Abstract
Cotton, the backbone of global textile production, demands sustainable agricultural practices to ensure fiber, food, and environmental security. Cotton crop play an essential role in farming economies; however, production is sometimes affected by various diseases that harm production. We proposed a methodology that uses formal modeling and verification for requirements confirmation to improve the monitoring and detection of cotton crop diseases. The correct information and requirements about disease symptoms can improve disease monitoring and prediction. The Temporal Logic of Action (TLA+) is used to construct a mathematical model to verify requirements by providing disease symptoms and then model checking to ensure correctness properties. Using model checking in TLA + ensures the reliability and correctness of disease symptom detection. We consequently used deep learning models to predict cotton diseases, i.e., Aphids, Armyworms, Bacterial Blight, Powdery Mildew, Target Spot, and Healthy leaf. Our results show that the Convolutional Neural Network (CNN) model achieved an overall accuracy of 98.7% with class-specific accuracy ranging from with F1-scores across all classes (e.g., 0.90 for Powdery Mildew and 0.87 for Army Worm).
Similar content being viewed by others
Introduction
In Pakistan, the agricultural sector is the backbone of the economy, and it relies on producing important crops such as cotton, wheat, rice and corn, which are influenced by strategic and environmental factors that determine yield production. Cotton is an important cash crop that supports the livelihoods of almost 150 million individuals across 75 countries. Pakistan is the fifth-largest cotton producer, contributing 5% to global production and supporting 1.7 million farmers1. Cotton contributes significantly to Pakistan’s economy, using 50% of its industrial workforce and 60% of its exports. Despite its importance, production has declined by 27.2% due to climate change, production costs, poor seed quality, and cotton crop diseases. Climate change, with uneven rainfall and temperature changes, has increased pests like whiteflies and pink bollworms, decreasing cotton yields1.
Remote sensing and machine learning monitor crop productivity, facilitating extensive and continuous observation of agricultural practices. This method2 enables the detection of crop and growth trends over agricultural areas, improving agriculture to maximize land use and resource efficiency. Nazeer et al.3 used a deep learning model that identifies disease susceptibility levels, enabling instantaneous actions to identify disease transmission. A deep learning model is needed to assess susceptibility levels to cotton leaf curl disease in some regions of cotton production, thus contributing to a proactive method for crop management.
Deep learning improved agricultural practices, crop classification, and health evaluation. Shahid et al.4used an ensemble of deep learning models, achieving 98% accuracy in predicting cotton crops from other plants via refined analysis. Technological progress and advanced computational models have acquired significant developments in the agricultural sector. Cotton is vital among numerous crops due to its use in different industries. However, cotton crops are susceptible to diseases, which can significantly impact overall yield and agricultural crop improvement5. Thrips are small, slim insects with fringed wings that feed on plant sap, causing damage by creating stippling and silvering on leaves6. Figure 1 presents the proposed approach for this work.
Jassids, tiny greenish insects; Whiteflies, small white insects, feed on sap, excrete honeydew, and transmit plant viruses7; control methods include insecticides and predatory insects. Pink Bollworm larvae damage cotton bolls, reducing yield; control involves using Bt cotton and insecticides8. The work9 presents machine learning (ML) for accurate cotton yield prediction, using weekly weather as input. Key predictors included minimum temperature and relative humidity. Amani and Marinello10 proposes a deep-learning model to optimize cotton cultivation soil parameters and agricultural variables. Analyzing 13 factors reduces costs and environmental impact while enhancing productivity.
Monitoring cotton crop diseases is critical for supporting sustainable agricultural practices and safeguarding farmers’ livelihoods. Diseases such as Aphids, Army worm, and Bacterial Blight threaten crop yield and quality. Effective detection and management strategies can play a key role in mitigating these risks, minimizing economic losses, and ensuring the reliability of cotton farming approaches.
Problem statement
Detecting cotton crop diseases is essential to mitigate yield losses that affect the agricultural economy and challenges to the agriculture sector. Existing methods for disease detection provide predictions and the monitoring of diseases; however, their reliability depends on verifying their requirements. The problem is to verify the requirements for ensuring correctness properties to improve reliability—a deep learning model for predicting cotton crop diseases based on images dataset.
Research objectives and research question
The objectives of this proposed approach are as follows:
-
Verify the correctness and reliability using TLA+, a robust formal specification and modeling language.
-
Predict cotton crop diseases using a deep learning algorithm based on an image dataset.
-
Evaluations of the Deep Learning model are needed to establish its effectiveness in accurately predicting cotton crop diseases.
-
Facilitate informed decision-making for agronomists and farmers by understanding cotton crop health and enabling crop management practices.
-
R. Q. 1.
How does Temporal logic of action (TLA+) improve correctness and reliability using formal modeling and verification methods to identify cotton crop diseases?
-
R. Q. 2.
How can deep learning models, Convolutional Neural Networks (CNN), be used for early and accurate prediction of cotton diseases based on image data?
Literature review
Machine learning and deep learning advancements have transformed cotton agriculture by solving farmers’ challenges. Da Silva Andrea et al.11 presented a remote sensing and machine learning framework for estimating plant height in cotton fields. This method improves monitoring capabilities, providing efficient growth management in agriculture sectors. Feng et al.12 proposed a technique for detecting and counting cotton saplings with UAV multispectral images. The technology facilitates crop evaluation, enabling farmers to make decisions regarding resource distribution throughout the growth phases.
The classification of cotton leaf diseases has significantly advanced due to deep learning methodologies, essential for disease control, as these diseases lessen yields by 20% each year. A robust approach to cotton plant disease detection using an improved Deep Convolutional Neural Network, achieving an accuracy of 97.98% on a dataset of 2293 images13,14. It explores the impact of data splits, pooling layers, and epoch sizes, outperforming prior methods in accuracy and efficiency. A comprehensive review of machine learning and deep learning techniques for crop pest detection and classification used RF, SVM, CNN, and DBN. It demonstrates their potential to enhance crop productivity and protection by enabling automated, large-scale monitoring with reduced human error and effort15.
The glass-box approach, the Explainable Boosting Machine (EBM), is used for cotton yield prediction across the continental United States16. Key findings focus on precipitation, EVI, and LAI as dominant features, contributing 78% to the model’s feature importance, followed by pairwise feature interactions (16%) and static features (6%).
AI-driven hydroponics system using an Extra Convolutional Neural Network optimized with the Levy Flight Carnivorous Plant algorithm (ExCNN-LFCP) and an IoT cloud server. The system automates and optimizes hydroponic farming by precisely managing environmental parameters, improving effectiveness and sustainability. Sensor data is processed to predict optimal conditions, enabling remote monitoring and centralized control. Experimental results show superior performance, achieving 99.1% accuracy and 97.3% recall17. An automated black gram plant disease (BPLD) detection system, ITL-CHB, using an Improved Inception-V3 Transfer Learning model optimized with the Crossover Honey Badger (CHB) algorithm. The system enhances disease classification accuracy using data augmentation techniques to address data imbalance. Feature extraction is performed using the pre-trained Inception-V3 model, optimized through CHB. Performance evaluation demonstrates 98% accuracy, outperforming traditional methods like SVM, EfficientNet, ResNet-50, and CNN18.
Big data and transfer learning methodologies have improved the scalability of cotton crop surveillance. Stephen et al.19demonstrated a big-data-based cotton plant monitoring system using pre-trained CNN architectures to enhance plant health monitoring. MobileNetV3Large outperformed ResNet18, GoogLeNet, and InceptionV3, achieving 93.9% accuracy, 96.12% specificity, and 97.48% precision. The system classifies 11 cotton plant regions and provides farmers with an understanding of harvesting, yield, and plant health, including wildflower detection and disease identification. Smartphone image collection and a smart application ensure practical, real-world usability for precision agriculture. Aarthi et al.20 illustrated a DenseNet-121 pre-trained model with transfer learning to detect cotton leaf diseases, achieving a classification accuracy of 91%. Compared to traditional CNN methods, this approach improves disease detection efficiency and offers farmers a scalable approach to monitoring crop health. The model supports yield prediction, resource management, and crop protection decision-making.
This work21 uses machine learning techniques to classify surface soil types and evaluate soil physicochemical properties using spectroradiometer and satellite imagery. Applied in Phulambri Tehsil, Maharashtra, models like PLSR, SVM, and SAM achieved 95% classification accuracy for three major soil classes. These findings demonstrate the potential of machine learning in precision farming, enabling efficient soil management and improved agricultural productivity. The advancement of artificial intelligence has transformed different sectors, with agriculture becoming a vital sector experiencing harmful changes22.
Ashapure et al.23 review the applications of machine learning in agriculture, presenting its role in addressing challenges like resource limitations and climate change. Key areas include soil parameter prediction, crop yield forecasting, disease and weed detection, and livestock management through IoT-enabled systems.
Livieris et al.24 presented a multiple-input neural network model for cotton yield prediction, using soil, cultivation, and yield management data processed individually. The architecture efficiently handles mixed data, reduces overfitting, and offers flexibility with low computational cost. Tested on data from cotton farms in Central Greece, the model outperformed traditional and state-of-the-art neural network models, demonstrating its effectiveness in improving yield forecasting and benefiting agricultural economics. This work25 includes robotic capsicum harvesting using YOLOv8s for detection (mAP: 0.967) and YOLOv8s-seg for peduncle detection (mAP: 0.790). The Real Sense D455 camera achieved precise localization, while a tracking algorithm attained 94.1% accuracy. An Android app enhanced usability with performance and a 95% success rate, showcasing the potential for efficient agricultural automation. Wang et al.26 propose the White Bolls Index (WBI) for automatic cotton mapping, achieving over 82% accuracy across four regions. WBI distinguishes cotton based on boll opening intensity and identifies the onset of the boll opening stage using Sentinel-2 time series data, offering a scalable and efficient solution for crop monitoring.
This research introduces an ontology-based knowledge map model for dynamically managing and using data mining results in crop farming. The proposed system27 architecture for knowledge modeling, extraction, and exploitation has been implemented for crop management, demonstrating its effectiveness and potential for broader applications in agriculture. Haider et al.28 presents a crowd-sourced platform to gather agricultural knowledge and data for wheat disease identification. By combining Decision Trees (DT) and deep learning models like CNNs and validating results with domain experts, the approach achieved significant accuracy improvements: DT (28.5%) and CNN (4.3%, reaching 97.2%).
Narcisse et al.29demonstrated the development of ontoSYSPARCOTCI, a domain ontology for cotton plant health surveillance in Côte d’Ivoire, addressing pest challenges and enhancing systematic monitoring. Using the NeOn methodology, the ontology integrates resources and scenarios for structured knowledge representation. Results include a conceptual model in UP/UML and a hierarchy of concepts in OWL created with Protégé, enabling semantic reasoning and knowledge sharing for improved pest management.
The formal method checks the model’s correctness and reliability using formal verification techniques. Model checking is the technique for system verification. The model verifies whether the property is the variable of the system or not. Formal verification will help check the correctness of the proposed design based on its correctness properties. TLA + is a language used in the formal specification of concurrent and distributed systems, which involves verifying and checking the correctness30.
Gaps in previous research
Previous research on cotton leaf disease detection has made significant strides using deep learning models such as Deep CNN (97.98% accuracy), MobileNetV3Large (93.9%), and YOLOv8s. However, key gaps remain. Previous studies rely on small or imbalanced datasets, lack generalization across diverse environments, and often neglect real-world deployment challenges due to high computational costs. Moreover, formal verification methods ensure model reliability and availability in critical agricultural real-world applications. Our work addresses these issues by using a robust, different dataset, optimizing computational efficiency, and incorporating formal verification steps.
State of the art
Formal modelling and verification
Formal modeling and verification based on mathematics, logical construction, and formal language define and verify requirements that confirm the specifications. Formal modeling and verification are essential in safety-critical systems, offering a systematic mathematical framework that ensures correctness and reliability.
Formal verification is rigorous and ensures the correctness properties of the system’s safety. Formal methods are used in requirement verification and design to safeguard correctness properties using state, transition, proof obligations, theorem proving and inductive logic31. A model provides a basis to improve and verify the system rigorously, and it is also significant in safeguarding the system’s correctness. It takes input and systemically checks whether the system holds the correct safety properties.
Formal modeling involves the development of mathematical verification of a system’s architecture and dynamics, enabling potential states and transitions that the system experiences. The method eliminates ambiguities in specifications and ensures the system’s functionality corresponds with its requirements. Verification involves rigorously demonstrating that the model complies with fundamental qualities, such as safety and liveness, which ensure that the system avoids undesirable situations and reliably executes its intended activities.
This work systematically verifies that these criteria are maintained across scenarios using formal verification techniques like theorem proving and model checking. The model checker systematically inspects the model’s state space, ensuring designated properties, such as invariants or temporal properties, are designated. A Formal verification and modeling technique helps detect undetected errors in conventional testing methods.
Temporal logic of actions (TLA+)
Temporal logic of action (TLA+) is a formal specification and modeling language based on temporal logic and mathematical foundation. Leslie Lamport, TLA+, created a software development tool to ensure the correctness of concurrent and distributed systems32. It uses mathematical logic to describe system behaviour and allows designers to formulate and verify their design methods. Formal modeling and verification with the TLA + and PlusCal language are effective measures for ensuring that the concurrent and distributed systems are correct. TLA + is a language used for modeling behaviour, and the implementation process is more accessible than Pluscal. Formal verification via model checking ensures that a system verifies its requirements32. Figure 2 presents Workflow diagram for Temporal logic of action.
Model checking
Model checking33,34,35,36,37,38,39,40,41is one of the most practical formal approaches for systematic, automatic, and exhaustive verification. “It is a computer-assisted method for analyzing dynamical systems that state-transition systems can model“35 A mathematical model of the system is formed, and a comprehensive evaluation of the model is executed. It comprises checking all states and transitions in the model, i.e., an exhaustive model analysis. It generates an abstract demonstration of a system with all the possible transitions and states.
The correctness property of the model is verified through model-checking methods. The model takes input and systemically checks whether the system safeguards the system’s property. It is used to verify projects, i.e., the verification of software systems for spacecraft, nuclear reactors, aeroplanes, subway trains, and satellites. Its goal is to improve the reliability of verification by checking correctness properties.
“Model checking is an automated technique that, given a finite-state model of a system and a formal property, systematically checks whether this property holds for (a given state in) that model. Model-based verification techniques are based on models describing the possible system behaviour mathematically, precisely and unambiguously. The accurate modeling of systems leads to the identification of incompleteness, ambiguities, and inconsistencies in informal system specifications. A system model is accompanied by algorithms that systematically explore all states of the system model. It provides the basis for verification techniques ranging from an exhaustive exploration (i.e. model checking) to experiments with a restrictive set of scenarios in the model (i.e. simulation), or in reality (i.e. testing)“36.
Model checking uses temporal logic, i.e. linear-time temporal logic (LTL)44,45,46 and computation-tree temporal logic (CTL)42,43 for stating, checking, and verifying behavioural properties. Model checking is the mathematical verification of a system, and its result involves an exhaustive systematic investigation of the mathematical model47.
Correctness property
Safety property is correctness property. Correctness properties provide detailed system verification. The safety property is an invariant that asserts that “something bad never happens, that an acceptable state of affairs is maintained.” Calegari and Szasz48have defined safety property “S = {a1, a2,., an} as a deterministic process that asserts that any trace, including actions in the alphabet of S, is accepted by S. ERROR conditions are like exceptions which state what is not required, as in complex systems we specify safety properties by directly stating what is required”. According to49 “a safety property is a property that can specified by a safety formula of the form □p (i.e. temporal operator □ meaning always). This formula states that the property p holds throughout the computation”.
Justification for TLA+ model
TLA + selected for formal modeling and verification due to its strength in specifying and reasoning about system behaviours over time, particularly in concurrent or reactive systems like automated disease detection pipelines. TLA + allows us to rigorously define correctness properties—such as consistency, safety, and liveness- which are critical when deploying models in real agricultural environments. By applying model checking through TLA+, we systematically explore all possible states and transitions, ensuring that the detection process adheres to predefined rules under varying conditions. This approach enhances the reliability and robustness of our system by preemptively identifying potential design flaws before real-world deployment.
Convolutional neural network (CNN)
Zohar and Amir50 thoroughly studied the coefficients and other CNN structures used in computer vision. CNN, a deep learning model, can solve problems involving grid-like data structures such as images and videos51. An essential part of a CNN consists of kernels, strides, padding, pooling, and flattening.
The convolutional filter kernel is essential for extracting features from input data in convolutional neural networks. The convolution operation takes a kernel and slides it across the input by conducting element-wise multiplication and summing the outputs. The feature maps capture the fundamental patterns via edges or textures, which help the network acquire hierarchical representations from the input. The movement spacing in CNNs, determined by the stride parameter, is the kernel’s distance when it moves over the input during the convolution process. Upon the prolongation of each step, the spatial down-sampling appears, decreasing the size of the output feature map, which, in turn, enhances calculation speed. Reducing the length of each step maintains the spatial details, resulting in larger maps of distinctive features. The selection of stride directly affects the balance between spatial resolution and computational workload, affecting the network’s capacity to identify complex patterns in the input.
In CNNs, padding adds extra pixels around the input data, usually with zero values. The main objective of padding is to avoid spatial dimension loss following convolution. It ensures that the convolution process considers the entire input space, which is significant at the corners where information could be lost. Padding is critical for preserving spatial information and maximizing the features for the overall performance and effectiveness of the network. Pooling layers in CNNs is vital for reducing the size of the feature maps produced by convolutional layers. Max pooling and average pooling are prevalent methods for selecting average values from a cluster of adjacent pixels. Combining reduces complexity while maintaining translation invariance and preserving crucial information in feature maps. By pooling, learners discard less essential details and concentrate on the critical attributes to achieve more effective hierarchical feature learning. The following stages of the convolutional layer and the pooling layer result in the output matrix flattened into a linear vector with several dimensions. Those low-dimensional vectors are put in fully connected networks and converted to a more familiar neural network structure. The flattened layer is fundamental for the operation of CNNs because it provides a format for the spatial features of the acquired image, more processing, and making decisions. It links the convolutional, fully connected layers, thus ensuring that when the network learns subtle patterns and correlations from the input information, it will identify complex tasks like image classification.
The convolution operation is a fundamental aspect of Convolutional Neural Networks (CNNs) and is crucial in feature extraction. Mathematically, the equation represents the sliding of a filter (or kernel) over the input image or previous feature map. The kernel weights are multiplied by the corresponding pixel values in the receptive field, and the sum of these products is computed for each kernel position. This sum is then passed through an activation function, often a nonlinear function such as ReLU or Sigmoid, to introduce non-linearity into the network. The result of this operation is a feature map that captures spatial hierarchies of the input data, helping the network to detect patterns such as edges, textures, and shapes in images. Adding a bias term further enhances the model’s flexibility by shifting the activation, allowing the network to fit the data better.
Batch normalization (BN) is a technique employed to accelerate the training of deep neural networks by mitigating internal covariate shift, which refers to the changes in the distribution of layer inputs during training. The equation for batch normalization normalizes the input data by subtracting the batch mean and dividing it by the batch standard deviation, effectively standardizing the activations within each mini-batch. This step ensures that the network learns with consistent data distribution, reducing the chances of vanishing or exploding gradients. After normalization, the data is scaled and shifted by learned parameters γ (gamma) and β (beta), allowing the model to recover any necessary shifts or scaling that might have been lost in the normalization process. This operation stabilizes the training process and enables faster convergence, often resulting in improved model performance.
Max pooling is a downsampling operation commonly used in CNNs to reduce the spatial dimensions of the feature maps while retaining the essential information. The equation for max pooling describes the process of selecting the maximum value from each patch of the feature map, typically within a fixed-size window (e.g., 2 × 2 or 3 × 3). The primary goal of max pooling is to introduce spatial invariance by reducing the impact of small translations, rotations, and distortions in the input data. This operation reduces the computational burden of the network and helps mitigate overfitting by forcing the network to focus on the most prominent features rather than being sensitive to noise or minor variations. The downsampling effect achieved by max pooling enables deeper networks by reducing the number of parameters, thereby improving the model’s efficiency.
A neural network’s dense (or fully connected) layer aggregates information from all previous layers and maps it to the output. The equation for a dense layer involves multiplying the input vector by a set of weights and adding a bias term, which determines the activation level of each neuron in the layer. The output is then typically passed through an activation function, such as ReLU, Sigmoid, or Softmax, to introduce non-linearity and allow the model to learn complex patterns. In the context of a CNN, dense layers are typically used in the final stages of the network, where they aggregate the learned features from the convolutional and pooling layers to produce the final classification or regression output. The weights in the dense layer are learned through backpropagation during training, allowing the network to adjust and improve the mapping from input data to output predictions.
The softmax function is used primarily in the output layer of neural networks for multi-class classification problems. It converts the raw output scores logits into probabilities, ensuring that the sum of all predicted probabilities equals one. The softmax equation exponentiates each logit and normalizes it by dividing it by the sum of the exponentiated logits. This normalization process transforms the logits into a probability distribution, where each value represents the likelihood of a specific class. By applying softmax, the model provides a clearer interpretation of its output, enabling it to make probabilistic decisions. The class with the highest probability selected as the predicted label. Softmax is particularly advantageous in classification tasks with multiple possible categories, as it provides a coherent framework for decision-making, making it easy to identify the most probable class among many alternatives.
Recurrent neural network (RNN)
Recurrent Neural Network (RNN) produced to respond to problems from the sequences of input recognition52. Vanilla RNN aims to learn from input data encoded in the past but finds it hard to keep track of the information over a long period. Long Short-Term Memory (LSTM) is based on specific strategies that can catch and hold long-term associations more efficiently. Bidirectional RNNs can read input text in both forward and backward directions, allowing them to understand the contextual information from the prior and forward parts of the text at one time. In ESNs (echo state networks), the recurrent connections follow the simplest framework, whereas hierarchical RNNs with numerous layers provide an opportunity to learn complex representations from the data. Temporal resolutions divided among different neurons in Clockwork RNNs, and attention-based RNNs use mechanisms over a specific sequence segment. Every variant intended to address a challenge with various strategies combined with the ability of RNNs to perform tasks, including natural language processing, speech recognition, and time series analysis53.
Here, W_hh, W_xh, and W_hy are the weight matrices for the hidden-to-hidden, input-to-hidden, and hidden-to-output connections, respectively. b_h and b_y are the hidden state and output bias terms, respectively.
Long short-term memory (LSTM)
Long short-term memory (LSTM) is a progressive version of the RNN with a structure to overcome the difficulties of long-term patterns in sequential input collection57. LSTMs address the vanishing gradient problem that limits the capability of used RNNs to sustain information over long intervals. LSTMs include memory cells and three gating mechanisms, i.e. input, forget, and output gates, enabling the network to keep, discard, and produce information selection.
Here, W_f, W_i, W_C, and W_o are the weight matrices for the forget gate, input gate, cell state candidate, and output gate. b_f, b_i, b_C, and b_o are the corresponding bias terms. LSTMs can acquire and preserve significant contextual information over long periods, which makes them efficient at tasks that include sequences. LSTMs have demonstrated their significance in capturing complex relationships within sequential data, making them a fundamental component in various deep-learning applications54.
Materials and methods
Temporal logic of action (TLA+)
The first part of the work results concentrates on the TLA + model used to validate the safety properties for cotton disease monitoring. The TLA + specification confirms that the monitoring procedure complements safety properties, improves results and categorizes important diseases. Exploiting a model checker to analyze the TLA + specification enabled us to assess the system’s behavioural correctness across several scenarios.
Requirement elicitation of symptoms for disease monitoring
Requirement elicitation is a fundamental step in the development process, gathering and eliminating stakeholders’ requirements55. The first stage involves identifying, analyzing, and documenting the requirements for effective results. By eliciting clear, detailed information, requirement elicitation helps ensure that all project elements align with stakeholder goals. It involves techniques like interviews, surveys, observations, and making matrices to organize information effectively56,57. For the cotton crop case study, requirement elicitation includes listing diseases and their symptoms in a table for disease management. Table 1 demonstrates the Requirement Elicitation of Observed Symptoms. The elicitation helps stakeholders, such as farmers and researchers, understand the conditions and symptoms that need an efficient solution design and approach.
Requirement verification using temporal logic of action (TLA+)
The verification method confirmed that the safety properties specified in the TLA + model have been ensured effectively. The platform corrects transitions between states, ensuring that a suitable response (such as generating an alert) activates disease detection. Moreover, formal verification guarantees no chance of incorrect identification within the defined safety restrictions.

The CottonDiseasePrediction module, created through PlusCal, aims to forecast cotton plant diseases based on user-provided symptoms. The model incorporates constants such as SymptomInputs, denoting the symptoms exhibited by a cotton plant; SymptomsByDisease, which associates each disease with its respective symptoms; and DiseaseNames, which contains a list of possible diseases. The disease identification mechanism evaluates the user’s reported symptoms against the recorded symptoms of each condition to determine the most accurate match.

The algorithm starts establishing several factors, including DiseaseMatches (an array to monitor the match count for each disease), MostLikelyDisease (to retain the disease with the highest match), and HighestMatchCount (to record the maximum number of matching symptoms). During the CountMatches phase, the algorithm examines each disease, calculates the intersection between user-provided symptoms and established symptoms, and tallies the matches—the match count for each disease recorded in the DiseaseMatches array. Upon completion of all match calculations, the algorithm re-initializes the loop variable to determine which disease displays the highest match.

During the DetermineDisease phase, the algorithm evaluates the match counts for all diseases to identify the most corresponding symptoms. Should a disease exhibit a higher match count than the highest, the algorithm adjusts MostLikelyDisease and HighestMatchCount, respectively. The disease with the highest probability and its corresponding match count are displayed, indicating the condition that most accurately aligns with the given symptoms.
Invariants and model checking
The invariants InvariantNonNegativeMatches and InvariantMatchLimit ensure that the matching process remains logically sound. InvariantNonNegativeMatches guarantees that the match count for any disease (DiseaseMatches[j]) is never negative. InvariantMatchLimit ensures that the match count not exceed the total number of input symptoms (Cardinality(SymptomInputs)), maintaining realistic bounds on the comparisons. Figure 3 represents the Temporal State of Action for Disease Symptoms, and Fig. 4 shows the State Space Actions of Module Cotton Disease.
Safety property for requirement correctness
The Safety property ensures that the match count for any disease (DiseaseMatches[j]) never exceeds the current highest match count (HighestMatchCount), maintaining consistency during the disease identification process.
Deep learning models
Experimental setup and deployment
This work used an input image size of 256 × 256 for CNN models to ensure enough detail was captured from the cotton leaf images while keeping computation manageable. However, models like VGG19 and MobileNet are pretrained on the ImageNet dataset, which uses a default input size of 224 × 224, so their architectures are optimized for that size—hence, resizing input images accordingly when using these models is important. A batch size of 32 was consistently used across models as it balances training speed and GPU memory usage well. On GPUs, batch size doesn’t usually cause problems as long as it fits in memory, and keeping it consistent helps ensure stable and comparable training performance across models.
Dataset description
The dataset includes images for six common cotton leaf diseases—Aphids, Army Worm, Bacterial Blight, Powdery Mildew, and Target Spot—and healthy leaves for comparison. The dataset includes 3601 images. It is specifically designed to focus on diseases affecting only the leaves, without reference images for stems, buds, flowers, or bolls. The targeted approach makes it ideal for training machine learning models to identify diseases, helping in detection practices for cotton crops. Figure 5 shows the dataset sample images for cotton crop dataset.
Deep learning model details
The convolutional neural network (CNN) model used for monitoring and predicting cotton leaf diseases follows a layered architecture designed to extract and classify image features effectively. The input layer receives a batch of 32 RGB images, each with a size of 256 × 256 pixels. The model starts with convolutional layers (Conv2D) with 3 × 3 kernels and ReLU activation, progressively increasing the number of filters from 32 to 64 to capture intricate features. MaxPooling2D layers follow each convolutional layer, reducing the spatial dimensions by half, thus downsampling the images. This sequential pattern of convolution and pooling continues for five layers, with the feature maps shrinking in size while retaining the most significant information. After flattening the pooled feature maps into a 1D vector, a dense layer with 64 neurons and ReLU activation is applied, followed by the output layer uses softmax activation to classify the input into one of the six disease categories. Table 2 shows deep learning model details.
Justify hyperparameter choices
The choice of hyperparameters in this CNN model for cotton leaf disease classification is critical for ensuring both model performance and stability. The learning rate is set to the default value of 0.001, providing a balanced training approach and ensuring the model converges smoothly without overshooting the minima. The architecture comprises 5 convolutional layers (Conv) and 5 pooling layers (Pool), which allow the network to learn complex hierarchical patterns from the input data. The 3 × 3 filter size is chosen as it is standard for CNNs, offering a good trade-off between capturing spatial features and reducing the number of parameters, thus minimizing computational costs. The number of filters increases from 32 to 64 as the network deepens, enabling richer feature extraction while maintaining a manageable model size. ReLU activation is used in the convolutional layers to mitigate the vanishing gradient problem, while Softmax is employed in the output layer to handle multi-class classification. A batch size of 32 is selected to balance GPU memory usage and the stability of the training process. The Adam optimizer, known for its adaptive learning rates, is assumed to be used, which is well-suited for most CNN-based tasks. Table 3 shows hyperparameter choices justification.
Hyperparameters chosen based on extensive experimentation: a learning rate of 0.001 ensures stable convergence, dropout rates of 0.3 and 0.5 prevent overfitting, and filter sizes 3 × 3 maintain spatial hierarchy. CottonLeafNet was selected for its accuracy and computational efficiency balance, outperforming heavier models in resource-limited agricultural settings.
This work uses TensorFlow and Keras libraries for model construction, using the functionalities provided by these libraries to handle data processing and augment training data effectively58,59 . The network begins with a convolutional layer with 32 filters and several layers with increased filter sizes. Each convolutional layer uses a (3 × 3) kernel and ReLU activation to ensure the model learns non-linear patterns60. Max-pooling layers are combined after each convolutional layer to reduce the spatial dimensions, which helps decrease computation and prevents overfitting by extracting dominant features.
Following the convolutional and pooling layers, the model flattens the extracted feature maps into a one-dimensional vector, passing through a fully connected (dense) layer with 64 neurons. This dense layer further refines the learned features before the final output layer. The output layer contains six neurons, each representing a class in the cotton crop classification problem, and uses a softmax activation function. This setup assigns probabilities to each class, enabling the model to output the likelihood that an image belongs to a specific category.
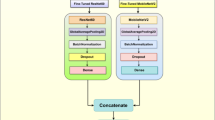
Figure 6 represents the architecture of a CNN used for image classification tasks. It starts with an input layer, passing images into the network. The ConvLayer applies convolution operations using filters (e.g., 3 × 3), creating feature maps. These are normalized with BatchNormalization and activated using ReLU to introduce non-linearity. MaxPooling reduces the spatial dimensions while retaining essential features. The convolution, normalization, and pooling processes are repeated to deepen the network. Flattening converts feature maps into a vector passed through FullyConnectedLayers for dense feature extraction. Dropout prevents overfitting, and the OutputLayer uses softmax for final class probabilities.
Experiments with 100, 150, 300 epochs and analysis results
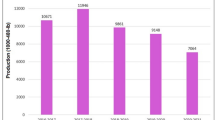
Figure 7 illustrates performance comparisons across epochs. The CNN model trained for 50, 100, and 300 epochs to assess the effect of training duration on model performance. At 50 epochs, the model achieved an accuracy of 94.53%, with a precision of 94.31%, recall of 95.04%, and F1 score of 94.46%. Training for 100 epochs improved the accuracy to 96.61%, and further training for 300 epochs yielded the best performance with an accuracy of 98.7%, precision of 98.63%, recall of 98.7%, and F1 score of 98.65%. These results demonstrate that increasing the number of epochs can significantly enhance the model’s ability to learn and generalize, provided overfitting is controlled.
RNN such as LSTM as a comparison and report findings
When comparing the performance of the CNN and LSTM models, it is evident that the CNN significantly outperformed the LSTM in classifying cotton leaf diseases. The CNN model achieved an overall accuracy of 98.7%, with F1-scores across all classes (e.g., 0.90 for Powdery Mildew and 0.87 for Army Worm), indicating reliable and consistent predictions. In contrast, the LSTM model only reached an accuracy of 48%, with lower precision and recall scores across most classes, particularly struggling with classes like Target Spot and Bacterial Blight. This suggests that CNN, better suited for image data due to its ability to capture spatial features, is more effective and stable than LSTM for this classification task.
Ablation studies to analyze the impact of data augmentation and hyperparameters
The ablation study reveals that data augmentation significantly improves model performance, as removing it led to a drop in accuracy. Interestingly, eliminating image rescaling resulted in the highest test accuracy, suggesting that the rescaling process might have negatively affected input quality. Changes in hyperparameters such as optimizer, kernel size, and the number of filters had little to no impact on accuracy. Figure 8 shows Ablation study Impact of Preprocessing and hyperameters. The study highlights that data preprocessing techniques, particularly augmentation and rescaling, play a more critical role in model performance than minor architectural changes.
Computational efficiency and real-world applicability
Running the model on a GPU significantly accelerated training and evaluation, enabling efficient experimentation with different configurations and ablation studies. Computationally, this demonstrates the practicality of deploying deep learning models in real-time scenarios where speed is crucial, such as in mobile or IoT-based plant disease detection systems. In real-world applications, the model’s ability to identify cotton diseases can assist farmers in making timely decisions, reducing crop loss, and improving yield. The study also highlights how tuning data preprocessing and architecture can enhance model reliability, making it suitable for deployment in agricultural monitoring tools. By using a deep learning model, the network learns hierarchical features that enable it to differentiate between understated differences in plant diseases and pests. Figure 9 demonstrates the sample prediction of cotton crop diseases. The CNN model is used in this study to analyze visual features such as colour variations, texture anomalies, and irregularities in leaf structures. These competence features of CNNs in agricultural disease detection provide a solution for detection. Such models can help farmers take preventive measures to improve crop health and yield. These predictions also demonstrate the probability of further scalability across various plant species and environmental conditions.
.
Discussion and results
Disease monitoring model acceptance criteria
The Disease Monitoring Model is constructed on well-defined requirements and formal verification to ensure its reliability and correctness in monitoring cotton crop disease. Our previous work in confirming the correctness properties of safety for autonomous robotic multi-agent systems61,62,63 and flood monitoring systems64,65,66,67,68 have helped us in the formal modeling and verification of cotton crop disease detection. Formal modeling constructs the model behaviour in identifying diseases based on input symptoms. The model ensures correctness and reliability throughout construction by formal requirements into functional and non-functional categories.
Functional requirements, which are written in the table, define the critical tasks of the model, such as initializing key variables, iterating through diseases, calculating symptom matches, and identifying the most likely disease. Table 4 presents the acceptable criteria for the disease detection model. The associated acceptance criteria confirm that the implementation follows these defined behaviors. For example, DiseaseMatches and MostLikelyDisease are initialized appropriately, while iterations through DiseaseNames ensure accurate match calculations and disease identification.
Non-functional requirements and efficient handling of various disease symptoms ensure that the model performance remains robust even when processing multiple diseases and symptoms. Input and output requirements validate that the input symptom set and disease list are well-formed, while output specifications confirm that the system provides clear, actionable information regarding identified diseases and matching symptom counts.
Formal verification plays a critical role in validating the model requirements. Probable errors, such as failing to identify diseases with zero matches, are addressed by confirming logical reliability and correctness. The structured acceptance criteria are a benchmark, ensuring the model meets stakeholders’ prospects while maintaining scalability, reliability, correctness, and clarity in disease identification.
Performance evaluation of deep learning model
Why accuracy varies across diseases
Accuracy varies across cotton leaf disease classes due to multiple interrelated factors. As the Table 5 Classification report the visual similarity between diseases, such as Bacterial Blight and Target Spot, often leads to model confusion due to overlapping symptoms, whereas distinctly manifested diseases like Powdery Mildew—characterized by white powdery lesions—yield higher precision (0.98). Minor class imbalances and differences in image quality or intra-class variability also impact performance; for instance, Bacterial Blight showed lower precision (0.62), possibly due to noisy or blurred samples. In some cases, recall and precision trade-offs indicate class-specific misclassification tendencies: Aphids had high recall (0.89) but lower precision (0.75), suggesting over-prediction, while Army Worm demonstrated substantial precision (0.90) but moderate recall (0.84), implying under-detection. Additionally, overlapping symptoms among diseases and healthy leaves (e.g., discolouration, necrotic spots) contribute to inter-class confusion, affecting recall and precision. Finally, the diversity of test samples, including variations in lighting or leaf maturity, particularly in the Healthy class, influences the model’s ability to generalize, underscoring the need for high-quality, diverse datasets.
Confusion matrix
The provided confusion matrix illustrates the performance of a deep learning model for predicting cotton leaf diseases. The matrix classifies five categories: Aphids, Army Worm, Bacterial Blight, Healthy, Powdery Mildew, and Target Spot. Each row represents the actual class, while the columns correspond to the predicted classes. High values along the diagonal indicate correct predictions, with Bacterial Blight (66) and Target spot (47) having the highest accuracy in classification. Misclassifications are apparent in non-diagonal cells, such as the 7 misclassified Armyworms predicted as Aphids and the 6 misclassified Powdery Mildew predicted as Aphids. Figure 10 presents confusion matrix for cotton crop diseases leaf and healthy leaf The model performs well overall, as evidenced by the significant concentration of correct classifications, but there is still room for improvement in differentiating specific disease categories.
ROC curve
The ROC curve for the multi-class classification model demonstrates its effectiveness in predicting cotton leaf diseases. Each class, including Aphids, Armyworm, Bacterial Blight, Healthy, Powdery Mildew, and Target Spot, represents a distinct line with corresponding AUC (Area Under the Curve) values. The model shows strong classification performance, with most classes achieving AUC values above 0.96, indicating high predictive accuracy. The highest AUC values are observed for Aphids, Armyworms, and Healthy and Powdery Mildew (AUC = 0.98), reflecting their well-separated distributions regarding true and false positive rates. Bacterial Blight shows a slightly lower AUC value of 0.96, suggesting a marginal decrease in classification performance for this category. Figure 11 shows ROC Curve for multiple classes of cotton diseases The Chance line (dashed) represents random guessing, further shows the model’s superior performance across the categories. Overall, the ROC curve illustrates a robust classification ability, with minimal misclassification across the cotton leaf disease categories.
Confidence intervals (95%)
The 95% confidence interval for accuracy, ranging from 0.286 to 0.380, indicates that with 95% confidence, the true accuracy of the model falls within this range. This means that if you repeatedly test the model on different datasets or under slightly varying conditions, 95% of the time, the accuracy would fall between 28.6% and 38.0%. The relatively wide range suggests considerable uncertainty in the model’s performance, possibly due to factors such as high variability in the data or insufficient model training. While the model’s accuracy may still be helpful, this range highlights the importance of gathering more data or refining the model to increase the precision and reliability of the performance estimate.
K-fold cross-validation (k = 10) for generalization
To ensure robust generalization of the proposed model, a 10-fold cross-validation conducted. Several enhancement techniques incorporated into the model architecture and training process. Firstly, data augmentation strategies such as random flips, rotations, and zooming were employed to artificially expand dataset diversity, reducing the risk of overfitting and enhancing the model’s ability to generalize to unseen data. Batch normalization was applied after each convolutional layer to stabilize learning by normalizing activations, improving training dynamics and model robustness. Additionally, dropout was introduced in the dense layer before the output to mitigate overfitting by randomly disabling neurons during training, forcing the network to learn more generalized features. L2 regularization further discouraged large weight magnitudes, promoting simpler, more generalizable models. The training was also optimized using early stopping, which halted learning when validation loss ceased to improve, and a learning rate scheduler (ReduceLROnPlateau) dynamically reduced the learning rate upon stagnation in validation performance. The k-fold validation results demonstrated encouraging performance, with the best validation accuracy reaching 82.64% (Fold 5) and an average accuracy of 80.56% across all folds. The accuracy range (0.7603–0.8264) indicated consistent performance with low variance, suggesting that the model reliably captures the distinguishing features of cotton leaf diseases and generalizes well across different data partitions.
Compare performance with state-of-the-art models (VGG19, Alex-Net, Mobile-Net, Efficient-Net)
This study conducted a comparative analysis of the baseline CNN achieved a commendable performance with an accuracy of 95.57% and a precision, recall, and F1-score of 0.96, indicating reliable but suboptimal classification capability. In contrast, MobileNet demonstrated superior performance, attaining 99.74% accuracy with perfect scores (1.00) across all key metrics, including precision, recall, and F1-score. Although VGG19 initially yielded moderate results, post-fine-tuning, its performance significantly improved and aligned with MobileNet’s, reaching 99.74% accuracy and perfect evaluation metrics. Despite the comparable accuracy, MobileNet’s lightweight architecture offers efficiency, particularly for resource-constrained or mobile environments, making it preferable for real-time field deployment. Table 6 shows comparison with previous studies. These findings underscore the effectiveness of transfer learning and highlight that, when fine-tuned appropriately, pre-trained models like MobileNet and VGG19 can significantly outperform conventional CNNs in classification accuracy and generalization ability.
Conclusion
This work focuses on the significance of formal modeling and formal verification for improving the reliability and correctness of cotton crop disease monitoring and detection methods. Using the TLA + improved a robust mathematical model that systematically verified the requirements for disease symptom detection. Using model checking, we ensured the constructed model’s correctness, centered on validating safety properties. The safety property verified to confirm the method’s ability to match disease symptoms with the highest confidence correctly. This approach guarantees a consistent and reliable framework for disease monitoring. Moreover, deep learning models, i.e. CNN, LSTM, and RNN demonstrated the effectiveness of deep learning in predicting cotton crop diseases. The CNN model achieved an overall accuracy of 98.7%, with class-specific accuracy metrics ranging from with F1-scores across all classes (e.g., 0.90 for Powdery Mildew and 0.87 for Army Worm).
Contributions
-
I.
Formal verification of disease symptoms using TLA+: This Work presents TLA + as a formal approach for verifying requirements in agricultural disease detection. The application of formal verification ensures the robustness of the monitoring framework. We showed model checking using TLA + to verify critical symptom detection parameters, thereby increasing confidence in the accuracy of disease detection under various crop conditions.
-
II.
Formal methods enhanced reliability and correctness: This Work is set apart using formal verification, resulting in a reliable and scalable framework for crop monitoring. This method rectifies a critical shortcoming in the agricultural industry by ensuring correctness in symptom requirements detection when applying formal Modeling and verification to crop management methods.
-
III.
Deep learning model for disease prediction: The CNN, LSTM, RNN model is trained to predict six situations, including Aphids, Armyworms, Bacterial Blight, Powdery Mildew, Target Spot, and Healthy crop states.
-
IV.
Prospective for field deployment and practical impact: This approach corresponds with actual agricultural needs, providing a practical choice for evaluating cotton crop health. The framework’s robust predictive accuracy and reliability indicate significant potential for agricultural use, improving disease management procedures and resulting in more crop yields.
Future work
Short-term objectives
In the short term, the aim is to ensure the effective performance of the disease detection framework for cotton crops. Deep learning model to identify specific diseases, like Aphids, Armyworms, Bacterial Blight, Powdery Mildew, and Target Spot, while recognizing healthy crop conditions. Furthermore, we use TLA + to verify an understanding of each disease’s symptoms, utilizing model checking to validate the correctness of symptom detection. Collaborating with agricultural professionals during requirement collection to improve and ensure it matches users’ requirements in agrarian environments.
Long term objectives
In the long term, we propose to increase the impact of our cotton crop monitoring system by broadening its applicability to other vital crops, including rice, wheat, and corn, and to determine diseases affecting crops by training it on novel, crop-specific disease datasets. The proposed approach provides a structure applicable to various crops it is used for a broader spectrum of farmers and agricultural sectors, improving its reliability across various farming communities. A monitoring approach aims to transform conventional agrarian methods, enhancing both productivity and environmental conservation in the long term. The strategy coincides with global objectives for sustainable agriculture and food security through robust crop management practices.
Data availability
The dataset analysed during the current study is available in the “Cotton Plant Disease” dataset, https://www.kaggle.com/datasets/dhamur/cotton-plant-disease/dataAll coding files, including deep learning models and TLA+ toolbox modules, are publicly available on GitHub at https://github.com/abdulpk/MuhammadMusa for further use and analysis.
References
Ashraf, S., Ch, K. M., Ashraf, I. & Akbar, N. A phenomenological inquiry into farmers’ experiences growing cotton in punjab, Pakistan. Sci. Rep. 14 (1), 13846 (2024).
Hasan, S. S., Goheer, M. A., Uzair, M. & Fatima, S. Modeling and monitoring cotton production using remote sensing techniques and machine learning: a case study of Punjab, Pakistan. Environ. Dev. Sustain. 1–22 (2024).
Nazeer, R. et al. Detection of cotton leaf curl disease’s susceptibility scale level based on deep learning. J. Cloud Comput. 13 (1), 50 (2024).
Shahid, M. F. et al. An ensemble deep learning models approach using image analysis for cotton crop classification in AI-enabled smart agriculture. Plant. Methods. 20 (1), 104 (2024).
Papageorgiou, E. I., Markinos, A. T. & Gemtos, T. A. Fuzzy cognitive map based approach for predicting yield in cotton crop production as a basis for decision support system in precision agriculture application. Appl. Soft Comput. 11 (4), 3643–3657. https://doi.org/10.1016/j.asoc.2011.01.036 (2011).
Khan, M. & Damalas, C. A. Farmers’ knowledge about common pests and pesticide safety in conventional cotton production in Pakistan. Crop Prot. 77, 45–51. https://doi.org/10.1016/j.cropro.2015.07.014 (2015).
Fue, K. G., Porter, W. M., Barnes, E. M. & Rains, G. C. An extensive review of mobile agricultural robotics for field operations: focus on cotton harvesting. AgriEngineering 2 (1), 150–117. https://doi.org/10.3390/agriengineering2010010 (2020).
Ülkü, M. A., Akgün, M., Venkatadri, U., Diallo, C. & Chadha, S. S. Managing environmental and operational risks for sustainable cotton production logistics: system dynamics modelling for a textile company. Logistics 4 (4), 34. https://doi.org/10.3390/logistics4040034 (2020).
Kashyap, G. R. et al. Machine learning ensembles, neural network, hybrid and sparse regression approaches for weather based rainfed cotton yield forecast. Int. J. Biometeorol. 1–19. https://doi.org/10.1007/s00484-024-02661-1 (2024). https://link.springer.com/article/
Amani, M. A. & Marinello, F. A deep Learning-Based model to reduce costs and increase productivity in the case of small datasets: A case study in cotton cultivation. Agriculture 12 (2), 267. https://doi.org/10.3390/agriculture12020267 (2022).
da Silva Andrea, M. C., de Oliveira Nascimento, J. P. F., Mota, F. C. M. & de Souza Oliveira, R. Predictive framework of plant height in commercial cotton fields using a remote sensing and machine learning approach. Smart Agricultural Technol. 4, 100154 (2023).
Feng, Y. et al. Cotton seedling detection and counting based on UAV multispectral images and deep learning methods. Remote Sens. 15 (10), 2680 (2023).
Pandey, B. N., Singh, R. P., Pandey, M. S. & Jain, S. Cotton leaf disease classification using deep learning based novel approach. In 2023 International Conference on Disruptive Technologies (ICDT) pp. 559–561 (IEEE, 2023).
Rai, C. K. & Pahuja, R. Classification of diseased cotton leaves and plants using improved deep convolutional neural network. Multimedia Tools Appl. 82 (16), 25307–25325 (2023).
Chithambarathanu, M. & Jeyakumar, M. K. Survey on crop pest detection using deep learning and machine learning approaches. Multimedia Tools Appl. 82 (27), 42277–42310 (2023).
Celik, M. F., Isik, M. S., Taskin, G., Erten, E., & Camps-Valls, G. Explainable artificial intelligence for cotton yield prediction with multisource data. IEEE Geoscience and Remote Sensing Letters 20, 1–5 (2023).
Senthil Pandi, S., Reshmy, A. K., Elangovan, D. & Vellingiri, J. AI-driven environmental monitoring for hydroponic agriculture: ExCNN-LFCP approach. Earth Sci. Inf. 18 (1), 1–25 (2025).
Senthil Pandi, S., Senthilselvi, A., Prabavathi Raman, P. & Manju, I. Early detection and classification of black gram plant leaf diseases using ITL-CHB method. IETE J. Res. 1–14. https://doi.org/10.1080/03772063.2024.2425775 (2024).
Stephen, A., Arumugam, P. & Arumugam, C. An efficient deep learning with a big data-based cotton plant monitoring system. Int. J. Inform. Technol. 16 (1), 145–151 (2024).
Arathi, B. & Dulhare, U. N. Classification of cotton leaf diseases using transfer learning-densenet-121. In Proceedings of Third International Conference on Advances in Computer Engineering and Communication Systems: ICACECS 2022 393–405 (Singapore: Springer Nature Singapore, 2023).
Vibhute, A. D. & Kale, K. V. Mapping several soil types using hyperspectral datasets and advanced machine learning methods. Results Opt. 12, 100503 (2023).
Sharma, A., Jain, A., Gupta, P. & Chowdary, V. Machine learning applications for precision agriculture: A comprehensive review. IEEE Access. 9, 4843–4873 (2020).
Ashapure, A. et al. Developing a machine learning based cotton yield Estimation framework using multi-temporal UAS data. ISPRS J. Photogrammetry Remote Sens. 169, 180–194. https://doi.org/10.1016/j.isprsjprs.2020.09.015 (2020).
Livieris, I. E., Dafnis, S. D., Papadopoulos, G. K. & Kalivas, D. P. A Multiple-Input neural network model for predicting cotton production quantity: A case study. Algorithms 13 (11), 273. https://doi.org/10.3390/a13110273 (2020).
Paul, A., Machavaram, R., Kumar, D. & Nagar, H. Smart solutions for capsicum harvesting: unleashing the power of YOLO for detection, segmentation, growth stage classification, counting, and real-time mobile identification. Comput. Electron. Agric. 219, 108832 (2024).
Wang, N., Zhai, Y. & Zhang, L. Automatic cotton mapping using time series of Sentinel-2 images. Remote Sens. 13 (7), 1355. https://doi.org/10.3390/rs13071355 (2021).
Ngo, Q. H., Kechadi, T. & Le-Khac, N. A. Knowledge representation in digital agriculture: A step towards standardized model. Comput. Electron. Agric. 199, 107127 (2022).
Haider, W., Rehman, A. U., Durrani, N. M. & Rehman, S. U. A generic approach for wheat disease classification and verification using expert opinion for knowledge-based decisions. IEEE Access. 9, 31104–31129 (2021).
Narcisse, T. K. N. G. et al. Contribution to the construction of an ontology for the phytosanitary surveillance of cotton in Côte d’Ivoire. In 2023 IEEE Multi-conference on Natural and Engineering Sciences for Sahel’s Sustainable Development (MNE3SD) 1–8 (IEEE, 2023).
Lamport, L. Specifying systems: the TLA + language and tools for hardware and software engineers. (2002).
Woodcock, J., Larsen, P. G., Bicarregui, J. & Fitzgerald, J. Formal methods and experience. ACM Comput. Surveys (CSUR), 41(4), 1–36 (2009).
Cousineau, D. et al. TLA + proofs. In FM 2012: Formal Methods: 18th International Symposium, Paris, France, August 27–31, 2012. Proceedings 18147–154 (Springer Berlin Heideberg, 2012). https://link.springer.com/chapter/. https://doi.org/10.1007/978-3-642-32759-9_14
Clarke, E. M. et al. Model Checking. 2nd edn (MIT press, 2018).
Clarke, E. M., Henzinger, T. A., Veith, H. & Bloem, R. Handbook of Model Checking 1st edn (Springer International Publishing AG, part of Springer Nature, 2018).
Clarke, E. M., Henzinger, T. A. & Veith, H. Introduction to model checking. In Handbook of Model Checking, 1st ed. 1–26 (Springer International Publishing AG, part of Springer Nature, 2018). https://doi.org/10.1007/978-3-319-10575-8_1
Baier, C. & Katoen, J. P. Principles of Model Checking Representation and Mind Series. (The MIT Press, 2008).
Clarke, E. M., Grumberg, O. & Peled, D. A. Model Checking (MIT Press, 1999).
Katoen, J. Concepts, algorithms, and tools for model checking, Arbeitsberichte des Instituts für Mathematische Maschinen und Datenverarbeitung, University of Erlangen–Nürnberg, Erlangen, Germany, vol. Band, no. 1 (1999).
Clarke, E. M., Emerson, E. A., Jha, S. & Sistla, A. P. Symmetry reductions in model checking. In: Computer Aided Verification. CAV 1998. Lecture Notes in Computer Science, vol.1427 (eds Hu A.J. & Vardi M.Y.) 147–158 (1998).
Clarke, E. M., Enders, R., Filkorn, T. & Jha, S. Exploiting symmetries in temporal logic model checking. Formal Methods in System Design, 9, 77–104 (1996)
Clarke, E. M., Emerson, E. A. & Sistla, A. P. Automatic verification of finite state concurrent systems using Temporal logic. ACM Trans. Program. Lang. Syst. 8 (2), 244–263 (1986).
Emerson, E. A. Temporal and modal logic. In Handbook of Theoretical Computer Science Vol. B (ed. van Leeuwen J.) 995–1072. (Cambridge, MA, USA: MIT Press, 1990). http://dl.acm.org/citation.cfm?id=114891.114907
Valmari, A. A stubborn attack on state explosion. Formal Methods Syst. Des. 1(4), 297–322 (1992). https://doi.org/10.1007/BF00709154
Vardi, M. & Wolper, P. An automata-theoretic approach to automatic program verification (preliminary report). In Proceedings of 1st IEEE Symposium on Logic in Computer Science 332–344 (IEEE, 1986).
Kupferman, O., Vardi, M. Y. & Wolper, P. An automata-theoretic approach to branching-time model checking. J. ACM. 47(2), 312–360 (2000).
Gerth, R. R. D., Kuiper, P., & Penczek, W. A partial order approach to branching time logic model checking. In Proceedings Third Israel Symposium on the Theory of Computing and Systems 130–139 (1995).
Westergaard, M. & Verbeek, H. Cpn tools. http://cpntools.org/].
Calegari, D. & Szasz, N. Verification of model transformations: a survey of the state-of-the-art. Electronic Notes in Theoretical Computer Science, vol. 292, pp. 5–25, proceedings of the XXXVIII Latin American Conference in Informatics (CLEI). http://www.sciencedirect.com/science/article/pii/S1571066113000042 (2013)
Magee, J. & Kramer, J. Concurrency: State Models and Java Programs 2nd edn (John Wiley and Sons, 2006).
Zohar, M. & Amir, P. Temporal Verification of Reactive Systems: Safety (Springer-Verlag New York, Inc., 1995).
Bhatt, D. et al. CNN variants for computer vision: history, architecture, application, challenges and future scope. Electronics 10 (20), 2470 (2021).
Nguyen, L. H. et al. Spatial-temporal multi-task learning for within-field cotton yield prediction. In The Pacific-Asia Conference on Knowledge Discovery and Data Mining 343–354 (Springer, Cham, 2019) https://doi.org/10.1007/978-3-030-16148-4_27
Sherstinsky, A. Fundamentals of recurrent neural network (RNN) and long short-term memory (LSTM) network. Phys. D: Nonlinear Phenom. 404, 132306 (2020).
Banerjee, I. et al. Comparative effectiveness of convolutional neural network (CNN) and recurrent neural network (RNN) architectures for radiology text report classification. Artif. Intell. Med. 97, 79–88 (2019).
Yu, Y., Si, X., Hu, C. & Zhang, J. A review of recurrent neural networks: LSTM cells and network architectures. Neural Comput. 31 (7), 1235–1270 (2019).
Lim, S., Henriksson, A. & Zdravkovic, J. Data-driven requirements elicitation: a systematic literature review. SN Comput. Sci. 2(1), 16 (2021). https://link.springer.com/article/10.1007/s42979-020-00416-4
Murugesh, S. & Jaya, A. Retracted article: an integrated approach towards automated software requirements elicitation from unstructured documents. J. Ambient Intell. Humaniz. Comput. 12 (3), 3763–3773. https://doi.org/10.1007/s12652-019-01667-7 (2021).
Nalchigar, S., Yu, E. & Keshavjee, K. Modeling machine learning requirements from three perspectives: a case report from the healthcare domain. Requirements Engineering, 26, 237–254. (2021). https://link.springer.com/article/10.1007/s00766-020-00343-z
Ertam, F. & Aydın, G. Data classification with deep learning using Tensorflow. In 2017 International Conference on Computer Science and Engineering (UBMK) 755–758 (IEEE, 2017).
Gulli, A. & Pal, S. Deep Learning with Keras (Packt Publishing Ltd., 2017).
Zou, D., Cao, Y., Zhou, D. & Gu, Q. Gradient descent optimizes over-parameterized deep ReLU networks. Mach. Learn. 109, 467–492 (2020).
Akhtar, N., Guyadec, Y. L. & Oquendo, F. Formal specification and verification of multi-agent robotics software systems: a case study. Proceedings of the International Conference on Agents and Artificial Intelligence (ICAART 09). Porto, Portugal, January 19–21. (INSTICC Press, 2009).
Akhtar, N. Contribution to the formal specification and verification of a multi-agent robotic system. PhD thesis, Ecole Doctorale, Laboratory VALORIA (2010).
Akhtar, N., Guyadec, Y. L. & Oquendo, F. Formal requirement and architecture specifications of a multi-agent robotic system. J. Comput. 4(4), 75–80 (2012).
Akhtar, N. & Khan, S. Formal architecture and verification of a smart flood monitoring system-of-systems. Int. Arab. J. Inform. Technol. (IAJIT) 16(2), 211–216 (2019).
Akhtar, N. et al. Hierarchical coloured petri-net based multi-agent system for flood monitoring, prediction, and rescue (fmpr). IEEE Access. 7, 180544–180557 (2019).
Akhtar, N., Rehman, A. & Khan, D. M. Formal verification of safety and liveness properties using coloured Petri-Nets: A flood monitoring, warning, and rescue system. J. Inform. Communication Technol. Rob. Appl. (JICTRA). 09(01), 80–88 (2018).
Rehman, A., Akhtar, N. & Alhazmi, O. H. Formal modeling, proving, and model checking of a flood warning, monitoring, and rescue system-of‐systems. Sci. Program. 2021(1). https://doi.org/10.1155/2021/6685978 (2021).
Author information
Authors and Affiliations
Contributions
Abdul Rehman: Conceptualized the study, developed the TLA+ formal models, and performed model verification for disease symptom requirements and reviewed, and implemented deep learning models CNN, LSTM, RNN for disease prediction.Dr. Nadeem Akhtar : Designed and implemented the deep learning framework, including CNN model training and evaluation for disease prediction and formal verification of correctness properties for this work.Dr. Omar H Alhazmi: Conducted data collection, preprocessing, and analysis of results, and contributed to manuscript writing and revisions.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rehman, A., Akhtar, N. & Alhazmi, O.H. Monitoring and predicting cotton leaf diseases using deep learning approaches and mathematical models. Sci Rep 15, 22570 (2025). https://doi.org/10.1038/s41598-025-06985-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-06985-9
Keywords
This article is cited by
-
A Quantum-Enhanced Deep Learning Framework for Precise Leaf Disease Detection in Tomato and Cotton Crops
Iranian Journal of Science and Technology, Transactions of Electrical Engineering (2025)