.
To address the inherent limitations of both CNNs and Transformers, the Mamba architecture16, based on state space models (SSMs), has emerged recently as a promising alternative. Mamba not only effectively models long-range dependencies but also achieves linear computational complexity with respect to input sequence length. Owing to its state-space formulation and hardware-aware design, Mamba exhibits remarkable efficiency in processing sequential data, particularly demonstrated in natural language processing tasks17. Several recent studies have extended Mamba into computer vision, including U-Mamba18 and Vision Mamba19, showcasing its potential for efficient global context modeling. However, existing Mamba-based approaches still overlook specific challenges in 3D medical imaging, such as effectively modeling spatial relationships across multiple directions and efficiently capturing multi-frequency, high-resolution spatial information, which are critical to accurate segmentation.
Considering these unmet challenges, there is a clear need for an architecture specifically tailored to 3D medical image segmentation that combines efficient long-range dependency modeling with effective spatial and frequency-domain feature representation.
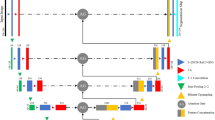
The lrchitecture of CDA-Mamba: an encoder-decoder framework with mamba blocks. The MFGC module (Fig. 2) enhances feature representation by incorporating multi-frequency information, capturing both fine details and global structures. The Tri-Directional Mamba module (Fig. 3) models volumetric dependencies by sequentially processing three orthogonal views. Viewing Figs. 1, 2 and 3 together clarifies the interactions between these components.
Motivated by these limitations, we propose CDA-Mamba (Cross-Directional Attention Mamba), a novel architecture specifically designed to overcome these challenges by effectively modeling multi-directional spatial dependencies and selectively integrating computationally intensive self-attention mechanisms at critical semantic stages. Our core contributions can be summarized as follows:
-
1.
Multi-frequency gated convolution (MFGC) module for enriched feature representation we introduce the MFGC module as a critical preparatory step before Mamba processing. Standard convolutions often struggle to simultaneously capture the varying frequencies of spatial information present in complex 3D medical scans. The MFGC module explicitly extracts features across multiple frequency bands. Crucially, it employs a sophisticated gating mechanism to dynamically integrate these multi-frequency features with spatial information. This gating process is designed to selectively emphasize channels that carry salient diagnostic information while actively suppressing irrelevant noise and redundant features. This design enables the model to capture both fine-grained details and broad structural contexts, improving its ability to effectively process complex medical images.
-
2.
Tri-directional mamba module for comprehensive volumetric dependency modeling to holistically capture global context within 3D feature maps, we propose a novel tri-directional Mamba application. Directly applying sequence models to flattened 3D data can lead to the loss of crucial spatial relationships, while processing full 3D volumes with other methods is often computationally prohibitive for long-range dependency modeling. Our approach innovatively decomposes the 3D feature volume into three orthogonal sets of 1D sequences—along the axial, sagittal, and coronal axes. Each set of sequences is processed by a dedicated Mamba block. This tri-directional scanning enables the model to efficiently capture long-range dependencies from three distinct spatial perspectives (height, width, and depth). This multi-axial modeling provides a more robust and comprehensive understanding of the anisotropic nature of volumetric data, which is vital for accurately segmenting structures with complex 3D morphologies..
-
3.
Selective self-attention integration for balanced efficiency and global context we propose a judicious integration of self-attention mechanisms to augment Mamba’s capabilities without incurring excessive computational overhead. While Mamba is efficient for long-range dependencies, the quadratic complexity of self-attention makes its widespread use in deep networks computationally burdensome, especially for high-resolution 3D data. However, self-attention excels at capturing an all-to-all global context. Instead of uniformly applying self-attention, we strategically integrate self-attention blocks only into the final two, higher-semantic layers of the Mamba encoder. These deeper layers typically encode more abstract and global features, where a comprehensive understanding of context is most beneficial.
Through these architectural innovations, CDA-Mamba strikes an optimal balance between efficiency and accuracy, establishing itself as a promising solution for 3D medical image segmentation. Extensive experiments on the BraTS202320 and AIIB202321 datasets validate its effectiveness.
Related work
State Space Models (SSMs) have recently demonstrated remarkable success across various domains22,23,24,– 25,23,24,. However, their application to 3D medical image segmentation remains largely unexplored. This paper introduces CDA-Mamba (Cross-Dimensional Attention Mamba), a novel hybrid architecture specifically designed for this task.
Challenges in 3D Medical Image Segmentation: Effectively modeling long-range spatial dependencies and managing the computational burden associated with high-resolution volumetric data pose significant challenges for 3D medical image segmentation. While Transformer models have advanced the field by leveraging self-attention mechanisms to capture global context, their quadratic complexity limits their efficiency and practicality when processing high-resolution 3D medical images26. This necessitates the exploration of more efficient alternatives, such as SSM-based architectures.
Exploration of Mamba-based Vision Models: The Mamba architecture16, an efficient SSM, has witnessed some exploration in computer vision. Approaches like U-Mamba18, Vision Mamba19, and its variants27,28 have demonstrated Mamba’s potential for feature extraction and global context modeling. However, existing methods generally lack optimization for the specific challenges inherent in 3D medical image segmentation. SegMamba29 proposed a model integrating a U-shaped structure with Mamba for modeling global volumetric features at different scales, incorporating a gated spatial convolution module. Nevertheless, it fails to leverage the combined strengths of Convolutional Neural Networks (CNNs) and Transformers, nor does it effectively integrate cross-dimensional information. We propose CDA-Mamba, which introduces a novel multi-dimensional feature fusion strategy to combine Mamba’s efficiency with the advantages of cross-dimensional features, as shown in Fig. 1. Moreover, by selectively integrating self-attention modules, CDA-Mamba enhances the modeling of long-range spatial dependencies while preserving computational efficiency.
Multi-Frequency Analysis and Attention Mechanisms: Concurrent research efforts have explored the integration of multi-frequency techniques and attention mechanisms, aiming to enhance the extraction of both local and global context from fine-grained to coarse-grained information30,31. Specifically, the 2D DCT32,33 has been widely employed in computer vision for its compression capabilities and ability to extract frequency statistics, thereby improving representational power. MADGNet34 demonstrated progress in 2D medical image segmentation by combining multi-frequency information with multi-scale features, enhancing the model’s ability to detect subtle variations in lesion characteristics. However, this approach is not directly applicable to 3D images. To address this limitation, we develop a Multi-Frequency Gated Convolution (MFGC) tailored for 3D images.
Method
Multi-frequency gated convolution module
When processing 3D medical images, although the Mamba architecture exhibits linear time complexity and efficiently captures long-range dependencies, it is primarily based on state space models (SSMs) and is inherently designed for sequential modeling. This characteristic poses certain limitations in capturing local spatial details and multi-frequency features. For instance, the Mamba structure struggles to adequately extract high-frequency information in medical images, such as fine edge structures, texture details, and contrast variations between different tissues. However, medical imaging modalities such as MRI and CT often exhibit rich high-frequency variations, which are crucial for accurate segmentation. To address this limitation, it is essential to extract effective spatial and frequency-domain features before feeding them into the Mamba architecture for medical image segmentation. Given that medical images typically exhibit more pronounced high-frequency variations due to their imaging modalities, we introduce the Multi-Frequency Gated Convolution (MFGC) module to incorporate multi-frequency information, complementing it with the proposed 3D Multi-Frequency Channel Attention (3D MFCA) module. These components work together to enhance feature extraction, ensuring that both spatial and frequency-domain information are effectively captured and utilized to improve segmentation performance. Specifically, by leveraging the 3D Discrete Cosine Transform (3D DCT)32,33, the 3D MFCA module captures feature distributions in both the spatial and frequency domains, generating channel attention maps that adaptively enhance important channels while suppressing irrelevant ones. This fusion of multi-frequency information significantly strengthens the network’s ability to discern fine details and reduce noise, thereby improving segmentation accuracy. The overall structure of MFGC is illustrated in Fig. 2.
The input tensor is processed through two parallel branches. The first branch applies two consecutive 3D convolutions, each followed by instance normalization and ReLU activation, capturing local spatial patterns with deeper receptive fields. The second branch utilizes a single 3D convolution combined with instance normalization and ReLU. After processing through the dual branches, the resulting features are concatenated along the channel dimension and subsequently fused using a \(1 \times 1 \times 1\) convolution.
To further enhance the fused representation, we adopt a new 3D Multi-Frequency Channel Attention (3D MFCA) mechanism. Consider an input feature map at scale s for the \(i_{\text {th}}\) sample, whose spatial dimensions are \(\left( D_s, H_s, W_s\right)\), and whose channel dimension is \(C_s\). Denote this feature map by
within the 3D space, suppose a set of Discrete Cosine Transform (DCT) frequency indices is selected, \(\{(z_k, u_k, v_k) \mid k = 1, 2, \dots , K\}\), where K denotes the number of chosen frequency components. We introduce the 3D DCT basis function:
where \(0 \le d < D_s\), \(0 \le h < H_s\), and \(0 \le w < W_s\). By multiplying the input feature map channel-wise with this basis and summing over the spatial dimensions, one obtains
where the notation “ : ” indicates operation along the channel dimension. The resulting \(X_{i}^{s,k}\) captures the channel-wise projection onto the \(k_{th}\) 3D DCT frequency component, thereby encoding the frequency-domain properties of the original feature map.
Each basis function corresponds to a different frequency component. By performing element-wise multiplication between the basis functions and the input feature maps, followed by summation along the spatial dimensions, we obtain the projection of each channel in the corresponding frequency domain. Since high-frequency components capture sharp edges and fine details, while low-frequency components represent the overall structure, incorporating frequency-domain features effectively enhances the network’s sensitivity to subtle details and its ability to capture structural information.
To capture diverse statistical characteristics, global average pooling, global max pooling, and global min pooling (often implemented via negative inversion followed by max pooling) are applied to each \(X_{i}^{s,k}\). These yield:
These statistics from all K frequency components are then aggregated (e.g., via mean pooling or other feasible strategies) to obtain three global statistics:
Subsequently, \(\{Z_{\text {avg}}^s, Z_{\text {max}}^s, Z_{\text {min}}^s\}\) are further processed to generate the channel attention map. These transformations can be mathematically represented as:
where \(W_1 \in {\mathbb {R}}^{\frac{C_s}{r} \times C_s}\), \(W_2 \in {\mathbb {R}}^{C_s \times \frac{C_s}{r}}\). here, r denotes the channel reduction ratio, while \(\delta (\cdot )\) represents a non-linear activation function and \(\sigma (\cdot )\) denotes the Sigmoid function. The summation over \(d \in \{\text {avg}, \text {max}, \text {min}\}\) indicates the element-wise addition of the transformed representations derived from each pooling statistic, followed by normalization using the Sigmoid operation.
Finally, the resulting channel attention vector \(M_i^s\) is broadcast to match the original feature map’s spatial dimensions and multiplied element-wise with \(X_i^s\) along the channel axis:
where \(\otimes\) denotes channel-wise multiplication. By emphasizing significant channels and diminishing the influence of less relevant ones, the network can better focus on features crucial to the target task, thereby improving its performance in medical image segmentation. Furthermore, in the Multi-Frequency Gated Convolution module, a residual connection is introduced following the 3D MFCA block to retain the original feature information and mitigate potential performance degradation.
Tri-directional mamba module
To comprehensively capture the spatial dependencies in 3D medical images, we design the Tri-Directional Mamba Module, which effectively models volumetric relationships by sequentially processing three orthogonal perspectives of the input tensor, as illustrated in Fig. 3.
In terms of architecture, given an input feature map \({\widetilde{X}}_i^s \in {\mathbb {R}}^{C_s \times D_s \times H_s \times W_s}\), the module first establishes a skip connection to preserve the original input and ensures that the channel dimension \(C_s\) aligns with the model’s internal parameter dimension. The module then iteratively processes the three orthogonal axes depth, height, and width. For each axis, the tensor is reshaped and permuted to designate the selected axis as the sequential dimension, resulting in a flattened tensor.
To ensure feature stability and effectiveness, a LayerNorm operation is applied before passing the features into the Mamba block. The Mamba block integrates convolutional operations with a hidden state of predefined dimensions, enabling the joint modeling of local features and long-range dependencies. This design allows the module to simultaneously capture both spatially localized structures and global contextual information. The computation within the Tri-Directional Mamba Module can be formalized as follows:
here, \({\widetilde{X}}_i^s\) denotes the input feature map, while \(f_d(\cdot )\), \(f_h(\cdot )\), and \(f_w(\cdot )\) are axis-specific flattening functions corresponding to the depth, height, and width dimensions, respectively. \(\text {LN}(\cdot )\) represents the LayerNorm operation, and the Mamba block processes the normalized tensors to capture both local and global dependencies.
After processing through the Mamba block, the outputs are permuted back to their original shapes. The three transformed tensors are then aggregated via element-wise summation to integrate multi-axis contextual information. Finally, the fused output is combined with the input feature map in a residual manner, ensuring the retention of input information and facilitating gradient flow. The tri-directional orthogonal processing strategy of this module comprehensively captures feature dependencies in 3D space. This approach not only enhances the model’s ability to handle complex volumetric data but also leverages the strengths of the Mamba architecture for efficient feature extraction and modeling. By preserving computational efficiency, this design markedly enhances the model’s capacity to capture multi-frequency spatial dependencies, making it especially well-suited for tasks such as medical image segmentation.
Self-attention in CDAMamba encoder
Our feature encoder is built upon the CDAMamba block, which is capable of extracting multi-scale and multi-frequency features. Self-attention blocks are strategically integrated into the last two layers of the CDAMamba encoder, rather than being applied across the entire network, as illustrated in Fig. 1. This strategy of employing self-attention mechanisms at lower resolutions enables the capture of fine-grained details in both short and long range spatial dependencies, while significantly reducing the high computational cost associated with their universal application. Building on insights from previous studies7,14,35, we employ a CNN-based decoder with skip connections to generate the segmentation results.
Experiment results
Experiment settings
In this study, we evaluate the model’s segmentation performance on two distinct medical image segmentation datasets: BraTS202320 and AIIB202321. The raw data were split into 70% for training, 10% for validation, and 20% for testing. For the experiments, 3D ROI (Region of Interest) cropping was applied with a window size of \(128 \times 128 \times 128\) to ensure efficient processing of high-resolution medical imaging data. For the BraTS2023 dataset, each sample includes four imaging modalities: native T1, post-contrast T1-weighted (T1Gd), T2-weighted (T2), and T2 Fluid Attenuated Inversion Recovery (T2-FLAIR). Consequently, the input tensor has 4 channels. The output tensor also has 4 channels, corresponding to 3 segmentation classes—WT (whole tumor), ET (enhancing tumor), and TC (tumor core)—along with the background. In contrast, the AIIB2023 dataset is a publicly available airway segmentation dataset consisting of high-resolution computed tomography (HRCT) scans. For this dataset, the input tensor has 1 channel, and the output tensor has 2 channels, representing the segmentation of airway structures. The model optimization was performed using the SGD optimizer with Nesterov momentum acceleration39. Compared to the AdamW optimizer, the SGD optimizer demonstrated superior robustness and generalization performance for our tasks. To enhance the model’s generalizability across different data distributions, we incorporated a variety of data augmentation techniques, including random flipping, random cropping, mirroring, gamma correction, and elastic distortions. All experiments were conducted on a server equipped with two NVIDIA V100 GPUs.
Comparison with state-of-the-art models
We compared CDAMamba with seven state-of-the-art (SOTA) segmentation methods, including nnUNet3, TransUNet36, UNETR13, Swin-UNETR37, Swin-UNETR v214, MedNeXt38, and SegMamba29. As shown in Table 1, CDAMamba achieved the highest average Dice Similarity Coefficient (DSC) across all segmentation categories in the BraTS2023 dataset, demonstrating its outstanding segmentation performance. Specifically, CDAMamba attained an average DSC of 91.44%, outperforming the second-best method, SegMamba, by 0.12%. For the WT (Whole Tumor) category, CDAMamba achieved a DSC of 93.84%, surpassing SegMamba and Swin-UNETR v2 by 0.24% and 0.46%, respectively. Similarly, for the TC (Tumor Core) and ET (Enhancing Tumor) categories, CDAMamba obtained DSC scores of 92.71% and 87.76%, which are 0.06% and 0.05% higher than those of SegMamba. The superior average DSC achieved by CDAMamba on BraTS2023, along with its leading performance in individual WT, TC, and ET categories, underscores the efficacy of its hybrid design. For instance, the notable 0.24% and 0.46% DSC improvement in the WT category over SegMamba and Swin-UNETR v2 can be attributed to the Tri-Directional Mamba module’s comprehensive modeling of 3D volumetric context, allowing for better delineation of the entire tumor extent. Concurrently, the MFGC module’s focus on multi-frequency information likely contributes to the precise segmentation of the TC and ET, which often exhibit heterogeneous textures and subtle boundary details that benefit from enhanced high-frequency feature extraction.
Compared to transformer-based models such as TransUNet and UNETR, CDAMamba consistently outperformed them in all categories. This demonstrates the advantage of integrating multi-scale and multi-frequency features into the model architecture, enabling more accurate segmentation of complex anatomical structures.
On the AIIB2023 dataset, CDAMamba once again achieved the best performance, with an IoU of 88.72%, a DLR (Dice for Large Regions) of 71.01%, and a DBR (Dice for Branch Regions) of 61.53%, exceeding the second-best method, SegMamba, by 0.13%, 0.80%, and 0.25%, respectively. These improvements underscore CDAMamba’s capability to handle intricate airway structures, particularly in challenging branch regions.
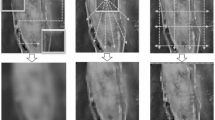
Figure 4 presents the ground truth (GT) alongside segmentation results from nnUNet, TransUNet, UNETR, Swin-UNETR, Swin-UNETR v2, MedNeXt, SegMamba, and the proposed CDAMamba. Transformer-based models (e.g., TransUNet and UNETR) demonstrate improved boundary delineation but still exhibit noticeable inconsistencies in regions with complex shapes or low contrast. SegMamba also shows competitive performance. Compared to other state-of-the-art methods, these visualizations highlight the capability of CDAMamba to accurately delineate tumor boundaries and capture fine-grained details.
CDAMamba outperformed all other models across both datasets, a success attributed to the innovative integration of multi-scale and multi-frequency features. This integration empowers the model to capture fine-grained details and broader structural information, making it highly effective for segmenting complex medical imaging data.
Ablation study
To evaluate the contribution of each module in the proposed CDAMamba architecture, we conducted an ablation study by systematically removing or altering specific components of the model. This analysis aims to identify the significance of each module in achieving accurate segmentation and robust performance. In the ablation study, we made the following modifications to the baseline CDAMamba model:
-
1.
Removal of the Multi-Frequency Gated Convolution (MFGC) Module: To evaluate the impact of integrating high- and low-frequency information.
-
2.
Exclusion of the Tri-Directional Mamba Module for feature fusion: To assess the contribution of the Tri-Directional Mamba mechanism.
-
3.
Elimination of the Self-Attention mechanism: To analyze the role of Self-Attention in the model.
To ensure consistency, all experiments were conducted under identical training and testing conditions, using the BraTS2023 dataset with the same hyperparameter settings.
The ablation study results in Table 2 highlight the impact of removing key components from the CDAMamba model. In Configuration 1, removing the Multi-Frequency Gated Convolution (MFGC) module led to a decline in average performance (90.19% vs. 91.44%) with the most significant drop in the Tumor Core (TC) region (89.76% vs. 92.71%). This underscores the MFGC module’s critical role in enhancing high-frequency feature extraction, which is particularly essential for accurately segmenting tumor cores. Given the smaller size and higher structural complexity of the tumor core (TC) region, it relies more on fine-grained texture and boundary details. The MFGC module facilitates the integration of multi-frequency information, preserving both global context and local high-frequency details, thereby improving segmentation accuracy, especially for TC. In Configuration 2, excluding the Tri-Directional Mamba module caused the largest performance drop (89.74% Avg), particularly in the Enhancing Tumor (ET) (85.87% vs. 87.76%) and TC (91.23% vs. 92.71%) regions. This demonstrates the module’s importance in feature fusion, enabling the model to handle complex tumor structures with consistency and accuracy. Its absence significantly reduced the model’s ability to capture intricate relationships among features. For Configuration 3, the removal of the Self-Attention mechanism led to a smaller but notable decline in performance (90.35% Avg), with the greatest impact in the Enhancing Tumor (ET) region (87.76% to 86.12%). This highlights the Self-Attention mechanism’s role in focusing on fine details and distinguishing subtle differences in low-contrast areas. Overall, the study demonstrates the necessity of each component for achieving robust and accurate tumor segmentation.
In addition to segmentation accuracy, computational efficiency is another critical factor determining the practical deployment of deep models in clinical settings. To comprehensively evaluate the practicality of CDA-Mamba, we compare its inference speed (in seconds per case) and segmentation accuracy (Dice score) against several state-of-the-art methods under identical experimental settings.
All experiments were conducted using the same input resolution of \(128^3\), on an NVIDIA Tesla V100S GPU with 32 GB memory. The inference time is measured by averaging the processing time across multiple test cases, ensuring a fair comparison of computational efficiency. As shown in Table 3, CDA-Mamba achieves significantly better segmentation accuracy (Dice score of 91.44%) while maintaining competitive inference speed (1.98s per case), outperforming most previous Transformer-based methods (TransUNet36, UNETR13, Swin-UNETR37) and CNN-based methods (nnUNet3). Notably, CDA-Mamba provides superior segmentation performance comparable to SegMamba29 but with faster inference speed, highlighting its practical advantage, especially in resource-constrained clinical environments.
To evaluate the impact of the self-attention mechanism at different layers of the CDA-Mamba model, we conducted comparative experiments. Specifically, we employed a variety of strategies to append self-attention modules after the TDAMamba blocks and analyzed the model’s performance based on the average Dice coefficient. The experimental results on the BraTS2023 Dataset are presented in Table 4. As shown in the table, the models with self-attention applied after all TDAMamba blocks (SA after all TDAMamba blocks) and after the last two TDAMamba blocks (SA after the last 2 TDAMamba blocks) achieved comparable performance, with average Dice coefficients of 91.42 and 91.44, respectively. This suggests that integrating self-attention into the later stages of the model effectively enhances segmentation performance. In contrast, models incorporating self-attention after the first and second TDAMamba blocks (SA after 1st & 2nd TDAMamba blocks) or after the first and third TDAMamba blocks (SA after 1st & 3rd TDAMamba blocks) exhibited relatively lower performance. This further corroborates that incorporating self-attention in the early layers yields limited performance gains while incurring increased computational overhead. Overall, the experimental results demonstrate that selectively applying self-attention in the later layers of the CDA-Mamba model significantly improves segmentation performance while avoiding unnecessary computational costs.
Conclusion
In this work, we introduced CDA-Mamba, a novel segmentation model specifically designed for 3D medical image analysis. By integrating the Multi-Frequency Gated Convolution (MFGC) module, the Tri-Directional Mamba mechanism for comprehensive feature fusion, and the Selective Self-Attention Integration strategy, CDA-Mamba effectively addresses the critical challenges of multi-dimensional feature integration, spatial-frequency feature fusion, and fine-grained detail extraction. A comprehensive ablation study clearly demonstrated the pivotal roles of each proposed component in achieving state-of-the-art segmentation performance. Competitive experimental results on both the BraTS2023 and AIIB2023 datasets validated the effectiveness and efficiency of the proposed architecture. Particularly, CDA-Mamba exhibits an optimal balance between segmentation accuracy and computational efficiency, making it well-suited for clinical applications involving volumetric medical data. Despite its promising capabilities, CDA-Mamba still faces certain limitations. First, the current architecture has not yet been validated extensively on extremely high-resolution medical images or multi-modal medical data, potentially constraining its generalizability across diverse clinical scenarios. Second, although CDA-Mamba improves computational efficiency over transformer-based methods, further optimization is required for edge computing environments with stringent hardware constraints.
Future research directions include extending CDA-Mamba to handle multi-modal and higher-resolution medical imaging data, integrating domain adaptation techniques to enhance generalization across different imaging modalities and clinical sites, and further optimizing the model for deployment on edge devices and other resource-constrained clinical environments.
References
De Fauw, J. et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat. Med. 24(9), 1342–1350 (2018).
Ouyang, D. et al. Video-based AI for beat-to-beat assessment of cardiac function. Nature 580(7802), 252–256 (2020).
Isensee, F. et al. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211 (2021).
Umirzakova, S., Mardieva, S., Muksimova, S., Ahmad, S. & Whangbo, T. Enhancing the super-resolution of medical images: Introducing the deep residual feature distillation channel attention network for optimized performance and efficiency. Bioengineering 10(11), 1332. https://doi.org/10.3390/bioengineering10111332 (2023).
Khan, A. A., Mahendran, R. K., Perumal, K. & Faheem, M. Dual-3DM3AD: Mixed transformer based semantic segmentation and triplet pre-processing for early multi-class alzheimer’s diagnosis. IEEE Trans. Neural Syst. Rehabil. Eng. 32, 696–707. https://doi.org/10.1109/TNSRE.2024.3357723 (2024).
Chen, C. et al. Ma-sam: Modality-agnostic SAM adaptation for 3D medical image segmentation. Med. Image Anal. 98, 103310 (2024).
Lee, H. H., Bao, S., Huo, Y. & Landman, B. A. “3D UX-Net: A large kernel volumetric ConvNet modernizing hierarchical transformer for medical image segmentation,” arXiv preprint arXiv:2209.15076, Sep. (2022).
Vaswani, A. Attention is all you need (Advances in Neural Information Processing Systems, 2017).
Dosovitskiy, A. “An image is worth 16x16 words: Transformers for image recognition at scale,” arXiv preprint arXiv:2010.11929, (2020).
Perera, S., Navard, P. & Yilmaz, A. “SegFormer3D: An efficient transformer for 3D medical image segmentation,” in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 4981–4988, (2024).
Shen, Y. et al. “FastSAM3D: An efficient segment anything model for 3D volumetric medical images,” in Medical Image Computing and Computer Assisted Intervention – MICCAI 2024, M.G. Linguraru, et al., Eds., (2024).
Alhussen, A., Haq, M. A., Khan, A. A., Mahendran, R. K. & Kadry, S. XAI-RACapsNet: Relevance aware capsule network-based breast cancer detection using mammography images via explainability O-net ROI segmentation. Expert Syst. Appl. 261, 125461 (2025).
Hatamizadeh, A., Nath, V., Tang, Y., Yang, D., Roth, H. R. & Xu, D. “UNETR: Transformers for 3D medical image segmentation,” in 2022 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, pp. 1748–1758, (2022).
He, Y., Nath, V., Yang, D., Tang, Y., Myronenko, A. & Xu, D. “SwinUNETR-V2: Stronger Swin Transformers with stagewise convolutions for 3D medical image segmentation,” in International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 416–426, Cham: Springer Nature Switzerland, (2023).
Gong, H., Kang, L., Wang, Y., Wan, X. & Li, H. “nnMamba: 3D biomedical image segmentation, classification and landmark detection with state space model,” arXiv preprint arXiv:2402.03526, (2024).
Gu, A. & Dao, T. “Mamba: Linear-time sequence modeling with selective state spaces,” arXiv preprint arXiv:2312.00752, (2023).
Ren, L., Liu, Y., Lu, Y., Shen, Y., Liang, C. & Chen, W. “Samba: Simple hybrid state space models for efficient unlimited context language modeling,” arXiv preprint arXiv:2406.07522, (2024).
Ma, J., Li, F. & Wang, B. “U-Mamba: Enhancing long-range dependency for biomedical image segmentation,” arXiv preprint arXiv:2401.04722, (2024).
Zhu, L., Liao, B., Zhang, Q., Wang, X., Liu, W. & Wang, X. “Vision Mamba: Efficient visual representation learning with bidirectional state space model,” arXiv preprint arXiv:2401.09417, (2024).
Kazerooni, A. F. et al. The brain tumor segmentation (BraTS) challenge 2023: Focus on pediatrics (CBTN-CONNECT-DIPGR-ASNR-MICCAI BraTS-PEDs), arXiv preprint arXiv:2305.XXXX, (2024).
Nan, Y. et al. Hunting imaging biomarkers in pulmonary fibrosis: Benchmarks of the AIIB23 challenge. Med. Image Anal. 97, 103253 (2024).
Behrouz, A. & Hashemi, F. “Graph Mamba: Towards learning on graphs with state space models,” in Proceedings of the 30th ACM SIGKDD Conference on Knowledge Discovery and Data Mining, pp. 119–130, (2024).
Zubic, N., Gehrig, M. & Scaramuzza, D. State space models for event cameras, in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 5819–5828, (2024).
Li, K., Li, X., Wang, Y., He, Y., Wang, Y., Wang, L. & Qiao, Y. VideoMamba: State space model for efficient video understanding, in European Conference on Computer Vision, Cham: Springer, pp. 237–255, (2025).
Qiao, Y., Yu, Z., Guo, L., Chen, S., Zhao, Z., Sun, M., Wu, Q. & Liu, J. VL-Mamba: Exploring state space models for multimodal learning, arXiv preprint arXiv:2403.13600, (2024).
Takahashi, S. et al. Comparison of vision transformers and convolutional neural networks in medical image analysis: A systematic review. J. Med. Syst. 48(1), 1–22 (2024).
Hatamizadeh, A. & Kautz, J. MambaVision: A hybrid Mamba-transformer vision backbone, arXiv preprint arXiv:2407.08083, (2024).
Liu, J. et al. Swin-UMamba: Mamba-based UNet with ImageNet-based pretraining. International Conference on Medical Image Computing and Computer-Assisted Intervention Cham: Springer Nature Switzerland, pp. 615–625 (2024).
Xing, Z., Ye, T., Yang, Y., Liu, G. & Zhu, L. SegMamba: Long-range sequential modeling Mamba for 3D medical image segmentation. International Conference on Medical Image Computing and Computer-Assisted Intervention Cham: Springer Nature Switzerland, pp. 578–588 (2024).
Alessio, S. M. & Alessio, S. M. Discrete wavelet transform (DWT), in Digital Signal Processing and Spectral Analysis for Scientists: Concepts and Applications, pp. 645–714 (2016).
Khayam, S. A. The discrete cosine transform (DCT): Theory and application. Michigan State Univ. 114(1), 31 (2003).
Hu, J., Shen, L. & Sun, G. Squeeze-and-excitation networks, in Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 7132–7141 (2018).
Qin, Z., Zhang, P., Wu, F. & Li, X. FcaNet: Frequency channel attention networks, in Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 783–792 (2021).
Nam, J.-H., Syazwany, N. S., Kim, S. J. & Lee, S.-C. Modality-agnostic domain generalizable medical image segmentation by multi-frequency in multi-scale attention, in Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 11480–11491 (2024).
Hatamizadeh, A., Nath, V., Tang, Y., Yang, D., Roth, H. R. & Xu, D. Swin UNETR: Swin Transformers for semantic segmentation of brain tumors in MRI images, in Brainlesion: Glioma, Multiple Sclerosis, Stroke, and Traumatic Brain Injuries. BrainLes 2021, A. Crimi and S. Bakas, Eds., Lecture Notes in Computer Science, vol. 12962. Cham: Springer, (2021).
Chen, J. et al. TransUNet: Transformers make strong encoders for medical image segmentation, arXiv preprint arXiv:2102.04306, (2021).
Hatamizadeh, A. et al. Swin UNETR: Swin Transformers for semantic segmentation of brain tumors in MRI images. International MICCAI Brainlesion Workshop , pp. 272–284 (2021).
Roy, S. et al. MedNext: Transformer-driven scaling of ConvNets for medical image segmentation, in International Conference on Medical Image Computing and Computer-Assisted Intervention , pp 405–415 (2023).
Xie, X., Zhou, P., Li, H., Lin, Z. & Yan, S. Adan: Adaptive Nesterov momentum algorithm for faster optimizing deep models. IEEE Trans. Pattern Anal. Mach. Intell., (2024).
Acknowledgements
This research was partially supported by the Key Scientific Research Project of Higher Education Institutions in Henan Province under Grant 25B520011, the 2025 International Science and Technology Cooperation Project of Henan Province under Grant 252102520053, the 2024 Nanyang Science and Technology Key Project under Grant 24KJGG115, and the 2025 Doctoral Special Research Project of Nanyang Normal University under Grant 2025ZX017. Furthermore, it received funding from the Natural Science Foundation of Henan Province of China under Grant 232300420095. The authors sincerely express their gratitude to these funding agencies for their generous support, which has been instrumental in the successful completion of this work.
Author information
Authors and Affiliations
Contributions
JiaShu Xu and YiHua Lan designed the methodology. Jiashu Xu and YingQi Zhang implemented the algorithm. JiaShu Xu and Chi Zhang prepared the figures. All authors contributed to writing and reviewing the manuscript.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xu, J., Lan, Y., Zhang, Y. et al. CDA-mamba: cross-directional attention mamba for enhanced 3D medical image segmentation. Sci Rep 15, 21357 (2025). https://doi.org/10.1038/s41598-025-06462-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-06462-3