Abstract
The human Brain-specific Serine/Threonine Kinase 2 (BRSK2), alternatively known as Synapses of Amphids Defective (SAD)-A, is mainly expressed in the brain, and required for neuronal polarization and differentiation. This gene contains the longest 5′ untranslated region (5′ UTR) pentanucleotide short tandem repeat (STR), (CGGCT)6, in human. We hypothesized that this exceptional length may confer selective advantage in cognitive functioning in human. The region spanning (CGGCT)6 was sequenced in a sample of 339 unrelated individuals, consisting of cases affected by late-onset neurocognitive disorder (NCD) (N = 163) and matched controls (N = 176). Consequently, we mapped CGGCT motifs and STRs across the human genome and obtained the phylogenetic tree of the BRSK2 sequence spanning the CGGCT STR in 19 species belonging to several orders of mammals, including Rodents, Carnivora, Artiodactyls, Perissodactyla, and Primates. We found that (CGGCT)6 was part of a complex island of 17 consecutive CGGCT motifs/STRs, ranging from 1 to 6-repeats, stretching the BRSK2 core promoter and 5′ UTR. Across the human genome, the CGGCT island was unique with respect to density, complexity, and repeat length of CGGCT motifs and repeats. This island was flanked by a 5′ UTR CGG STR in its downstream. The evolution of the CGGCT island mainly coincided with the phylogenetic distance of the species studied, and the CGG STR was primate-specific, suggesting directional, rather than random evolution of this complex sequence. While (CGGCT)6 was strictly monomorphic in the human samples studied, a 7-repeat of this motif was detected in the controls only. In another CGGCT repeat inside the CGGCT island, there was a significant excess of homozygosity for a long allele (4-repeat) in the controls (Mid-P = 0.02). At the same locus, a 3-repeat allele was detected in the NCD group only. Additionally, alleles were detected at the extreme short and long lengths of the CGG STR in the NCD group only. Probable diagnosis in the patients harboring divergent genotypes spanned Alzheimer’s disease and vascular dementia. We report a novel genomic feature, consisting of a CGGCT motif/STR island, and a CGG STR in BRSK2 that coincide with directional evolution of several orders of mammals. Several polymorphic and rare alleles were divergently distributed in the NCD and control groups across this region, which may reflect a possible link with cognitive functions in human.
Similar content being viewed by others
Introduction
Short tandem repeats (STRs), also known as microsatellites/simple sequence repeats, are comprised of hypermutable repeated copies of 1-6-bp motifs that are spread ubiquitously across vertebrate genomes, and are linked to vast biological, evolutionary, and pathological implications1,2,3,4,5,6,7,8,9,10,11,12.
In a genome-wide study of the 5′ untranslated region (UTR) of the protein-coding genes, we previously reported numerous STR loci in this interval, a portion of which were proposed to be of possible relevance to human evolution/fitness because of their exceptional length13. The human Brain-specific Serine/Threonine Kinase 2 (BRSK2), also called Synapses of Amphids Defective-A (SAD-A), contains the longest pentanucleotide STR, (CGGCT)6, in its 5′ UTR (Transcripts: ENST00000308219.13 and ENST00000528841.6). BRSK2 is located on chromosome 11p15.514, and is mainly expressed in the brain, and to a lesser extent in pancreas (https://www.proteinatlas.org/ENSG00000174672-BRSK2/tissue). BRSK2 is required for neuronal polarization and differentiation15and is involved in the control of the maturation of nerve terminals in the mammalian peripheral and central nervous systems16. Progressive loss of neuronal polarity is a major histopathological event in neural aging and neurodegenerative diseases, such as Alzheimer’s disease (AD), which precedes death and disappearance of nerve cells17,18.
Based on the significant functions of BRSK2 in the human brain, possible breach in neurodegenerative processes, and the location and exceptional length of (CGGCT)6 in human, we sequenced the genomic region spanning this repeat in a sample of Iranian subjects, consisting of late-onset neurocognitive disorders (NCDs) and controls. Consequential to our findings of a CGGCT island in BRSK2, as a novel genomic feature, we also mapped CGGCT motifs/STRs across the human genome. Furthermore, the phylogeny of this region was studied across several orders of mammals.
Results
Identification of a novel genomic feature, consisting of a CGGCT motif/STR Island spanning the core promoter and 5′ UTR of the human BRSK2 gene
Sequencing of the region spanning the (CGGCT)6 resulted in the identification of a novel genomic feature, consisting of an island of 17 consecutive CGGCT motifs and STRs of these motifs, ranging from 1 and 6-repeats (Fig. 1). In its downstream, this island was flanked by a CGG STR (Fig. 1). This complex sequence spanned the core promoter and 5′ UTR of the human BRSK2.
A novel genomic feature spanning the core promoter and 5′ UTR of human BRSK2. An island of consecutive CGGCT motifs (blue highlights) spanned the core promoter and 5′ UTR of BRSK2. This island was flanked at its downstream, by a CGG STR (green highlight), Underlines represent loci, at which polymorphisms and/or rare variants were detected in our subsequent sequencing results. The red sequence represents the 5′ UTR.
The BRSK2 CGGCT Island mainly coincided with the phylogenetic distance of several orders of mammals
The BRSK2 CGGCT island was dynamically conserved across several orders of mammals (Suppl. 1), each species having their own species-specific composition for this island. In the 5′ UTR, CGGCT reached maximum length of 6-repeats in human and chimpanzee. (CGGCT)6 was the initial target of the present research. The Old World monkeys, such as macaque, had 5-repeats, and in the New World monkeys, such as marmoset, CGGCT > 2-repeats were not detected at this locus. The above findings support a directional trend for the elongation of this specific CGGCT STR in primate evolution.
Moreover, the phylogenetic tree of the CGGCT island (input sequence available in Suppl. 1) mainly coincided with the evolutionary distance of several orders of mammalian species (Fig. 2), further supporting directional, rather than random evolution of this island. It should be noted that we refer to directionality as “change in the same direction over evolutionary time”.
Phylogenetic tree of the BRSK2 CGGCT island in several orders of mammals. The phylogenetic tree mainly coincided with the evolutionary distance of these species, indicating that the evolution of this region was directional i.e. change in the same direction over evolutionary time, rather than random. The input data were the sequences spanning the island, as provided in Suppl. 1.
Across the human genome, the BRSK2 CGGCT Island was unique with respect to the density and complexity of the CGGCT motifs/STRs
To examine whether this island was unique across the human genome, we obtained a whole-genome map of the CGGCT motifs/STRs (Suppl. 2). With respect to density and complexity, the BRSK2 CGGCT island was unique, genome-wide (Table 1). Moreover, the BRSK2 (CGGCT)6 was the second longest (CGGCT) STR, genome-wide, only preceded by a (CGGCT)35 in the promoter of the long non-coding RNA gene, SMIM2-AS1 (Transcript: ENST00000444663.7 SMIM2-AS1-202) (Table 1). (CGGCT)35 was human-specific and repeats of ≥ 4-repeats were not detected for this STR in any other species.
The flanking CGG STR was primate-specific
This 5′ UTR CGG STR was detected of ≥ 2-repeats in primates, and not any other order of mammals (Fig. 3). While lemur strains lacked the CGG STR, this STR was detectable and conserved throughout New and Old World monkeys, and great apes.
The BRSK2 CGG STR across several primates. (CGG) ≥ 2-repeats (green highlights) were detected in primates, and not in any other order of mammals. Multiple sequence alignment was obtained from the Ensembl Genome Browser 112 (https://asia.ensembl.org/index.html).
The BRSK2 CGGCT Island and CGG STR harbor various regulatory element binding sites in human
The CGGCT island and CGG STR contain binding sites for numerous transcription factors (TFs), such as POLR2A, RNF2, L3MBTL2, NRF1, and CBX1 (Fig. 4). Many of these elements may bind to more than one transcript isoform of human BRSK2, because this complex region spans at least two transcript isoforms of the gene.
TF-binding sites in the human BRSK2 CGGCT island and flanking CGG-repeat sequence. Two transcript isoforms of BRSK2 span this compound sequence. The horizontal bars illustrate the binding sites of various TFs, as indicated on the left, based on ENCODE ChIP-seq data (hg19). The position and length of each bar represent the genomic location and extent of binding, respectively. The shading of the bars, ranging from light gray to black, signifies the strength or confidence of the binding interaction, with darker shades indicating stronger or more reliable binding events.
A 7-repeat allele at the (CGGCT)6 locus was detected in the control group only
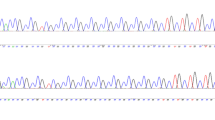
The (CGGCT)6 STR (underlined red sequence with blue highlight in Fig. 1) was strictly monomorphic in the human samples studied (Fig. 5A), except for one instance of a 6/7 heterozygote individual in the control group only (Fig. 5B). The individual harboring this allele was an 85-year-old male, with no history of cognitive impairment, MMSE = 28, and AMTS = 9.
A CGGCT repeat in the human BRKS2 CGGCT Island harbored divergent genotypes in the NCD and control groups
Inside the BRSK2 CGGCT island, a CGGCT repeat (underlined black sequence with blue highlight in Fig. 1), harbored a 4-repeat allele (the longest detected allele at this locus), homozygous genotype of which was in significant excess in the control group vs. NCD cases (Mid-P = 0.022) (Fig. 6A and B). At the same locus, a 3-repeat allele was detected in a 2/3 genotype, in one individual in the NCD group only (Fig. 6C). The NCD patient harboring the 2/3 genotype was a 65-year-old male with AMTS = 5, and cognition impairment in history and interviews. This patient was diagnosed with vascular dementia (VD).
A CGGCT repeat inside the human BRSK2 island harbored divergent alleles in the NCD and control groups. This CGGCT repeat mainly consisted of 2- and 4-repeat alleles, of which the 4-repeat allele and its homozygosity were in excess in the control group A) and B). At the same locus, a 3-repeat was detected in one patient with VD C).
At the CGG STR, alleles were detected at the extreme ends of the allele distribution curve in the NCD group only
At the CGG STR, alleles were detected at the extreme short and long ends of this STR in the NCD group only (Fig. 7A). While the allele range of this STR was between 8 to 9-repeats in the control group, this range was between 6 to 11-repeats in the NCD group (Fig. 7B). The patients harboring the extreme alleles received the diagnosis of probable late-onset AD.
Discussion
Here we report a novel genomic feature, consisting of an island of CGGCT motifs and STRs, stretching the core promoter and 5′ UTR of the human BRSK2 gene, which was unique with respect to the density and complexity of CGGCT across the human genome. This island was flanked by a down-stream CGG-repeat, also located in the 5′ UTR. Divergent polymorphic and rare alleles were detected across the BRSK2 CGGCT island and CGG STR in the NCD cases and controls. This complex sequence harbored binding sites for numerous TFs.
The sequence of the CGGCT island mainly coincided with the phylogenetic distance of several orders of mammals, indicating directional, rather than random evolution of this island. It should be noted that, here, directional evolution refers to change in the same direction over evolutionary time. The phylogenetic tree of this region across these mammals was mostly in line with the studies on the common ancestries across mammals19,20. In the 5′ UTR part of the island, CGGCT reached maximum repeat length of 6 in human and chimpanzee. The initial targeting of this region for sequencing was because of this (CGGCT)6, which was the longest 5′ UTR pentanucleotide STR in human13. Our genome-wide map of CGGCT in human revealed that (CGGCT)6 is the second longest repeat of this motif, genome-wide, only preceded by the SMIM2-AS1 (CGGCT)35. Remarkably, (CGGCT)35 was human-specific, and repeats of ≥ 4-repeats were not detected for this STR in any other species. The SMIM2 gene is almost exclusively expressed in male tissues (https://www.proteinatlas.org/ENSG00000139656-SMIM2/tissue).
Motif islands, such as the CGGCT island in BRSK2, are an emerging topic that may correlate with evolution and speciation. Another prime example of motif islands includes islands of GGC and GCC, of evolutionary relevance to humans21. Not only may the specific motifs across these islands, but also the repeat length of each motif across the islands be of biological and evolutionary relevance.
Similar to the CGGCT island, evolution of the CGG STR was directional, rather than random, evidenced by longer repeats in Old World monkeys and apes, in comparison with the more distantly related New World monkeys, and lack of this repeat in other species. It is possible that a restricted range of alleles at this locus links to normal cognitive functioning in humans. The above stems from our findings of extreme alleles in the NCD group only. Recent findings have shed light on a link between this type of STR and cognitive impairment spectrum disorders22,23,24,25.
STRs, whether polymorphic or rare, correlate with evolutionary processes, and adaptive and complex traits, such as cognition2,3,4,7,8,26,27. They bind TFs to tune eukaryotic gene expression28. Sequencing of several STRs in the regulatory regions of several other genes have led to the identification of rare divergent alleles in late-onset NCD21,29,30,31,32,33reinforcing the hypothesis that late-onset NCDs, such as AD and VD, at least in part, unambiguously link to a collection of rare variants across the human genome.
Point mutations may also be divergently occurring in the studied region in NCDs. For example, a G to T transversion mutation (the red underlined single nucleotide in Fig. 1) was detected in one individual affected by NCD (Fig. 8). This mutation was not detected in the control group and resulted in differential patterns of regulatory elements binding to the mutant vs. wild type nucleotide through bioinformatics analysis using TFBIND online software34. The NCD patient harboring this mutation was an 80-year-old male with a 7-year history of declining cognition impairment and AMTS = 5 at the time of interview, and diagnosed with probable late-onset AD. This patient harbored (CGGCT)6/6, (CGGCT)2/2, and (CGG)9/9 genotypes at the three loci introduced in the previous sections.
BRSK2 is mainly expressed in the brain, and the protein encoded by this gene has an essential role in neuronal polarization. Considering the directional evolution and human-specific composition of the CGGCT island and CGG STR complex, the location of this complex in the regulatory region of a biologically important gene in synaptic polarization, and several divergent genotypes across the region in the NCD and control groups, it is possible that this complex may be involved in the higher order brain functions. A link between BRSK2 and other cognitive disorders e.g. autism spectrum disorder (ASD) has been reported by several groups35,36. Interestingly, there are commonalities between AD and ASD at various genetic, pathological, and clinical levels37.
It should be noted that this is a pilot study, necessitating further exploration and validation in larger sample sizes of various neurological characteristics and phenotypes. Functional studies are also warranted to unveil how this complex region and the recruited regulatory elements may impact the expression and function of BRSK2.
Conclusion
We report a novel genomic feature, consisting of a unique island of CGGCT motifs and STRs, and a flanking CGG-repeat in the regulatory region of the brain-specific gene, BRSK2, which is dynamically conserved across several orders of mammals, and its evolution mainly coincides with the phylogenetic distance of these species. Several divergent polymorphic and rare alleles and genotypes were detected across this region in the late-onset NCD and control groups, which warrant exploring the region in larger sample sizes and a spectrum of neurological disorders.
Materials and methods
Subjects
Three hundred-thirty-nine Iranian subjects of ≥ 60 years of age, consisting of late-onset NCD patients (N = 163) (age range 60–90) and controls (N = 176) (age range 60–92) were recruited from the provinces of Qazvin and Rasht. Diagnosis of the NCD cases was as previously described38. Briefly, diagnosis of NCD cases was based on the DSM-5 diagnostic criteria39. The Persian version of the Abbreviated Mental Test Score (AMTS)40,41 was implemented, and a AMTS < 7 was an inclusion criterion for NCD, medical history and records were reviewed in all participants, and CT-scans were taken when possible. Furthermore, in several subjects, the Mini-Mental State Exam (MMSE) Test42 was implemented in addition to the AMTS. An MMSE score of < 24 was an inclusion criterion for NCD. The onset of symptoms in the NCD group was ≥ 60 years. The control group was selected based on cognitive AMTS of > 7 and MMSE > 24, and lack of major medical history. The cases and controls were matched based on age, gender, and residential district. The subjects’ informed consent was obtained (from their guardians where necessary) and their identities remained confidential throughout the study. The experimental protocols were approved by the Ethics Committee of the Social Welfare and Rehabilitation Sciences and were consistent with the principles outlined in an internationally recognized standard for the ethical conduct of human research. All methods were performed in accordance with the relevant guidelines and regulations.
Allele and genotype analysis
Genomic DNA was extracted from peripheral blood, using a standard salting out method. PCR reactions for the amplification of the human BRSK2 CGGCT island and CGG repeat were set up with the following primers:
Forward: CGTTCGTACAGGCTCGTGTC.
Reverse: GGTAGGGCCCAACATACTGC.
PCR reactions were carried out in a final volume of 20 µl, with a GC-TEMPase 2x master mix (Amplicon), in a thermocycler (Peqlab-PEQStar) under the following program: touchdown PCR: 95 ◦C for 5 min, 20 cycles of denaturation at 95 ◦C for 45 s, annealing for 45 s at 65◦C (-0.5 decrease for each cycle) and extension at 72 ◦C for 1 min, and 30 cycles of denaturation at 95 ◦C for 40 s, annealing at 55 ◦C for 45 s and extension at 72 ◦C for 1 min, and a final extension at 72 ◦C for 10 min. Genotyping of every sample included in this study was performed following Sanger sequencing, using the forward primer, in an ABI 3130 DNA sequencer. The Chromas software was used to analyze and score the sequences.
Sequencing through repeats can be notoriously difficult, especially for longer sequences. In our work, the PCR products used for sequencing were reasonable in size (~ 700 bp). Furthermore, the repeats in the BRSk2 gene were not excessively long i.e., the CGGCT motifs ranged from 1 to 7-repeats, and the CGG repeat ranged between 6 and 11 repeats. We used Sanger sequencing (rather than fragment analysis) in every sample, which is currently the most reliable method available for repeat scoring. In addition, several samples were randomly selected, and the process of PCR and sequencing were repeated in these samples to check for reproducibility.
Statistical analysis
OpenEpi (https://www.openepi.com) was used to analyze the allele and genotype data of the human samples studied.
Trans-species analysis of the BRSK2 CGGCT Island and flanking CGG repeat
The promoter and 5′ UTR sequences of the BRSK2 gene were screened in 19 species of mammals (Suppl. 1), spanning Rodents (Rat, Mouse, Shrew Mouse), Carnivora (Dog, Cat), Artiodactyla (Cow, Goat), Perissodactyla (Horse), and Primates (Mouse Lemur, New and Old World monkeys, and great apes), based on Ensembl Genome Brower 112 (https://asia.ensembl.org/index.html).
MUSCLE (https://ngphylogeny.fr/workflows/oneclick/)43,44,45 was used to draw the phylogenetic tree of the BRSK2 CGGCT island in the selected species. The input sequences are provided in Suppl. 1.
Extraction algorithm for the human whole-genome CGGCT motif/STR map
A Java software package was developed for the detection of tandem repeats, as previously described (https://github.com/arabfard/Java_Di_STR_Finder)46. Briefly, to extract CGGCT motifs and repeats, along with their corresponding genomic locations across the human genome, we utilized the latest version of the human genome assembly (GRCh38. p14) obtained from the UCSC Genome Browser (https://hgdownload.soe.ucsc.edu). The program initiated its search from the first nucleotide of the genome, continuously scanning for occurrences of CGGCT. It employed a window frame consisting of 5 nucleotides to identify instances of the CGGCT core sequence, followed by recording the count and location of the occurrences. It then searched for new CGGCT motifs, starting from the next nucleotide. To validate the results, the final list of identified CGGCTs underwent random manual evaluation, using the Ensembl Genome Browser 112 (https://asia.ensembl.org/index.html). The precise location of each CGGCT was determined as follows: The output was organized and classified in an Excel file (Suppl. 2), where the start and end points of each CGGCT were determined across the human genome.
Data availability
Sequence chromatograms obtained during this study are stored at the following links: NCDs: https://figshare.com/articles/dataset/_b_N_b_b_ovel_genomic_feature_in_the_brain-specific_BRSK2_NCDs_b_/28388054DOI: 10.6084/m9.figshare.28388054Controls: https://figshare.com/articles/dataset/_b_N_b_b_ovel_genomic_feature_in_the_brain-specific_BRSK2_Controls_b_/28387988DOI: 10.6084/m9.figshare.28387988.
Abbreviations
- AD:
-
Alzheimer’s Disease
- AMTS:
-
Abbreviated Mental Test Score
- ASD:
-
Autism Spectrum Disorder
- BRSK2:
-
Brain-specific Serine/Threonine Kinase 2
- MMSE:
-
Mini-Mental State Exam
- NCD:
-
Neurocognitive Disorder
- SAD-A:
-
Synapses of Amphids Defective (SAD)-A
- STR:
-
Short Tandem Repeat
- TF:
-
Transcription Factor
- UTR:
-
Untranslated region
- VD:
-
Vascular Dementia
References
Nikkhah, M., Rezazadeh, M., Khorshid, K., Biglarian, H. R., Ohadi, M. & A. & An exceptionally long CA-repeat in the core promoter of SCGB2B2 links with the evolution of apes and old world monkeys. Gene 576, 109–114. https://doi.org/10.1016/j.gene.2015.09.070 (2016).
Mohammadparast, S., Bayat, H., Biglarian, A. & Ohadi, M. Exceptional expansion and conservation of a CT-repeat complex in the core promoter of PAXBP1 in primates. Am. J. Primatol. 76, 747–756. https://doi.org/10.1002/ajp.22266 (2014).
Afshar, H. et al. Evolving evidence on a link between the ZMYM3 exceptionally long GA-STR and human cognition. Sci. Rep. 10 https://doi.org/10.1038/s41598-020-76461-z (2020).
Watts, P. et al. Stabilizing selection on microsatellite allele length at arginine vasopressin 1a receptor and Oxytocin receptor loci. Proc. Royal Soc. B: Biol. Sci. 284, 20171896. https://doi.org/10.1098/rspb.2017.1896 (2017).
Hannan, A. J. Tandem repeats mediating genetic plasticity in health and disease. Nat. Rev. Genet. 19, 286–298. https://doi.org/10.1038/nrg.2017.115 (2018).
Ranathunge, C. et al. Microsatellites as agents of adaptive change: an RNA-Seq-Based comparative study of transcriptomes from five Helianthus species. Symmetry 13, 933 (2021).
Fotsing, S. F. et al. The impact of short tandem repeat variation on gene expression. Nat. Genet. 51, 1652–1659. https://doi.org/10.1038/s41588-019-0521-9 (2019).
Afshar, H. et al. Natural selection at the NHLH2 core promoter exceptionally long CA-Repeat in human and Disease-Only genotypes in Late-Onset neurocognitive disorder. Gerontology 66, 514–522. https://doi.org/10.1159/000509471 (2020).
Maddi, A. M. A., Kavousi, K., Arabfard, M., Ohadi, H. & Ohadi, M. Tandem repeats ubiquitously flank and contribute to translation initiation sites. BMC Genomic Data. 23, 59. https://doi.org/10.1186/s12863-022-01075-5 (2022).
Arabfard, M., Kavousi, K., Delbari, A. & Ohadi, M. Link between short tandem repeats and translation initiation site selection. Hum. Genomics. 12, 47. https://doi.org/10.1186/s40246-018-0181-3 (2018).
Emamalizadeh, B. et al. The human RIT2 core promoter short tandem repeat predominant allele is species-specific in length: a selective advantage for human evolution? Mol. Genet. Genomics. 292, 611–617. https://doi.org/10.1007/s00438-017-1294-4 (2017).
Valipour, E. et al. Polymorphic core promoter GA-repeats alter gene expression of the early embryonic developmental genes. Gene 531, 175–179. https://doi.org/10.1016/j.gene.2013.09.032 (2013).
Namdar-Aligoodarzi, P. et al. Exceptionally long 5′ UTR short tandem repeats specifically linked to primates. Gene 569, 88–94 (2015).
Miura, K., Masuzaki, H., Ishimaru, T., Niikawa, N. & Jinno, Y. J. J. O. H. G. A hhai/bstui polymorphism in a novel G.ne at H.man chromosome 11p15. 5. J. Hum. Genet. 43, 283–284 (1998).
Kishi, M., Pan, Y. A., Crump, J. G. & Sanes, J. R. J. S. Mammalian SAD kinases are required for neuronal polarization. Science 307, 929–932 (2005).
Lilley, B. N. et al. J. P. O. T. N. A. O. S. SAD kinases control T.e maturation O. nerve T.rminals in T.e mammalian peripheral and central nervous systems. Proc. Natl. Acad. Sci. U S A. 111, 1138–1143 (2014).
Wu, Y. G. et al. The effects and potential of microglial polarization and crosstalk with other cells of the central nervous system in the treatment of alzheimer’s disease. Neural Regen Res. 18, 947–954 (2023).
Cid-Arregui, A., De Hoop, M. & Dottii, C. G. J. N. O. A. Mechanisms O. neuronal Polarity. Neurobiol. Aging. 16, 239–243 (1995).
Cannarozzi, G., Schneider, A. & Gonnet, G. A phylogenomic study of human, dog, and mouse. PLoS Comput. Biol. 3, e2 (2007).
Nikaido, M., Nishihara, H. & Okada, N. SINEs as credible signs to prove common ancestry in the tree of life: A brief review of pioneering case studies in retroposon systematics. Genes (Basel). 13 (6), 989. https://doi.org/10.3390/genes13060989 (2022). PMID: 35741751; PMCID: PMC9223172.
Tajeddin, N. et al. Novel Islands of GGC and GCC repeats coincide with human evolution. Gene 902, 148194 (2024).
Khamse, S. et al. A hypermutable region in the DISP2 gene links to natural selection and Late-Onset neurocognitive disorders in humans. Mol. Neurobiol. 61 (11), 8777–8786 (2024).
Annear, D. J., Vandeweyer, G., Sanchis-Juan, A., Raymond, F. L. & Kooy, R. F. J. G. R. Non-Mendelian inheritance patterns and extreme deviation rates of CGG repeats in autism. Genome Res. 32, 1967–1980 (2022).
Khamse, S. et al. R. A (GCC) repeat in SBF1 reveals a novel biological phenomenon in human and links to late onset neurocognitive disorder. Sci. Rep. 12, 15480 (2022).
Annear, D. J. & Kooy, R. F. J. E. T. I. L. S. Unravelling the link between neurodevelopmental disorders and short tandem CGG-repeat expansions. Emerg. Top. Life Sci. 7, 265–275 (2023).
Ranathunge, C. & Welch, M. E. J. B. Clinal variation in short tandem repeats linked to gene expression in sunflower (Helianthus annuus L). Biomolecules 14, 944 (2024).
Press, M. O., McCoy, R. C., Hall, A. N., Akey, J. M. & Queitsch, C. J. G. R. Massive variation of short tandem R.peats with functional consequences across strains of Arabidopsis thaliana. Genome Res. 28, 1169–1178 (2018).
Horton, C. A. et al. Short tandem repeats bind transcription factors to tune eukaryotic gene expression. Science 381, eadd1250 (2023).
Alizadeh, S. et al. G. A GCC repeat in RAB26 undergoes natural selection in human and harbors divergent genotypes in late-onset alzheimer’s disease. Gene 893, 147968 (2024).
Khamse, S. et al. A (GCC) repeat in SBF1 reveals a novel biological phenomenon in human and links to late onset neurocognitive disorder. Sci. Rep. 12, 15480. https://doi.org/10.1038/s41598-022-19878-y (2022).
Afshar, H. et al. Natural selection at the NHLH2 core promoter exceptionally long CA-repeat in human and disease-only genotypes in late-onset neurocognitive disorder. Gerontology 66, 514–522 (2020).
Afshar, H. et al. Evolving evidence on a link between the ZMYM3 exceptionally long GA-STR and human cognition. Sci. Rep. 10, 19454 (2020).
Jafarian, Z. et al. R. Natural selection at the RASGEF1C (GGC) repeat in human and divergent genotypes in late-onset neurocognitive disorder. Sci. Rep. 11, 19235 (2021).
Tsunoda, T. & Takagi, T. Estimating transcription factor bindability on DNA. Bioinformatics. (1999). Jul-Aug;15(7–8):622 – 30 https://doi.org/10.1093/bioinformatics/15.7.622. PMID: 10487870.
Viggiano, M. et al. Genomic analysis of 116 autism families strengthens known risk genes and highlights promising candidates. NPJ Genom Med. 9, 21 (2024).
Bacchelli, E. et al. Whole genome analysis of rare deleterious variants adds further evidence to BRSK2 and other risk genes in autism spectrum disorder. Res Sq, PMC10635364 (2023).
Khan, A. et al. Alzheimer’s disease and autistic spectrum disorder: is there any association? CNS Neurol. Disord Drug Targets. 15, 390–402 (2016).
Khamse, S. et al. Novel implications of a strictly monomorphic (GCC) repeat in the human PRKACB gene. Sci. Rep. 11, 20629. https://doi.org/10.1038/s41598-021-99932-3 (2021).
American Psychiatric Association, DSM-5 Task Force. Diagnostic and Statistical Manual of Mental Disorders: DSM-5™ 5th edn (American Psychiatric Publishing, Inc., 2013). https://doi.org/10.1176/appi.books.9780890425596
Foroughan, M. et al. Validity and reliability of a Bbreviated M ental T est S core (AMTS) among older I Ranian. Psychogeriatrics 17, 460–465 (2017).
Hodkinson, H. M. Evaluation of a mental test score for assessment of mental impairment in the elderly. Age Ageing. 1, 233–238. https://doi.org/10.1093/ageing/1.4.233 (1972).
Carpenter, C. R. et al. Accuracy of dementia screening instruments in emergency medicine: A diagnostic Meta-analysis. Acad. Emerg. Med. 26, 226–245. https://doi.org/10.1111/acem.13573 (2019).
Frédéric Lemoine, D. et al. Gascuel, NGPhylogeny.fr: new generation phylogenetic services for non-specialists, Nucleic Acids Research, Volume 47, Issue W1, 02 July 2019, Pages W260–W265. https://doi.org/10.1093/nar/gkz303
Thomas Junier, E. M. & Zdobnov The Newick utilities: high-throughput phylogenetic tree processing in the Unix shell. Bioinformatics 26, 1669–1670 (2010).
Lemoine, F., Entfellner, D. & Wilkinson, J. B. Renewing felsenstein’s phylogenetic bootstrap in the era of big data. Nature 556, 452–456 (2018).
Ohadi, M. et al. Novel crossover and recombination hotspots massively spread across primate genomes. Biol. Direct. 19, 70 (2024).
Author information
Authors and Affiliations
Contributions
MO conceived the project, scored the alleles and genotypes, analyzed the data, and wrote the original and edited manuscript. MA and HB performed the bioinformatics and statistical analyses and made the figures. SKh performed the molecular experiments. AD contributed to data collection and coordination. All authors read and agreed to the final version of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was in accordance with guidelines outlined by the Ethics Committee of the University of Social Welfare and Rehabilitation Sciences, Tehran, Iran. Written informed consent to participate in the study was obtained from all subjects or their legal guardian. This research was consistent with the principles outlined in an internationally recognized standard for the ethical conduct of human research. All methods were performed in accordance with the relevant guidelines and regulations. The identity of all individuals participating in this study remained confidential throughout the study.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ohadi, M., Bayat, H., Khamse, S. et al. A directionally evolved genomic feature in BRSK2 harbors divergent alleles in neurocognitive disorders. Sci Rep 15, 21888 (2025). https://doi.org/10.1038/s41598-025-07803-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-07803-y