Abstract
Alternative splicing is a fundamental mechanism of gene expression regulation that increases mRNA diversity and can be partially regulated by the circadian clock. Time-dependent production of transcript isoforms from the same gene facilitates coordination of biological processes with the time of day and is a crucial mechanism enabling organisms to cope with environmental changes. In this study, we determined the impact of future ocean acidification conditions on circadian splicing patterns in the brain of fish, while accounting for diel CO2 fluctuations that naturally occur on coral reefs. The temporal splicing pattern observed across a 24-hour period in fish from the control group was largely absent in those exposed to either stable or fluctuating elevated CO2 conditions. Splicing patterns were influenced not only by an overall increase in CO2 concentration but also by its stability, with 6am and 6pm emerging as key timepoints when the majority of aberrant splicing events were identified. We found that fish in fluctuating CO2 conditions exhibited increased temporal plasticity in splicing events compared to fish in stable CO2 conditions. This was especially notable for genes associated with neural functioning. Our findings suggest that natural temporal splicing patterns in fish brains are disrupted by elevated CO2 exposure, with CO2 stability also influencing molecular responses. The increased plasticity in temporal splicing activity observed in fish in fluctuating CO2 environments may provide greater flexibility in biological responses to external pH changes, potentially enabling them to better cope with future ocean acidification conditions.
Similar content being viewed by others
Background
Ocean acidification (OA), one of the aspects of global change, has been shown to have adverse effects on numerous marine organisms due to its impact on diverse biological processes1,2,3. While marine fishes were initially thought to be resilient to OA, many studies suggest that exposure to elevated CO2 can negatively affect various aspects of their physiology and behaviour1,4,5,6. Some of the most concerning detrimental effects of OA on coral reef fish, with significant ecological consequences such as sensory and behavioural abnormalities, have been observed at CO2 levels between 700 and 1000 µatms7,8,9,10,11,12. These levels are projected for the open ocean by the end of this century and are commonly used in many OA studies13. However, sensitivity to OA varies across species, with some showing little to no effect14,15. Such variability suggests that some fish species may be more resilient to rising CO2 levels than others. A similar variability across species has also been observed at the molecular level, with studies showing divergent gene expression patterns in response to elevated CO2 exposure16.
One of the most notable molecular pathways affected in fish brains upon exposure to elevated CO2 levels is the circadian rhythm16,17,18,19,20. The circadian clock synchronises biological processes with the external environment, enabling organisms to anticipate upcoming changes and adjust their physiology and behaviour accordingly22,23,24. Since the circadian rhythm governs almost all physiological processes25its perturbation under OA conditions could lead to variations in synaptic transmission and neuronal activity26. Such changes in the circadian rhythm could potentially provide greater flexibility in adjusting physiological and behavioural responses to OA. However, studies examining the effect of OA on circadian rhythm in fish have so far been conducted only at a single timepoint. Therefore, further studies incorporating multiple timepoints throughout a circadian day are needed to gain a more comprehensive understanding of the interplay between environmental pH variability and circadian rhythm in fish.
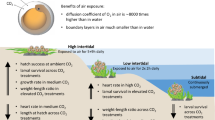
Furthermore, the majority of OA studies have been conducted at stable, elevated CO2 levels, thereby neglecting the temporal fluctuations in CO2 levels resulting from a variety of biological and physical processes in coral reefs, coastal zones, and shallow water habitats27,28,29. Coral reefs are highly dynamic and undergo daily cycles of CO2 variation resulting from photosynthesis/respiration, calcification/dissolution, and physical factors like flow rates and trajectory30,31,32. Studies incorporating diel CO2 oscillations have shown reduced severity of behavioural abnormalities observed in stable elevated CO2 environments33,34. Additionally, ecologically and evolutionarily important traits such as survival, growth, and metabolic function appear to be less affected by elevated CO2 in fluctuating compared to stable environments35,36,37. This disparity may be attributed to differences in the magnitude of circadian control of these biological processes in stable versus fluctuating environments21. In fact, the brain transcriptional profile of coral reef fishes differed significantly between those exposed to stable and fluctuating CO2 conditions with distinct regulation of circadian rhythm genes in each environment21. Therefore, one of the mechanisms potentially facilitating the increased flexibility of the biological responses of fish to pH changes in diel fluctuating environments could be the adjustment of circadian rhythm. As CO2 uptake by the oceans continues to rise, these natural CO2 fluctuations will intensify as seawater buffering capacity diminishes32,38. In light of the ongoing amplification of global environmental changes and heightened environmental variability, it is imperative to incorporate these fluctuations, which mirror the natural habitats of species, into experiments to accurately assess the biological and ecological consequences of OA.
While alterations in the expression levels of circadian rhythm genes have been repeatedly reported in fish upon exposure to OA conditions, there is limited information on the regulatory mechanisms that drive these patterns in fish. Alternative mRNA splicing, a crucial RNA-processing mechanism that enhances cellular proteomic complexity39,40has been linked to the circadian clock and its modulation41,42,43,44,45. Evidence suggests that clock regulated alternative splicing events play a role in the rhythmic expression of various genes, including neuro-specific genes and neurotransmitters46,47,48. Consequently, rhythmic splicing events could influence the physiological and behavioural responses of organisms to environmental variations, such as changes in CO2 levels. Indeed, the role of alternative splicing has been highlighted in driving adaptive evolution over both short and long timescales47 and alternative splicing has been implicated as a mechanism driving the response of coral reef fish to marine heatwaves48. Given its potential to rapidly generate phenotypic diversity, alternative splicing may serve as a key molecular mechanism for facilitating phenotypic plasticity in response to rapidly changing environments caused by climate change.
In this study, we examine the transcriptional patterns mediated by alternative splicing in driving the molecular response of fish to OA across their daily cycle. We assess the impact of environmental CO2 variation at four timepoints throughout a circadian (24-hr) day and aim to: (1) determine if the temporal patterns of splicing events in the brain in control conditions are altered by exposure to near-future predicted CO2 levels, and (2) gain a comprehensive understanding of splicing patterns under stable and fluctuating CO2 conditions by conducting pairwise comparisons between control and each CO2 condition separately for each time point. We reveal how splicing events may facilitate regulation of biological responses of fish to environmental CO2 variability across a circadian timeline.
Methods
Sample collection and experimental design
The experiment reported below was conducted in accordance with all institutional and national law regulations, as well as relevant guidelines and regulations of the Animal Research: Reporting In Vivo Experiments (ARRIVE guidelines)49. Ethics approval for the study was granted by James Cook University with the approval number A2418.
Late juvenile stage Acanthochromis polyacanthus (spiny chromis damselfish) were collected from the wild from Davies Reef (−18.829, 147.656) within the Great Barrier Reef, Australia, in July and August 2017. The fish were then transported to the Sea Simulator (SeaSim - an automated system of seawater tanks that allows precise control and manipulation of various environmental factors (https://www.aims.gov.au/about/facilities/national-sea-simulator)) at the Australian Institute of Marine Science (AIMS). The fish were habituated at SeaSim by housing four individuals in each tank for a duration of seven days. The fish were then divided into three treatment groups with different CO2 concentrations: control (400 µatms), stable elevated CO2 (1000 µatms), and diel fluctuating elevated CO2 (1000 ± 500 µatms) with a high CO2 peak at 2am and low CO2 peak at 2pm. The experimental setup consisted of two replicate 50 L tanks per treatment group, with each tank containing between 8 and 12 fish (Fig. 1; Supplementary Table S1). The experimental system is described in detail in50. Briefly, it consisted of a flowthrough system supplied with ultra-filtered seawater (0.04 μm), at 50 L h−1, sourced from the Great Barrier Reef lagoon to the National Sea Simulator facility at AIMS. Seawater was continuously monitored in real-time to ensure it remained within the parameters for seawater from the central Great Barrier Reef, with temperature, pH, and salinity measured regularly and total alkalinity measured weekly. The pCO2 levels was controlled using a custom Model Predictive Control running on a micro-programmable logic controller (Series S7-1500, Siemens, Australia), integrated with the general SeaSim control system. Pure CO₂ was delivered through Gas Mass Flow Controllers (GFC17 series, Aalborg, NY, USA) according to the profiling schedule and was dissolved in flow-through water using membrane contactors (Membrana Liqui-Cel, 3 M, USA). All tanks were maintained under a consistent photoperiod of 12/12hr (light/dark cycle) and temperature of 28.5ºC. All fish were fed once per day at 7:30 am. The control CO2 level represents current ocean CO2 concentration, the stable CO2 treatment reflects the end-of-century projection13and the fluctuating CO2 treatment mirrors levels seen in some tidal lagoons51 and is predicted to become more prevalent in the future due to the increasing effects of climate change38. The fish were exposed to the respective conditions for a period of 14 days, after which 4–5 individuals from each treatment group were sampled at 2am, 6am, 2pm, and 6pm, over a 48-hour period. In total, we had 20 individuals in control (400 µatms), 20 individuals in stable elevated CO2 (1000 µatms), and 18 individuals in diel fluctuating elevated CO2 (1000 ± 500 µatms) across the four timepoints. The fish were euthanized by cervical dislocation, after which their weight was measured and the brain tissue was dissected, snap-frozen in liquid nitrogen and stored at −80ºC. All the sampled fish were on average 3.41 ± 1.08 g in weight (Supplementary Table S2).
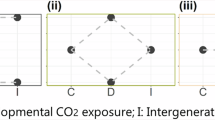
Schematic illustration of sample collection, acclimation, CO2 exposure treatments, brain dissection timepoints and types of splicing events analysed. The red circles represent the time points when the brain tissues were collected. A total of 4–5 fish brains were sampled at each time point from each treatment group, resulting in 20 samples from control, 20 from the stable CO2 treatment, and 18 from the fluctuating CO2 treatment across all four timepoints.
RNA extraction and sequencing
Whole frozen fish brains were homogenized in RLT buffer (Qiagen) using a TissueLyser with single use silicon beads, according to manufacturer’s recommendation based on tissue weight. Total RNA was extracted from all the samples (N = 58) using Qiagen RNeasy Micro Kit, following manufacturer’s instructions. On-column DNase digestion was performed following manufacturer’s instructions to remove any potential DNA contamination. The concentration and quality of the extracted RNA was measured using a Qubit and Bioanalyzer, respectively. RNA-Seq libraries were prepared for samples with RIN values above 8 using the Truseq-RNA Illumina kits v2. Paired-end RNA sequencing was then done on an Illumina Hiseq4000, with samples from different timepoints and treatments randomized across lanes, for 101 bp reads at Macrogen, Korea. An average of 37.7 ± 4.9 million read pairs were obtained across the 58 RNA libraries sequenced (Supplementary Table S3).
Sequence processing and splicing analysis
The quality of the raw RNA-Seq reads were determined using FastQC v0.11.552 and low quality reads and adapters were trimmed using Trimmomatic v0.3953 using the parameters ILLUMINACLIP: adapters.fa:2:30:15:8:true, SLIDINGWINDOW:4:20, MINLEN:32, HEADCROP:8. Further filtering was done using kraken v2.0.8-beta54 to identify and remove potential bacterial, fungal, and viral contaminant sequences using a confidence score of 0.3. An average of 34.9 ± 4.6 million high quality RNA-Seq reads were mapped to the Acanthochromis polyacanthus reference genome (ASM210954v1) using STAR v2.7.10b_alpha55 in two-pass mode with default parameters (Supplementary Table S3). rMATS v4.1.256 was used to classify the alternative splicing (AS) events observed in the transcript isoforms in the GTF annotation file into five types: alternative 5’ splice site (A5SS), alternative 3’ splice site (A3SS), skipped exon (SE), mutually exclusive exon (MXE), and retained intron (RI). Each AS event was required to be supported by an average of at least five reads mapped to both spliced junctions and exons and have an average percent spliced in (PSI) value > 0.1 across all samples. To further ensure the retention of only high-confidence AS events, a minimum of 29 samples were required to have non-zero counts for both inclusion junction reads (reads supporting the inclusion of an exon or splice site) and skipping junction reads (reads supporting the skipping of an exon or splice site).
Differential splicing (DS) events were identified using rMATS by performing pairwise comparisons of AS events, considering reads mapped to both splice junctions and exons (parameters: --readLength 90 --variable-read-length --cstat 0.05). Firstly, to determine the natural 24-hour temporal splicing pattern, we performed pairwise comparisons of AS events between specific timepoint pairs in the control group: 2am vs. 6am; 6am vs. 2pm; 2pm vs. 6pm; and 6pm vs. 2am. We then assessed whether these natural rhythmic splicing patterns were altered by increased CO2 concentration and its stability by comparing the AS events between the same four timepoint pairs in the stable and fluctuating elevated CO2 conditions. Secondly, we conducted pairwise comparisons of AS events between the control group and both stable and fluctuating elevated CO2 treatments, separately at each of the four time points to gain a more comprehensive understanding of the DS response in both the elevated CO2 conditions. Lastly, to identify diurnal splicing patterns specific to fluctuations in CO2 concentration, we compared AS events between 2am and 2pm (the high and low CO2 peaks), filtering out DS events present in the control group.
DS events reported by rMATS from all the pairwise comparisons were further analysed using the Mapping Alternative Splicing Events to pRoteins (MASER) R/Bioconductor package v1.14.0 in R v4.2.1. DS events with low coverage (< 5 average reads across splice junctions) were discarded, and only those with an FDR corrected p-value < 0.05 and |ΔPSI| > 0.1 were considered to be significant in each pairwise comparison, similar to what has been done in previous studies57,58. Subsequently, each significant differential splicing event was mapped to Ensembl transcripts using MASER’s “mapTranscriptsToEvents” function for functional characterization of the resulting isoforms. Specifically, we investigated whether the exons that were significantly differentially spliced encoded for protein domains or were located within the 5’ or 3’ untranslated region (UTR) of the mRNA transcript. The 5’ and 3’ UTRs contain various regulatory elements that are critical for post-transcriptional processes such as pre-mRNA processing, mRNA stability, and translation initiation59. Therefore, alterations in the sequence of these regulatory regions could have important functional implications. Furthermore, the ExPASy translate tool60 was used to translate the isoforms produced from each significant DS event into protein sequences. This facilitated the identification of premature stop codons in the isoforms, which could potentially be targeted for degradation via the non-sense-mediated decay (NMD) pathway or result in the production of truncated proteins61.
Differential expression analysis
To determine if the DS events translate to differences in gene expression, we performed differential gene expression analysis on all the pairwise comparisons described above. Specifically, the gene expression levels were quantified after mapping to the A. polyacanthus reference genome from Ensembl database (gene build 103) using Salmon62 and the differential gene expression analysis was carried out using the DESeq2 v1.28.163 package in R v4.0.3. For all pairwise comparisons, differentially expressed genes with an FDR < 0.05 were considered to be significant. The overlap between the differentially spliced and differentially expressed genes were determined for each pairwise comparison and visualized using Venn diagrams (https://bioinformatics.psb.ugent.be/webtools/Venn/).
Results
Overview of alternative splicing events in A. polyacanthus
The A. polyacanthus genome comprises of 35,487 distinct transcripts corresponding to 24,565 genes, with an average of 1.44 transcripts per gene. Across all our samples, a total of 5,689 alternative splicing events corresponding to 2,948 genes (14.7% of expressed genes) were identified with high confidence. These events resulted from five different types of alternative splicing: alternative 5’ splice site (70 events), alternative 3’ splice site (132 events), skipped exon (2,673 events), mutually exclusive exon (2,616 events), and retained intron (198 events; Supplementary Table S4).
Time course of differential splicing over a 24-hour period
Across the four timepoints there existed a clear temporal pattern in significant differential splicing (DS) events (FDR < 0.05 and ΔPSI > 0.1) within the control group, with exon skipping (SE) being the predominant splicing type across most comparisons (Fig. 2a, b); Supplementary Table S5). The period between 2am − 2pm had the greatest number of genes undergoing DS (25 genes) compared to the latter period of the day (four genes between 2pm – 2am; Fig. 2a). Notably, upon exposure to both stable and fluctuating elevated CO2 conditions, these natural rhythmic splicing patterns observed in the brain in control were largely absent (Fig. 2a). The number of genes with significant DS events was lower in both the CO2 treatments compared to control. The only exception was 2pm vs. 6pm in the fluctuating CO2 treatment, which had the highest number of genes with DS events (eight genes) for this time period (Fig. 2a). Furthermore, across the four time periods, both the stable and fluctuating CO2 treatments exhibited distinct splicing profiles involving different sets of genes compared to the control group (Fig. 2c). Notably, only one gene, unannotated ENSAPOG00000013961, was commonly DS in the same time period (2am vs. 6am) in both control and the fluctuating CO2 treatment. Similarly, another gene, BMAL1A, showed a common temporal splicing pattern in control and stable CO2 treatment (6am vs. 2pm), albeit BMAL1A did not achieve statistical significance in control (FDR = 0.057).
(a) Number of differentially spliced genes in the control, stable and fluctuating elevated CO2 treatment across a 24-hour period. (b) Distribution of the five splice types among the differentially spliced genes in the control, stable and fluctuating elevated CO2 treatments over the 24-hour period. A3SS: Alternate 3’ splice site, A5SS: Alternate 5’ splice site, SE: skipped exon, MXE: mutually exclusive exons, RI: retained intron. (c) The number and overlap of differentially spliced genes in the control, stable elevated and fluctuating elevated CO2 treatment across a 24-hour period.
Differential splicing patterns in stable or fluctuating CO2 environments
To pinpoint changes in the splicing patterns depending on the stability of CO2 conditions we compared alternative splicing events in control with stable and fluctuating CO2 treatments separately at each of the four time points. The number of genes with significant DS events (FDR < 0.05 and ΔPSI > 0.1) between control and the stable CO2 treatment were as follows: two genes at 2am, six genes at 6am, six genes at 2pm, and one gene at 6pm (Fig. 3a; Supplementary Table S6). At the same four time points, the fluctuating CO2 versus control comparison had a higher number of genes with significant DS events than the stable CO2 versus control comparison. Specifically, three genes at 2am, sixteen genes at 6am, ten genes at 2pm, and sixteen genes at 6pm showed significant DS events between control and the fluctuating CO2 treatment (Fig. 3a). Interestingly, only in the fluctuating CO2 treatment were genes involved in neuronal functioning DS across all the four timepoints.
(a) Number of differentially spliced genes between control and stable or fluctuating elevated CO2 treatment for each of the four time points. (b) The number of differentially spliced genes and the overlap between control and treatments with stable elevated and fluctuating elevated CO2 conditions across four time points. (c) Distribution of the five splice types among the differentially spliced genes in the stable and fluctuating CO2 treatments across the four time points. A3SS: Alternate 3’ splice site, A5SS: Alternate 5’ splice site, SE: skipped exon, MXE: mutually exclusive exons, RI: retained intron.
There was very little overlap among the genes undergoing DS between fish exposed to stable and fluctuating CO2 treatments at any of the four time points (Fig. 3b). Furthermore, across the four time points, the types of alternative splicing varied among the DS genes. Skipped exon was the most common splicing type, with five events in stable and 21 events in fluctuating elevated CO2 conditions. In contrast, alternative 3’/5’ splice sites were the least abundant in both CO2 treatments (Fig. 3c).
We found that only ~ 28% of the total significant DS events (FDR < 0.05 and ΔPSI > 0.1) in both the stable and fluctuating CO2 conditions led to what we defined as ‘aberrant splicing’, where the spliced exons either overlapped a protein domain, the 3’ or 5’ untranslated region (UTR), or the splicing event resulted in the generation of a premature stop codon. Among the DS events identified in the stable CO2 versus control comparison, aberrant splicing events were identified in three genes at 6am and one gene at 2pm (Table 1; Supplementary Figure S1). In the fluctuating CO2 versus control comparison, aberrant splicing was identified in one gene at 2 am, five genes at 6 am, two genes at 2 pm, and four genes at 6 pm (Table 1; Supplementary Figure S1). Notably, the majority (53%) of these aberrant splicing events resulted in the generation of premature stop codons in the respective isoforms (Supplementary Table S6).
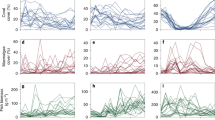
Across all the four timepoints assessed, 6am was found to be a critical time point in both the stable and fluctuating CO2 treatments, while 6pm was an additional pivotal timepoint in the fluctuating CO2 treatment, as majority of the aberrant splicing events was observed at these time points. Notably, the gene SRGAP3 which is involved in synaptic activity, showed a similar splicing pattern, with a decrease in Percent Spliced In (PSI) of the second mutually exclusive exon (MXE exon2), in both the stable and fluctuating CO2 treatments, albeit at different timepoints (6am in stable CO2 treatment but at 6pm in fluctuating CO2 treatment; Fig. 4(a)). The two other genes with aberrant splicing that was unique to the stable CO2 treatment at 6am were TFCP2, a transcription factor regulating genes encoding acid-sensing ion channels, and METTL3, a methyltransferase involved in post-translational mRNA methylation (Fig. 4(b)). In the fluctuating CO2 treatment aberrant splicing events were identified in a total of eight genes at 6am and 6pm (Fig. 4(c); Supplementary Table S6). These genes were associated with five broad functions, specifically neurogenesis and neuronal functioning (CEP20, MTMR11), RNA processing (EXOSC9, ENSAPOG00000001802), metabolism (GALCB, RSAD1), circadian rhythm (BMAL1A), and cell motility and intracellular transport (ENSAPOG00000011552).
Percentage of splicing inclusion (PSI) values of the spliced exons for (a) SRGAP3 in stable elevated CO2 and fluctuating elevated CO2, (b) genes in stable elevated CO2 alone, (c) genes in fluctuating elevated CO2 alone. SE: skipped exon; A3SS: alternative 3’ splice site; A5SS: alternative 5’ splice site; MXE: mutually exclusive exon; RI: retained intron. For SE and RI events, the PSI value refers to the inclusion level of the cassette exon or retained intron, respectively. For A3SS and A5SS, the PSI value refers to the inclusion of the longer exon. For MXE events, there are two sets of box plots with the first set referring to exon 1 PSI values and the second set referring to exon 2 PSI values. Alongside each gene is a schematic representation of the isoform structures from the differential splicing events. The blue rectangles represent the exons undergoing differential splicing, the red rectangles represent the exons adjacent to the spliced exon, and the connecting lines represent the introns. The purple rectangles represent the protein domains (Prosite and Pfam) overlapping the exons which was inferred from the Ensembl database. The longer green rectangles represent full-length transcript isoforms while the shorter green bar represents transcript isoforms with a premature stop codon. The unfilled rectangles represent 5’/3’ UTR regions. The arrows indicate gene orientation on DNA strands (positive or negative).
Differential splicing events between the high and low peaks of the diel CO2 cycle
In the fluctuating CO2 treatment, we found changes in splicing events between the high (2am) and low (2pm) CO2 peak that were absent in control. Specifically, between the two CO2 peaks, six genes exhibited significant DS (FDR < 0.05 and ΔPSI > 0.1): three SE events (SULF1, TJP2B, TJP2A), one mutually exclusive exon (MXE) event (ESRRB), and two retained intron (RI) events (MED22, unannotated ENSAPOG00000016345; Supplementary Table S7). The absence of these DS events in control suggests that they are specifically influenced by fluctuations in CO2 concentration. In two (ESRRB and TJP2B) of these six genes the splicing events potentially resulted in disruption of the reading frame of the respective isoforms and hence could be considered as ‘aberrant’. Specifically, skipping the second mutually exclusive exon (MXE exon2) in the gene ESRRB, involved in cellular energy metabolism, and skipping exon 11 in TJP2B, the gene encoding the tight junction protein B, which is part of the blood brain barrier, introduces a premature stop codon in the respective isoforms, potentially rendering them non-functional (Fig. 5). These skipping events are more prevalent at 2am in ESRRB and at 2pm in TJP2B. Interestingly, TJP2A, a paralogue of TJP2B, was also significantly DS between the CO2 peaks, however, both isoforms of TJP2A resulting from the splicing event have full open reading frames and hence retain their coding potential (Fig. 5).
Percentage of splicing inclusion (PSI) values of the spliced exons between the high (2am) and low (2pm) peak in the fluctuating CO2 treatment. SE: skipped exon; MXE: mutually exclusive exon. For SE events, the PSI value refers to the inclusion level of the cassette exon. For MXE events, there are two sets of box plots with the first set referring to exon 1 PSI values and the second set referring to exon 2 PSI values. Below each gene is a schematic representation of the isoform structures from the differential splicing events. The blue rectangles represent the exons undergoing differential splicing, the red rectangles represent the exons adjacent to the spliced exon, and the connecting lines represent the introns. The longer green rectangles represent full-length transcript isoforms while the shorter green bar represents transcript isoforms with a premature stop codon. The brown rectangle represents two functional isoforms of slightly different length of TJP2A. The arrows indicate gene orientation on DNA strands (positive or negative).
Comparison of differential gene expression and differential splicing across the 24-hour period
We found minimal overlap among the genes showing significant differences in splicing (DS; FDR < 0.05 and ΔPSI > 0.1) and expression (DE; FDR < 0.05) over a 24-hour period in control, as well as in both the stable and fluctuating elevated CO2 treatments, with 35 genes being commonly DS and DE in at least one of the pairwise comparisons in at least one of the three experimental conditions (Supplementary Table S8). Specifically in control, only four genes (SRCIN1, SORBS2, MTHFSD, ENSAPOG00000002603) in 2am vs. 6am, three genes (RBM5, CLASP2, SUN1B) in 6am vs. 2pm, and one gene (EGLN3) in 6pm vs. 2pm were commonly DS and DE. Similarly, in the stable CO2 treatment, one gene in 2am vs. 6am (ENSAPOG00000011664) and three genes (PRKCAA, BMAL1A, SLC25A12) in 6am vs. 2pm were commonly DS and DE. The fluctuating CO2 treatment showed the least overlap, with only one gene (NDRG4) commonly DS and DE in 6am vs. 2pm (Supplementary Figure S2).
Similarly, when considering the comparison between stable CO2 versus control and fluctuating CO2 versus control separately, for each individual timepoint, there was generally no overlap between the DE and DS genes, with only a total of four DS genes also being DE in at least one of the pairwise comparisons (Supplementary Table S9). Specifically, none of the DS genes were found to be DE in the stable CO2 treatment at any of the four timepoints (Supplementary Figure S3). In the fluctuating CO2 treatment, only one gene (TJP2B) at 2pm was both DS and DE (Supplementary Figure S3). Furthermore, only 2 genes (TJP2B and SULF1) were commonly DS and DE between the high and low CO2 peaks within the fluctuating CO2 treatment (Supplementary Figure S4). These findings suggest that majority of genes undergo regulation either by alternative splicing or differential expression alone. The detailed results of the differentially expressed genes are provided in Supplementary Tables S10-S12.
Discussion
In this study, we investigated alternative splicing (AS) changes at four time points during the day in the brain of A. polyacanthus when exposed to projected future ocean acidification scenarios, considering both stable CO2 concentration and diel fluctuations. We found that the temporal differential splicing (DS) events observed under ambient CO2 conditions were mostly absent when fish are exposed to future ocean acidification scenarios (both stable and fluctuating elevated CO2). Given the intricate interaction between circadian rhythms, environmental cues, and biological processes64the observed change in temporal regulation of splicing in the brain in elevated CO2 conditions could affect synaptic activity and neural functioning64,65,66. Furthermore, the limited overlap between genes showing temporal splicing and temporal expression in both stable and fluctuating CO2 conditions indicates multi-layered gene regulatory response in A. polyacanthus to environmental CO2 variations. AS and differential expression play nonredundant roles in driving the response to elevated CO2 conditions, potentially because these two processes are regulated by distinct genetic loci and influence different biological processes67. This decoupling of splicing and transcript expression could enhance flexibility in driving plastic responses to environmental changes. Several studies have reported that AS does not always result in changes in transcript expression levels, suggesting that this functional non-redundancy is a common phenomenon in plasticity and phenotypic diversification67. Furthermore, A. polyacanthus has a lower proportion of genes undergoing AS (14.7%) compared to other teleosts, where alternatively spliced genes ranges from 17% in zebrafish to 43% in Fugu68. Hence, splicing might play a minor role in driving the plastic responses of A. polyacanthus to environmental changes, as evidenced by the lower proportion of AS events compared to changes in gene expression. A similar pattern was also observed in this species in response to ocean warming48. While DS may not significantly alter transcript abundance, it could play a regulatory role in driving cellular responses to periodic CO2 fluctuations, whereas the DE genes are likely associated with different biological processes. In fact, splicing and gene expression were found to play distinct yet complementary roles in driving parallel adaptive evolution in Artic charr, with DS genes largely associated with central regulatory pathways69. A similar mechanism may be occurring here, as the key timepoints before CO2 changes (6am and 6pm) involved DS of genes with potentially broader regulatory implications, such as widespread RNA processing, circadian rhythm, and neural activity.
Different CO2 stability levels showed differential effects on splicing, as there was no substantial overlap among the DS genes between fish in stable and fluctuating CO2 conditions at all four time points analysed. This suggests distinct molecular responses depending on the stability of CO2 levels throughout the day. Among the four time points across the 24-hour period, 6am emerged as critical in both the stable and fluctuating CO2 treatments, while 6pm was key exclusively in the fluctuating CO2 treatment. These timepoints exhibited the highest number of DS genes with aberrant splicing events as well as the greatest diversity in the functions of these DS genes. In the stable elevated CO2 treatment, TFCP2 and METTL3 were of particular interest, both being DS at 6am when compared to control. TFCP2 is a transcription factor regulating genes encoding acid-sensing ion channels which control deviations from extracellular pH in fish70and METTL3 is a methyltransferase that potentially modulates the processing and stability of RNAs involved in diverse cellular processes under elevated CO2 conditions71. Given that the primary response mechanism of fish when exposed to stable elevated CO2 conditions is to regulate acid-base homeostasis2,3,21the DS of this gene might enable the fish to mitigate the risk of acidosis by maintaining physiological pH levels during the crucial early morning hours. In contrast, under fluctuating CO2 conditions, aberrant splicing events were observed in genes involved in metabolism (GALCB, RSAD1), RNA processing (EXOSC9, ENSAPOG00000001802), and neurogenesis and synaptic activity (CEP20) at 6am when compared to control, indicating changes in brain metabolic activity and neural functioning in response to an anticipated decline in CO2 levels during the day. This temporal change in the splicing of genes associated with various aspects of neuronal functioning and transcriptome-wide RNA processing might enable the fish to optimise neural function in response to periodic oscillations in CO2 levels35. Additionally, genes associated with the sensory system (MTMR11) and circadian rhythm (BMAL1A) exhibited DS exclusively at 6pm in the fluctuating CO2 treatment when compared to control. Changes in the expression of core circadian rhythm genes have been repeatedly reported to be involved in acclimation of fish to elevated CO2 environments16,17,20,21,72. Furthermore, alternative splicing of BMAL is shown to facilitate molecular adjustments to meet different physiological needs73,74,75. Given the central role of circadian rhythms in regulating physiological processes, including metabolism and synaptic activity76,77the observed DS event in BMAL1A, could be a potential response to coordinate physiological processes in anticipation of the expected increase in the CO2 levels during the night78. The distinct splicing responses observed in our study suggests that fish in fluctuating CO2 conditions have increased neural plasticity which might enable them to fine-tune physiological processes in anticipation of diel CO2 fluctuations, whereas this is limited under stable CO2 conditions. Indeed physiological performance for ectotherms was found to be greater under fluctuating environments compared to stable environments when faced with environmental changes79. These results illustrate the interplay between environmental cues and temporal splicing in shaping the molecular responses of fish, highlighting that certain time points are key and that response mechanisms vary depending on the stability of the CO2 concentration throughout the day.
In fluctuating elevated CO2 conditions, fish exhibited increased plasticity in temporal splicing activity with consistently greater number of DS events compared to the stable elevated CO2 treatment at all four time points. Specific to the fluctuating elevated CO2 treatment was the DS of genes involved in various aspects of neural functioning such as the sensory system, calcium channel activity, blood brain barrier permeability, and brain metabolism. This suggests intricate regulation of neural function throughout the day potentially in response to periodic CO2 fluctuations. Hence, our results suggest that fish in fluctuating CO2 conditions might be able to anticipate periodic changes in CO2 levels and alter their brain activity accordingly72,78. Further evidence of such coordination of splicing activity with diel CO2 fluctuations was observed from the direct comparison of splicing events between the high (2am) and low (2pm) CO2 peaks within the fluctuating elevated CO2 treatment. Of particular interest was the DS of the estrogen related receptor beta (ESRRB), a transcription factor that regulates numerous metabolic genes, and TJP2B, a tight junction protein part of the blood brain barrier, as the DS events in both genes affected the proportion of the respective functional isoforms between the high and low CO2 peaks. Estrogen related receptors (ERRs) play a key role in coordination of circadian rhythm and metabolic homeostasis80. Similarly, the circadian clock regulates the activity of various genes involved in blood brain barrier permeability and transport, including tight junction proteins, which play a crucial role in regulating the chemical microenvironment of the central nervous system, thereby influencing neural activity81,82. Interestingly, TJP2A, a paralog of TJP2B, was also significantly DS between the high and low CO2 peaks, but without affecting the reading frame of the resulting isoforms. This could be a compensatory mechanism to mitigate the potential loss of function in TJP2B at 2 pm, although the impact of the skipped exon on TJP2A’s function remains uncertain and needs further investigation.
While further research is needed to understand the downstream impacts and functional distinctions between the various transcript isoforms produced by the splicing events in the brain of the spiny chromis damselfish, the observed temporal variation in alternative splicing in fluctuating environments could stem from feed-forward mechanisms driven by the circadian clock21. Feed-forward processes enable organisms to adjust cellular functions in anticipation of periodic changes and coordinate physiological processes with external environmental cues78. In fluctuating environments, circadian shifts in gene expression, impacting downstream processes like growth, have been observed in fish species, with potential links to neuronal activity17,26,83. Notably, a study on A. polyacanthus found that circadian rhythm genes showed a much greater change in expression under fluctuating elevated CO2 conditions compared to stable CO2 conditions, suggesting enhanced flexibility in adjusting to periodic CO2 fluctuations21. Interestingly, in our study majority of DS events occurred at 6am and 6pm, the two timepoints immediately preceding changes in CO2 levels. These anticipatory changes in splicing activity might provide fish with increased flexibility in regulating molecular responses fluctuating elevated CO2 conditions.
Conclusion
Our findings reveal that temporal splicing patterns in the brain of A. polyacanthus are disrupted upon exposure to elevated CO2 conditions. Fish in fluctuating elevated CO2 environments exclusively demonstrated greater temporal splicing plasticity, particularly in genes associated with circadian rhythm and neural functioning. Given that a gene’s function can be influenced by the ratio between its various transcript isoforms with potentially different properties and functions, this temporal variation in splicing events could enable nuanced regulation of biological processes depending on the time of day73. This enhanced temporal regulation likely enables fish in fluctuating conditions to anticipate and adjust to periodic CO2 fluctuations by fine-tuning and synchronizing their internal biological processes with external environmental cues. Such adjustment involves multiple layers of gene regulation, as evidenced by the limited overlap between DE and DS genes, potentially offering enhanced flexibility in responding to environmental changes. However, further studies incorporating proteomics and detailed functional analysis of transcript isoforms are needed to fully understand the downstream impacts of these splicing events. Therefore, temporal regulation of molecular processes in fluctuating conditions might modulate downstream biological responses of coral reef fish, potentially enabling them to cope with elevated CO2 conditions expected due to OA through coordinated transcriptional and splicing responses.
Data availability
The raw RNA sequences are deposited in NCBI under BioProject ID: PRJNA658203.
Abbreviations
- A3SS:
-
Alternative 3’ splice site
- A5SS:
-
Alternative 5’ splice site
- DE:
-
Differential expression
- DS:
-
Differential splicing
- MXE:
-
Mutually exclusive exon
- OA:
-
Ocean acidification
- PSI:
-
Percent spliced in
- RI:
-
Retained intron
- SE:
-
Skipped exon
- SeaSim:
-
Sea Simulator
References
Cattano, C., Claudet, J., Domenici, P. & Milazzo, M. Living in a high CO2 world: a global meta-analysis shows multiple trait‐mediated fish responses to ocean acidification. Ecol. Monogr. 88, 320–335 (2018).
Ishimatsu, A., Hayashi, M., Lee, K., Kikkawa, T. & Kita, J. Physiological effects on fishes in a high-CO2 world. J. Geophys. Res. Oceans. 110, 2004JC002564 (2005).
Strader, M. E., Wong, J. M. & Hofmann, G. E. Ocean acidification promotes broad transcriptomic responses in marine metazoans: a literature survey. Front. Zool. 17, 7 (2020).
Munday, P. L., Jarrold, M. D. & Nagelkerken, I. Ecological effects of elevated CO2 on marine and freshwater fishes: From individual to community effects. in Fish Physiology vol. 37 323–368 (Elsevier, 2019).
Paula, J. R. et al. The past, present and future of cleaner fish cognitive performance as a function of CO2 levels. Biol. Lett. 15, 20190618 (2019).
Porteus, C. S. et al. Near-future CO2 levels impair the olfactory system of a marine fish. Nat. Clim. Change. 8, 737–743 (2018).
Ferrari, M. C. O. et al. Intrageneric variation in antipredator responses of coral reef fishes affected by ocean acidification: implications for climate change projections on marine communities. Glob Change Biol. 17, 2980–2986 (2011).
Heuer, R. M. & Grosell, M. Physiological impacts of elevated carbon dioxide and ocean acidification on fish. Am. J. Physiol. -Regul Integr. Comp. Physiol. 307, R1061–R1084 (2014).
Munday, P. L. et al. Replenishment of fish populations is threatened by ocean acidification. Proc. Natl. Acad. Sci. 107, 12930–12934 (2010).
Munday, P. L. et al. Effects of elevated CO2 on predator avoidance behaviour by reef fishes is not altered by experimental test water. PeerJ 4, e2501 (2016).
Nagelkerken, I. & Munday, P. L. Animal behaviour shapes the ecological effects of ocean acidification and warming: moving from individual to community-level responses. Glob Change Biol. 22, 974–989 (2016).
Welch, M. J., Watson, S. A., Welsh, J. Q., McCormick, M. I. & Munday, P. L. Effects of elevated CO2 on fish behaviour undiminished by transgenerational acclimation. Nat. Clim. Change. 4, 1086–1089 (2014).
Kroeker, K. J. et al. Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob Change Biol. 19, 1884–1896 (2013).
Clark, T. D. et al. Ocean acidification does not impair the behaviour of coral reef fishes. Nature 577, 370–375 (2020).
Clements, J. C., Sundin, J., Clark, T. D. & Jutfelt, F. Meta-analysis reveals an extreme decline effect in the impacts of ocean acidification on fish behavior. PLOS Biol. 20, e3001511 (2022).
Kang, J. et al. Rapid evolution fuels transcriptional plasticity to ocean acidification. Glob Change Biol. 28, 3007–3022 (2022).
Lee, D. W., Song, J. A., Park, H. S. & Choi, C. Y. Circadian rhythm disturbances due to exposure to acidified conditions and different photoperiods in juvenile Olive flounder (Paralichthys olivaceus). Ocean. Sci. J. 56, 198–206 (2021).
Nilsson, G. E. et al. Near-future carbon dioxide levels alter fish behaviour by interfering with neurotransmitter function. Nat. Clim. Change. 2, 201–204 (2012).
Schunter, C. et al. An interplay between plasticity and parental phenotype determines impacts of ocean acidification on a reef fish. Nat. Ecol. Evol. 2, 334–342 (2018).
Suresh, S., Mirasole, A., Ravasi, T., Vizzini, S. & Schunter, C. Brain transcriptome of gobies inhabiting natural CO2 seeps reveal acclimation strategies to long-term acidification. Evol. Appl. 16, 1345–1358 (2023).
Schunter, C., Jarrold, M. D., Munday, P. L. & Ravasi, T. Diel pCO2 fluctuations alter the molecular response of coral reef fishes to ocean acidification conditions. Mol. Ecol. 30, 5105–5118 (2021).
Ding, J. M. et al. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science 266, 1713–1717 (1994).
Wilkins, M. B. A Temperature-dependent endogenous rhythm in the rate of carbon dioxide output of Periplaneta americana. Nature 185, 481–482 (1960).
Villamizar, N. et al. Circadian rhythms of embryonic development and hatching in fish: A comparative study of zebrafish (Diurnal), Senegalese sole (Nocturnal), and Somalian cavefish (Blind). Chronobiol Int. 30, 889–900 (2013).
Richards, J. & Gumz, M. L. Mechanism of the circadian clock in physiology. Am. J. Physiol. Regul. Integr. Comp. Physiol. 304, R1053–R1064 (2013).
Dmitriev, A. V. & Mangel, S. C. A circadian clock regulates the pH of the fish retina. J. Physiol. 522 Pt 1, 77–82 (2000).
Duarte, C. M. et al. Is ocean acidification an open-ocean syndrome? Understanding anthropogenic impacts on seawater pH. Estuaries Coasts. 36, 221–236 (2013).
Anthony, K. R. N., Kleypas, A., Gattuso, J. & J. & Coral reefs modify their seawater carbon chemistry – implications for impacts of ocean acidification. Glob Change Biol. 17, 3655–3666 (2011).
Waldbusser, G. G. & Salisbury, J. E. Ocean acidification in the coastal zone from an organism’s perspective: multiple system parameters, frequency domains, and habitats. Annu. Rev. Mar. Sci. 6, 221–247 (2014).
Falter, J. L., Lowe, R. J., Zhang, Z. & McCulloch, M. Physical and biological controls on the carbonate chemistry of coral reef waters: effects of metabolism, wave forcing, sea level, and geomorphology. PLoS ONE. 8, e53303 (2013).
Hannan, K. D. et al. Diel pCO2 variation among coral reefs and microhabitats at Lizard island, great barrier reef. Coral Reefs. 39, 1391–1406 (2020).
McNeil, B. I. & Sasse, T. P. Future ocean hypercapnia driven by anthropogenic amplification of the natural CO2 cycle. Nature 529, 383–386 (2016).
Jarrold, M. D., Humphrey, C., McCormick, M. I. & Munday, P. L. Diel CO2 cycles reduce severity of behavioural abnormalities in coral reef fish under ocean acidification. Sci. Rep. 7, 10153 (2017).
Laubenstein, T. D., Jarrold, M. D., Rummer, J. L. & Munday, P. L. Beneficial effects of diel CO2 cycles on reef fish metabolic performance are diminished under elevated temperature. Sci. Total Environ. 735, 139084 (2020).
Cross, E. L., Murray, C. S. & Baumann, H. Diel and tidal pCO2 × O2 fluctuations provide physiological refuge to early life stages of a coastal forage fish. Sci. Rep. 9, 18146 (2019).
Hannan, K. D., Munday, P. L. & Rummer, J. L. The effects of constant and fluctuating elevated pCO2 levels on oxygen uptake rates of coral reef fishes. Sci. Total Environ. 741, 140334 (2020).
Jarrold, M. D. & Munday, P. L. Diel CO2 cycles and parental effects have similar benefits to growth of a coral reef fish under ocean acidification. Biol. Lett. 15, 20180724 (2019).
Shaw, E. C., McNeil, B. I., Tilbrook, B., Matear, R. & Bates, M. L. Anthropogenic changes to seawater buffer capacity combined with natural reef metabolism induce extreme future coral reef CO2 conditions. Glob Change Biol. 19, 1632–1641 (2013).
Kim, E., Magen, A. & Ast, G. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 35, 125–131 (2007).
Kornblihtt, A. R. et al. Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell. Biol. 14, 153–165 (2013).
Hughes, M. E., Grant, G. R., Paquin, C., Qian, J. & Nitabach, M. N. Deep sequencing the circadian and diurnal transcriptome of Drosophila brain. Genome Res. 22, 1266–1281 (2012).
Preußner, M. et al. Rhythmic U2af26 alternative splicing controls PERIOD1 stability and the circadian clock in mice. Mol. Cell. 54, 651–662 (2014).
Sanchez, S. E. et al. A Methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 468, 112–116 (2010).
Gizowski, C., Zaelzer, C. & Bourque, C. W. Clock-driven vasopressin neurotransmission mediates anticipatory thirst prior to sleep. Nature 537, 685–688 (2016).
Shinohara, K., Funabashi, T., Nakamura, T. J., Mitsushima, D. & Kimura, F. Differential regulation of pituitary adenylate cyclase-activating peptide receptor variants in the rat Suprachiasmatic nucleus. Neuroscience 110, 301–308 (2002).
Wang, Q., Abruzzi, K. C., Rosbash, M. & Rio, D. C. Striking circadian neuron diversity and cycling of Drosophila alternative splicing. eLife 7, e35618 (2018).
Singh, P. & Ahi, E. P. The importance of alternative splicing in adaptive evolution. Mol. Ecol. 31, 1928–1938 (2022).
Chan, S. K. N. et al. The alternative splicing landscape of a coral reef fish during a marine heatwave. Ecol. Evol. 12, e8738 (2022).
Percie du Sert. Reporting animal research: explanation and elaboration for the ARRIVE guidelines 2.0. PLoS Biol. 18, e3000411 (2020).
Jarrold, M. D. & Munday, P. L. Elevated temperature does not substantially modify the interactive effects between elevated CO2 and diel CO2 cycles on the survival, growth and behavior of a coral reef fish. Front. Mar. Sci. 5, 458 (2018).
Shaw, E. C., McNeil, B. I. & Tilbrook, B. Impacts of ocean acidification in naturally variable coral reef flat ecosystems. J. Geophys. Res. Oceans. 117, 2011JC007655 (2012).
Andrews, S. FastQC: a quality control tool for high throughput sequence data. Available online at: (2010). http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Wood, D. E. & Salzberg, S. L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 15, R46 (2014).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Shen, S. et al. rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-Seq data. Proc. Natl. Acad. Sci. 111, (2014).
Thacker, S., Sefyi, M. & Eng, C. Alternative splicing landscape of the neural transcriptome in a cytoplasmic-predominant Pten expression murine model of autism-like behavior. Transl Psychiatry. 10, 380 (2020).
Aktas Samur, A. et al. In-depth analysis of alternative splicing landscape in multiple myeloma and potential role of dysregulated splicing factors. Blood Cancer J. 12, 171 (2022).
Schuster, S. L. & Hsieh, A. C. The untranslated regions of mRNAs in cancer. Trends Cancer. 5, 245–262 (2019).
Gasteiger, E. ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 31, 3784–3788 (2003).
Simpson, C. G. et al. Monitoring changes in alternative precursor messenger RNA splicing in multiple gene transcripts. Plant. J. 53, 1035–1048 (2008).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14, 417–419 (2017).
Love, M. I., Huber, W. & Anders, S. Moderated Estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zheng, X., Zhang, K., Zhao, Y. & Fent, K. Environmental chemicals affect circadian rhythms: an underexplored effect influencing health and fitness in animals and humans. Environ. Int. 149, 106159 (2021).
DeWoskin, D. et al. Distinct roles for GABA across multiple timescales in mammalian circadian timekeeping. Proc. Natl. Acad. Sci. 112, (2015).
Lai, F. et al. Responses of neurogenesis and neuroplasticity related genes to elevated CO2 levels in the brain of three teleost species. Biol. Lett. 13, 20170240 (2017).
Verta, J. P. & Jacobs, A. The role of alternative splicing in adaptation and evolution. Trends Ecol. Evol. 37, 299–308 (2022).
Lu, J. et al. Alternative splicing in teleost fish genomes: same-species and cross-species analysis and comparisons. Mol. Genet. Genomics. 283, 531–539 (2010).
Jacobs, A. & Elmer, K. R. Alternative splicing and gene expression play contrasting roles in the parallel phenotypic evolution of a salmonid fish. Mol. Ecol. 30, 4955–4969 (2021).
Montalbano, G. et al. Acid-sensing ion channels in zebrafish. Animals 11, 2471 (2021).
Liu, S. et al. METTL3 plays multiple functions in biological processes. Am. J. Cancer Res. 10, 1631–1646 (2020).
Schunter, C. et al. Molecular signatures of transgenerational response to ocean acidification in a species of reef fish. Nat. Clim. Change. 6, 1014–1018 (2016).
McGlincy, N. J. et al. Regulation of alternative splicing by the circadian clock and food related cues. Genome Biol. 13, R54 (2012).
Schoenhard, J. A., Eren, M., Johnson, C. H. & Vaughan, D. E. Alternative splicing yields novel BMAL2 variants: tissue distribution and functional characterization. Am. J. Physiol. Cell. Physiol. 283, C103–C114 (2002).
Lee, J., Park, E., Kim, G. H., Kwon, I. & Kim, K. A splice variant of human Bmal1 acts as a negative regulator of the molecular circadian clock. Exp. Mol. Med. 50, 1–10 (2018).
Marcheva, B. et al. Circadian clocks and metabolism. In Circadian Clocks Vol. 217 (eds Kramer, A., Merrow, M. et al.) 127–155 (Springer Berlin Heidelberg, 2013).
Pantazopoulos, H., Gamble, K., Stork, O. & Amir, S. Circadian rhythms in regulation of brain processes and role in psychiatric disorders. Neural Plast. 1–3 (2018). (2018).
Bernhardt, J. R., O’Connor, M. I., Sunday, J. M. & Gonzalez, A. Life in fluctuating environments. Philos. Trans. R Soc. B Biol. Sci. 375, 20190454 (2020).
Niehaus, A. C., Angilletta, M. J., Sears, M. W., Franklin, C. E. & Wilson, R. S. Predicting the physiological performance of ectotherms in fluctuating thermal environments. J. Exp. Biol. 215, 694–701 (2012).
Dufour, C. R. et al. Genomic convergence among ERRα, PROX1, and BMAL1 in the control of metabolic clock outputs. PLoS Genet. 7, e1002143 (2011).
Hrvatin, S. et al. Single-cell analysis of experience-dependent transcriptomic States in the mouse visual cortex. Nat. Neurosci. 21, 120–129 (2018).
Pulido, R. S. et al. Neuronal activity regulates blood-brain barrier efflux transport through endothelial circadian genes. Neuron 108, 937–952e7 (2020).
Nozue, K. et al. Rhythmic growth explained by coincidence between internal and external cues. Nature 448, 358–361 (2007).
Funding
SS was supported through the HKU start-up to CS. The project was funded by the General Research Fund from the Research Grants Council of Hong Kong SAR (17300721). PLM was supported by the ARC Centre of Excellence for Coral Reef Studies and TR was supported by Okinawa Institute of Science and Technology (OIST).
Author information
Authors and Affiliations
Contributions
The experiment was designed and run by MJ with help from PLM and CS. Fish collections were performed by CS and MJ. Molecular lab work was performed by CS and sequenced by CS and TR. SS carried out the transcriptome analysis with input from CS. SS lead the writing of the manuscript with input from CS and all authors read, edited and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Suresh, S., Jarrold, M., Ravasi, T. et al. Ocean acidification induces changes in circadian alternative splicing profiles in a coral reef fish. Sci Rep 15, 22107 (2025). https://doi.org/10.1038/s41598-025-06786-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-06786-0