Abstract
Wearable Sensor (WS)-based monitoring systems detect minute patient movements/ demands and abnormalities through periodic sensing and imaging. Sensor data observed over different intervals is not constant or available based on operating sequences. Due to variations in data sequences, the analysis process becomes complex, resulting in less precise outputs. To address this problem, an Allied Data Disparity Technique (ADDT) is proposed in this article. This technique identifies the disparity in different monitoring sequences in coherence with the clinical and previous values. Based on the mean disparity, the data requirement for the WS sequence is decided. This decision uses multiple substituted and predicted values obtained from previous instances. Multi-Instance Ensemble Perceptron Learning is used in this decision process, where the substitution instances for clinical and previous outcomes are performed. The ensemble perceptron selects the maximum clinical value correlating sensor data to ensure high sequence prediction. The ensembles are updated based on the highest precision-based WS values for diagnosis. This diagnosis-focused coalition between clinical and predicted WS values is updated periodically for the new precision levels identified.
Similar content being viewed by others
Introduction
WS data analysis revolutionizes patient monitoring by allowing continuous, non-invasive measurement of physiological parameters such as heart rate, blood pressure, and temperature1. WSs collect real-time data that can be sent to healthcare specialists for remote monitoring and analysis. The steady data flow enables early diagnosis of problems, resulting in preventive care and fewer hospital visits2. WSs are especially beneficial in chronic disease care, where constant monitoring is essential for tracking a patient’s progress3. These systems provide information about patient behavior, daily routines, and general health state4. Incorporating WS data into healthcare operations makes patient care more personalized and adaptive, results in prompt treatments based on real-world evidence. Such real-time monitoring increases the security of patients and the quality of care provided outside of typical healthcare facilities5,6. Variation identification in WS data sequences is crucial for accurate health monitoring7. WSs are susceptible to oscillations caused by mobility, climatic conditions, or sensor positioning, which can inject noise and unpredictability into the data8. Detecting these fluctuations effectively is critical for detecting relevant patterns connected to a patient’s health status. Advanced analytical procedures are required to distinguish between typical oscillations and indications of medical problems9. Sequence alignment, disparity analysis, and predictive modeling assist in examining sensor data against clinical benchmarks to find major variations10. Precision health monitoring uses real-time detection of these variances to deliver meaningful insights to healthcare practitioners. Variation detection systems can improve diagnostic accuracy by combining data from various sensors and using prior health data. It contributes to more targeted interventions and personalized treatment programs, which improve overall patient outcomes11,12.
Machine learning (ML) approaches are transforming the analysis of variances in WS data and allowing for more precise and efficient interpretation of complicated datasets13. Deep learning algorithms, ensemble approaches, and neural networks are increasingly used to handle real-time sensor data. These techniques effectively detect subtle patterns and anomalies that typical analytic methods can ignore14. ML models can be trained on massive datasets to identify trends and predict probable health problems, enabling more proactive patient care15. Neural networks can use movement data to detect irregular heart rhythms or forecast falls in older adults16. ML minimizes the strain on healthcare personnel by automating deviation detection while improving health assessment accuracy. These systems also enhance over time as they learn from fresh data, allowing them to give more reliable, real-time insights for precision health monitoring17,18. The above discussion discusses the need for the proposed sensor data analysis. Thus, the contributions are listed below:
-
The following section discusses the works related to WS data analysis, patient data monitoring, and the role of WSs. Besides, the optimizations through learning methods that are exclusive to WS data analysis are also uncovered in this discussion.
-
The introduction, discussion, and explanation of the proposed allied data disparity technique that addresses the problem of sequence issues in WS data accumulated. This technique utilizes multi-instance ensemble perceptron learning (MIEPL) to identify the disparities with the feasibilities of substitution and precision-based diagnosis support.
-
Integrating a real-time health monitoring dataset provides information on temperature, respiratory, oxygen levels, pulse rate, etc., and sensor values to analyze the sequence of detecting disparities. The analysis for precision and accuracy under different clinical and substituted sequences is also presented.
The research outline involves the in-depth research analysis in Sect “Related works”, followed by data source description in Sect. 3, material and proposed method explanation in Sect. 4, results and discussion analysis in Sect. 5, and concludes with a research summary in Sect. 6.
Related works
Arslan et al.19 designed an ML algorithm-based healthcare monitoring model for comprehensive patient care. The designed model uses k-nearest neighbor and support vector machine algorithms to analyze the actual health status of the patients. The healthcare data is collected from wireless devices that gather information via wireless sensors. The designed model increases the accuracy level of healthcare monitoring systems. Shi et al.20 introduced a distributed sensor function network (DSFNet) for action recognition systems. The DSFNet model utilizes the convolutional neural network (CNN) algorithm, which extracts optimal features for recognition. The motion and movement ratio of the people is calculated according to the range and necessities. The introduced DSFNet maximizes the precision of the action recognition process.
Gong et al.21 developed a self-powered wearable method for intelligent healthcare data management systems. The method identifies the restriction that causes issues during the data management process. The developed method provides feasible analysis services that secure the data security range in healthcare systems. The developed analysis method elevates the quality of communication and data-sharing among users. Alruwaili et al.22 proposed a probabilistic transfer learning-based data transmission-monitoring model (PTL-DTM) from WSs. The proposed model tackles the risk thresholds and time-consuming factors during data transmission. A two-step algorithm divides the data that needs to be transferred via wireless sensors. Experimental results show that the proposed approach enhances the monitoring systems’ specificity, security, and accuracy.
Imran et al.23 designed a lightweight CNN and bidirectional gated recurrent unit (CNN-BiGRU) model for human activity recognition. The CNN-BiGRU model collects the inertial sensor data, which provides optimal patterns of activity recognition. The model uses data mining techniques to analyze the important features and patterns for further processes. The designed model maximizes the overall accuracy level of the activity recognition process. Zha et al.24 proposed an SCNN-LSTM model for prediction of signal-to-noise ratio (SNR). This model is used for wearable wireless sensor networks. A filtering layer is analyzed to gather relevant data for SNR prediction. The model extracts specific frequencies and parameters, eliminating the latency while performing prediction services. Compared with others, the proposed model enlarges the precision ratio of the SNR prediction process.
Tatli et al.25 developed a new method for prediabetes detection in unconstrained conditions. The developed method uses ML and signal-processing techniques to classify the early symptoms of the patients. The signals are collected via wearable devices, which extract necessary data for prediction. The collected data is also used to reduce the computational cost of the process. The developed method elevates the performance level of the systems. Nihar et al.26 introduced an indoor personalized heat stress prediction model for healthcare applications. A data-driven ML algorithm is implemented in the model to predict the patients’ actual and extreme heat stress levels. The activities and conditions of the patients are calculated to get relevant data for the prediction process. The introduced model increases the accuracy range of the prediction process.
Matsumura et al.27 developed a new real-time personal healthcare data analysis approach for multimodal WSs. Edge computing technology is used in the approach to analyze the operational and functional vital signs of the patients. The approach is also used to generalize the diagnosis services provided to the patients. The developed approach enhances the effectiveness ratio of the healthcare applications. Pini et al.28 proposed a novel healthy brain and child development method. The proposed method is used to collect and calculate the data on infant activity via wearable wireless sensors. The method analyzes the infant’s activity per movement, motion, and sleep ratios. The method also provides optimal information to enhance the health condition of the infants. The proposed method maximizes healthy brain and child development performance and efficiency range.
Kim et al.29 developed a shuffled efficient channel attention model for stress detection in healthcare systems. Multimodal WS data is used as input and collected via wearable devices. A deep neural network algorithm is employed in the model to analyze and extract the necessary data for prediction. This efficient channel attention model minimizes the computational latency and complexity of the detection process. The developed model achieves high accuracy in the stress level detection process. Matsumura et al.30 introduced a new ML model for step length estimation in older adults and patients with neurological disorders. The introduced model analyzes the data from a single lower-back inertial measurement unit. The data produces relevant features for estimation, which eliminates the latency range. The introduced model reduces the error rate of the estimation process.
Mahato et al.31 proposed a Hybrid Multimodal WS for comprehensive Health Monitoring Systems (HM-HMS). It is used to identify the actual health condition of patients via wearable devices. The proposed model calculates the exact health issues tracked by health monitoring systems. It decreases the severity of providing medical services to the patients. Experimental results show that the proposed model increases the precision of diagnosis services. Gashi et al.32 designed a multiple sclerosis classification method using mobile and WS data. Behavioral markers and ML techniques are used to classify the exact types of multiple sclerosis. The patients’ living conditions and healthcare growth are identified and calculated for further prediction processes. The designed method enlarges healthcare monitoring systems’ overall effectiveness and significance range.
Li et al.33 developed an ML-based data analytic technique for athlete player health monitoring systems. The developed technique uses optical sensors to gather players’ health-related data via WS devices. A naïve Bayesian convolutional vector network is used to assess the collected data. The assessed data is used as input to predict the exact physical health condition of the players. The developed technique maximizes the performance level of the systems. Zhang et al.34 designed a quantitative gait analysis method using WSs for Parkinson’s disease patients. The designed method categorizes the objective and comprehensive stage of the patient’s health condition. The method reduces the complexity range that occurs during disease diagnosis services. Compared with others, the designed method enhances the accuracy level of the analysis process. Table 1 presents a comparative analysis of previous studies, highlighting the models, techniques used, application domains, key contributions, and identified research gaps.
Traditional healthcare monitoring methods have severe constraints that restrict their generalizability and precision. Traditional ML-based models like Arslan et al.‘s use k-NN or SVM classifiers, which lack the depth and versatility of newer learning architectures. DSFNet20 cannot capture temporal relationships or perform multi-sensor fusion, limiting its use in dynamic monitoring. CNN-BiGRU23 and other frameworks lack tailored learning and patient-specific updates. SCNN-LSTM24 is susceptible to noise and signal anomalies, while prediabetes detection25 and disease-specific designs32 are too narrowly targeted. The suggested approach uses ADDT and MIEPL to close these gaps. ADDT intelligently detects sensor sequence variations and harmonizes them with clinical data to ensure coherent analysis despite irregular data flows. The ensemble perceptron technique selects the most clinically aligned instances from various data streams to improve prediction accuracy. This dual mechanism provides real-time adaptation, noise resistance, individualized diagnosis, and periodic model updates, providing a scalable, high-precision solution to past work’s inadequacies.
The proposed ADDT has been chosen above existing models like CNN, LSTM, and others, because it can handle irregular, noisy, and incomplete WS data in real-time clinical settings. ADDT actively detects disparities by comparing current sequences with clinical and historical data, allowing intelligent substitution of missing or inaccurate inputs. Its computational efficiency is excellent for real-time monitoring, unlike resource-intensive and uninterpretable LSTM. ADDT prioritizes high-relevance data for accurate diagnosis using ensemble-based precision weighting and clinical correlation, unlike decision trees and SVMs, which struggle with temporal inconsistencies and lack adaptable substitution methods. The transparent decision method improves interpretability and clinician trust, giving it a robust and context-aware solution for WS data processing when standard machine learning algorithms fail.
Data collection and description
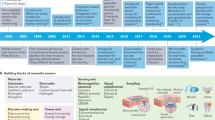
The data from the “Health Monitoring System” source (https://www.kaggle.com/datasets/nraobommela/health-monitoring-system?resource=download) is retrieved and used in this article to validate ADDT. This dataset provides two distinct pieces of information: WS sensed data and clinical values for temperature, heart rate, etc. This health monitoring for dehydration, medicine overdose, cold, cough, and respiratory imbalance uses temperature, heartbeat, pulse, and oximeter sensor values. The sensors operate at regular intervals, for which the sequence (interval) variations are identified. Based on the variations, the disparity is computed, and predicted substitutions are made. The dataset incorporating the proposed technique flow is illustrated in Fig. 1a.
WS data observation intervals and their corresponding sequences (per day) are referred to from the dataset used. The disparity detected is computed using the missing/erroneous sequences identified in any interval specified. Such sequences are extracted to substitute with previous observation values or clinical data through multiple instances. These instances generate total\(\:\:{\complement\:}_{sequences}\) (combination) values under the varying instances. From these instances, the ensemble perceptron detects the maximum correlating sensor value for diagnosis. From the dataset, a total of 4286 entries at an average of 714 counts for each sensor, with the individual sequences (in Fig. 1a) used to generate maximum possibilities. The values that increase the prediction accuracy are used for sequence substitution.
Proposed allied data disparity technique
The data analytic model is proposed to sense the clinical data from WS devices that acquire activities vital to humans. This observation takes place periodically, ensembles the clinical data, and detects the function that represents health monitoring in the early stages. In this execution step, the incomplete data are considered and provided incomplete data, which is detected using the data disparity technique. It ensures the prediction model observes the data in the stored dataset and generates the result. This approach is followed up with an MIEPL concept. Figure 1b depicts the workflow of the proposed ADDT architecture.
Input sensor and clinical data are preprocessed to eliminate noise, handle missing values, and normalize for consistency in the described model architecture for the ADDT method. Next, feature extraction extracts relevant and representative features from the processed data. A disparity detection module finds discrepancies and variations across numerous inputs. The system calculates the average deviation of these features using a mean disparity computation. The model begins an MIEPL phase with this disparity information, when many perceptrons simultaneously learn from different data subsets. Independent perceptrons contribute to a prediction pool. The model uses maximum correlation to pick perceptron outputs that agree most to improve accuracy. An ensemble updating method uses performance feedback to fine-tune the selected models. The intelligent decision-making procedure concludes with dependable diagnostic generation from improved ensemble outputs.
The diagnostically important factors, including heart rate variability, blood oxygen saturation trends, glucose fluctuations, and systolic/diastolic patterns, have been analyzed; feature extraction was performed on sensor and clinical data streams before the model was trained. Consideration of clinical relevance and statistical association with labelled diagnosis outcomes informed feature selection. The feature scales were normalized using z-score normalization as part of a preprocessing pipeline that included a moving average filter to decrease noise and clinical-value-based imputation to address missing values. The extracted features \(\:{v}^{{\prime\:}},{a}_{l},{x}^{{\prime\:}},{y}^{{\prime\:}}\) were then evaluated for disparity using the ADDT formulation, with \(\:{M}_{e}\) representing extracted clinical metrics and \(\:D{a}^{{\prime\:}}\) as sensor-derived sequences, these steps ensure that input features are coherent, denoised, and scale-invariant for the perceptron ensemble learning stage.
The evaluation runs through the data analytic method that identifies mean disparity for varying data sequences. The data sequences define the WS data variant and generate the periodic analysis of human health status. This approach evaluates human activities and vitals periodically and makes a diagnosis accordingly. It observes the data from WSs and evaluates them for sequence estimation. This technique detects the disparity in data sequences where the threshold maintenance is defined. The preliminary step is to analyze the incomplete clinical data, which is expressed in the following Equation.
Equation (1) computes the complete clinical data aggregate analysis \(\:\nabla\:\) derived from wearable sensors and helps identify gaps and disparities in clinical data records. This is used to analyze the incomplete clinical data, which refers to the dataset acquired from WS devices. The \(\:{v}^{{\prime\:}}\) are symbolized as detected human activities like walking and sitting, and \(\:{a}_{l}\) as vital signs for heart rate, and temperature. The data storage is\(\:\:Da{\prime\:}\) with healthy records, \(\:\:\beta\:\) is referred to as clinical data parameters like blood pressure, where \(\:\:y{\prime\:}\) is observed as a periodic detection value from the sensor. The sequences of data that are acquired from the sensor are\(\:\:Sq,\) the diagnosis of diseases is\(\:\:x{\prime\:},\) and\(\:\:{i}_{p}\) is labeled as incomplete data points. The term \(\:\sum\:_{\beta\:}D{a}^{{\prime\:}}-{i}_{p}\) Subtract incomplete values to factor in the impact of missing data. The analysis calculates a weighted product of clinical indicators and detected values, helping to estimate data reliability and health trends. From this observation step, the clinical data is acquired from the sensor device\(\:\:\left(\:Sq+Da{\prime\:}\right)*\sum\:_{{i}_{p}}\beta\:\). The examination takes place for the clinical data acquisition and detects incomplete data. The periodic observation is done for the data sequences where the activities are vital. The analysis is done for the data variant where the incomplete sequences are observed from the clinical data and the mapping with previous inputs. The overall representation of ADDT model is provided below:

Algorithm 1.
The process begins with initializing the sensor input data, followed by preprocessing to clean and structure the detected patterns. Key features are then extracted to facilitate deeper analysis. A loop mechanism is employed to iteratively evaluate sensor sequences, updating and refining the output at each step. The final output is generated based on maximum correlation and matched patterns, ensuring accurate and efficient sequence recognition.
The computation is done for the clinical data to detect the incomplete data sequences from the dataset. This phase examines the incomplete data that is extracted from the WS. The normal and abnormal values are detected from the attained clinical data, and it is mapped with the dataset\(\:\:\beta\:+\left({i}_{p}*{e}^{{\prime\:}}\right)<0\), in this case, greater than 0 resembles normal, or else it represents abnormal. On this basis, clinical data are used to examine the human activities and vital rate, which vary over the desired time interval. This process is carried out by diagnosing the activities and vitals, and it is matched with the dataset and generates the results. The diagnosis is done initially and extracted from the data sequences. Thus, this analysis step indicates the normal and abnormal values extracted from the data sequences acquired from the sensor device. The examination of activities and vitals is observed periodically, derived from the Eq. (2) below, calculates the periodic medical evaluation \(\:{M}_{e}\) analyzing historical and current data from sensor input aids in real-time decision-making.
The examination is done for human activities and vital functions, and the data sequences on WSs to capture periodic patient condition updates using real-time and historical data. The products and nested sequences show compounded health risks or events. The use of \(\:{p}^{{\prime\:}}\) as prediction-based features and the clinical data extraction is labeled as\(\:\:e{\prime\:}\) for correlation \(\:\:{T}_{0}\) is represented as detection timestamp. The examination is used to analyze the data extracted from the data sequence periodically\(\:\:\left[\left(Sq+{y}^{{\prime\:}}\right)*\nabla\:\right]\). The periodical data acquisition and the incomplete data from the sequenced sensor devices are evaluated. This data extraction phase is done by diagnosing the medical data and providing the necessary steps. Both the activities and vitals are used to observe the human status periodically. The main concept here is to detect the activities and vital signs periodically, and based on this, necessary action is taken. This process is considered by diagnosing the activities that indicate the data storage and whether it is normal or abnormal. The preprocessing of data for correlation is graphically illustrated in Fig. 2.
The data analysis rate increases and ensures a higher precision rate by properly detecting the patient’s normal and abnormal activities and vital signs. According to the threshold and update of the patient, the correlation factor\(\:\:\left({M}_{e}+\beta\:\right)\). The detection takes place from the extracted input features, and analysis takes place\(\:\:\left({v}^{{\prime\:}}+{a}_{l}\right)*\nabla\:\). The evaluation is considered for the data sequences associated with decision-making and improving the data analysis rate. The execution occurs by analyzing the clinical data and ensuring the higher threshold value (Fig. 2). The correlation between the activities and vital signs is observed for the data variant observed at different intervals. Based on the different intervals, incomplete data is acquired, and it is determined whether it is normal or abnormal, which is carried out with the prediction step discussed in the section below. Diagnosis uses varying input data and finds diseases at different time intervals. This detection step is used to find the data sequences in data storage and generates the output accordingly. From this observation step, the correlation and prediction are executed and elaborated in the following section, one by one. This case indicates that the clinical data observation for this disparity detection is carried out on the data. The disparity identification \(\:Ind\) used to quantify abnormality or data inconsistency in periodic clinical data is done periodically and expressed in Eq. (3) below. Followed by \(\:{e}_{r}\) as the error rate and residual from the model prediction, also applies \(\:{M}_{e},D{a}^{{\prime\:}},{v}^{{\prime\:}},{a}_{l},{x}^{{\prime\:}},{y}^{{\prime\:}}\) to validate coherence.
The identification is followed up in the above Equation, which examines the sensor device’s clinical data. This evaluation phase runs on the incomplete data and analyzes the desired value from the sequences. The sequences are considered from the examination, and it is represented as\(\:\:{M}_{e}\), the identification is\(\:\:Ind\), where the extraction is done for the evaluation of normal and abnormal data and acts accordingly. In this execution step, the activities and vital signs are monitored, and where there is coherence with the clinical data and maps with the dataset\(\:\:D{a}^{{\prime\:}}+\left({x}^{{\prime\:}}*\beta\:\right)\). This observation step relies on the disparity of the data and provides the disparity analysis for the coherent clinical data. By evaluating this step, the disparity is considered and provides the output based on the perception learning. This is processed under the prediction state and is observed periodically. The disparity detection flow is illustrated in Fig. 3.
The disparity detection process is illustrated in Fig. 3 by interlinking.\(\:\:{y}^{{\prime\:}}\) and\(\:\:Sq\). The aim is that\(\:\:\beta\:\) must balance\(\:\:\sum\:\left({v}^{{\prime\:}},{a}_{l}\right)\) in any\(\:\:{y}^{{\prime\:}}\) provided\(\:\:Sq\) is continuous. Therefore, the failing condition flow estimates the\(\:\:{i}_{p}\) as\(\:\:\left(\beta\:-{y}^{{\prime\:}}\right)\) in any sensed sequence such that\(\:\:Ind\) is the sum of\(\:\:\left(\nabla\:\:\text{a}\text{n}\text{d}\:{i}_{p}\right)\). Surpassing this output, the\(\:\:{g}_{x}\) and\(\:\:{o}_{r}\) mapping occurs for\(\:\:{i}_{p}\) estimation where the correlation between sensor and clinical data occurs. If the above condition is satisfied, then\(\:\:{e}_{r}\) is alone estimated, and the\(\:\:Sq\) is stored in\(\:\:{D}_{a}^{{\prime\:}}\). This\(\:\:{e}_{r}\) if found to be greater than\(\:\:{i}_{p}\), the mapping is further pursued for multiple input instances. In this step, the diagnosis is made for the correlation of the data sequences, and the output is given based on the identification. Identification plays a role in extracting data and finding the disparity in the sequences. If any disparity is detected, the signal is given to the healthcare and the activities and vitals are carried out on an interval basis. This interval of time is considered for the identification of incomplete data, which is mapped with the data\(\:\:{e}_{r}*\sum\:_{Ind}\left({p}^{{\prime\:}}+Da{\prime\:}\right)\). From this observation step, \(\:\left\{\sum\:\left({M}_{e}+D{a}^{{\prime\:}}\right)*\left({v}^{{\prime\:}}+{a}_{l}\right)\right\}\) is examined for the data variant. Based on this identification step, the sequence disparity is coherent with the dataset’s current and existing data. Hereafter, the identification of disparity is computed using the above Equation. The mean disparity is computed through decision-making described in the steps below:

Algorithm 2.
The algorithm works according to the Eq. (4) below, which is generated for the decision-making process.
The decision-making process is represented as\(\:\:{d}_{s}\), \(\:{g}_{x}\) is existing value \(\:\:{o}_{r}\) is the correlation mapping between the current and existing value generated from the medical center. Based on this method, the decision is made on the mean disparity, and the data requirement for the WS sequence is decided. This decision uses multiple substituted and predicted values obtained from previous instances. MIEPL is used in this decision process, where the substitution instances for clinical and previous outcomes are performed. Based on the above steps, the disparity detected for multiple data instances is analyzed in Fig. 4.
The disparity detection is less if it analyzes the incomplete sequence and substitutes medical data. This process indicates the monitoring of the vitals and activities of the patient periodically\(\:\:\left({v}^{{\prime\:}}+{a}_{l}\right)*p{\prime\:}\left({x}^{{\prime\:}}\right)\). In processing this detection, the time interval is considered to observe the disparity and provide the necessary action\(\:\:{e}_{r}\left(\beta\:*{M}_{e}\right)-Ind\). By evaluating this step, detection occurs by mapping carried out with the data storage. The data storage of clinical existing data is mapped and found whether it is normal or abnormal\(\:\:\left({m}_{0}+{b}_{a}\right)\). This analysis phase is done using a prediction method executed in the perceptron learning concept and shows lesser detection of disparity. Real-world wearable sensor data is typically uneven and asynchronously updated, but the ADDT framework is built to handle this and works without regularly spaced data. ADDT continuously compares real-time sensor measurements to clinical or historical values. When missing sequences or asynchronous gaps are found, the system triggers a substitution mechanism to extract the most contextually relevant instances, either temporal trend predictions or clinical data. These substitutions are filtered and weighted using ensemble precision scoring to integrate the most reliable and diagnostically helpful data. This dynamic handling of missing or delayed sequences allows ADDT to function well in extremely irregular real-world monitoring contexts.
Multi-instance ensemble perceptron learning
The proposed MIEPL is unique. It can analyse many sequences of sensor-clinical data (i.e., multi-instances) and produce reliable predictions by merging numerous perceptrons trained on different substitution instances. Our model treats every data sample as a collection of cases instead of the flat feature vector used by conventional perceptron models. This allows us to deal with data samples that fluctuate in time, such as sensor readings and intermittent clinical records. The “multi-instance” method allows the model to learn patterns from clinical data streams partially absent, asynchronous, or unevenly spaced by training and making decisions over similar but not identically structured inputs. By combining the predictions of several perceptron learners that have been trained on various weighted combinations of these input examples, the “ensemble” feature further improves dependability. Each perceptron evaluates the fused feature vector using dynamically assigned weights \(\:{w}_{0}\) from Eq. (7) and threshold calibration \(\:\mu\:\) from Eq. (5) ensures that even inconsistent or incomplete data subsets contribute to an accurate final decision.
The perceptron is a neural learning model that relies on the binary classification method for the attained input data, such as clinical data, and makes decisions accordingly. So, it relies on 0 or 1 cases where the threshold value is generated to observe the threshold value based on the input and weight. The comparison is taken for the weighted input with a threshold value and concludes if the input is greater than it generates the output as 1, or else it is 0. The value of \(\:\mu\:\) is selected through empirical validation using a validation dataset that contains known cases of both normal and abnormal clinical conditions. The \(\:\mu\:\) determines whether the weighted input derived from the sensor-clinical data fusion should be flagged as disperate/ anomalous (1) or acceptable (0). By guaranteeing that only sensor-clinical data inconsistencies that fall significantly below the decision threshold are highlighted, the calibrated µ value increases the accuracy of the ADDT framework and so improves forecast accuracy and system dependability.
The above Eq. (5) examines the threshold and maps with the clinical data. The threshold is labeled a\(\:\:\mu\:\), and the weight is\(\:\:{w}_{0}\) where the condition is satisfied with clinical data observation. In this execution step, data sequences are detected to analyze input. From this, the following parameters are discussed in the perceptron learning method.
-
1.
Extraction of Input features: It evaluates the characteristics and features of input clinical data, where multiple patterns and features are considered using an Eq. (6).
The feature of the input data is\(\:\:f{\prime\:}\), which includes the activities and vitals that are examined for the multiple patterns analysis.
-
2.
Initial Weight: For every initial input, the weights that generate the output are assigned, which is followed up based on the training section where it attains the optimal value given in Eq. (7).
The weights are assigned based on the input data, where the output is\(\:\:{o}_{u}\) that results during the training section.
-
3.
Summation function: A weighted sum of the attained input and distributed according to the respective weights.
The summation function is labeled as\(\:\:{s}_{f}\)where the assignment is carried forward for the clinical data.
-
4.
Activation function: This is used to observe whether the assigned weight is 0 or 1, which is mapped to the threshold value.
The activation function is\(\:\:{t}_{n}\); it generates the weight resulting in either 0 or 1 by mapping with the threshold value, which is discussed in Eq. (5).
-
5.
Bias Update: This factor is used to predict whether the model is in error, which is observed using a decision-making approach.
The bias is\(\:\:{b}_{i}\) It is used to predict the value, whether normal or abnormal, and takes place with the decision-making concept.
-
6.
Final Output: By processing in perceptron learning, output is generated based on the activation function that holds the threshold value, and it is formulated as.
The main factor here is the threshold value that indicates the activation function to perform, resulting in the output as 0 or 1. From the above parameter, the work for perceptron learning indicates the steps-wise computation and finds the output with the threshold value. The ensemble learning is diagrammatically depicted in Fig. 5 below.
The perceptron learning model for predictive outputs of\(\:\:Sq\) substitution is depicted in Fig. 5. The first input is the set of\(\:\:{e}^{{\prime\:}}\) (constant) and\(\:\:{g}_{x}\) (variable) for which independent\(\:\:{\omega\:}_{o}\) is assigned. This\(\:\:{\omega\:}_{o}\) is computed using Eq. (7) to ensure\(\:\:{S}_{f}\) meets the constraint\(\:\:\left({a}_{l}+{v}^{{\prime\:}}\right)\). If the constraint is satisfied, then the possibility of\(\:\:{e}^{{\prime\:}}\) is higher than the previous\(\:\:{g}_{x}\). Therefore, \(\:{d}_{s}\) for\(\:\:{M}_{e}\left({\omega\:}_{o}\right)\) is alone computed for\(\:\:{b}_{i}\left(\nabla\:\right)\) process. If these computations are sufficient for prediction, then the ensembles of\(\:\:{g}_{x}\) are confined to augment\(\:\:{e}^{{\prime\:}}\). Else,\(\:\:Ind\) was observed under\(\:\:{y}^{{\prime\:}}\)and\(\:\:{T}_{o}\) are used to verify\(\:\:{S}_{f}\) using\(\:\:\mu\:=0\) (minimum) and\(\:\:\mu\:=1\) (maximum) threshold outcomes. Therefore, the consecutive computation of the predictive output relies on\(\:\:Sq\) (presence) to ensure high predictive outputs are achieved. The substitution accuracy is graphically analyzed from this learning model as in Fig. 6.
The substitution accuracy shown in Fig. 6 is higher, which indicates the WSD for the data variant and observes the mapping for the vital activities.\(\:\:\left({v}^{{\prime\:}}+{a}_{l}\right)*\sum\:_{{M}_{e}}\left({y}^{{\prime\:}}+\beta\:\right)\). This detection occurs periodically, and data storage is used to identify the disparity. This substitution value is used to provide the threshold value, and it generates higher precision\(\:\:\mu\:>{u}_{b}\left(\gamma\:\right)+Sq\). Processing this step indicates the detection of normal and abnormal clinical data. The detection of maximum correlation is presented in the following algorithm.
Correlation of sensor input
The correlation is used to find a similar value from the clinical data and to observe a better diagnosis of human activities and vital signs. If the correlation is the same or equal, no emergency is observed, whereas if it is higher or lower, the emergency threshold is considered.

Algorithm 3.
The above algorithm states the correlation factor for the clinical data and finds the mapping with the threshold value, which is equated in the following derivative.
The correlation is derived from the threshold and decision-making process, which detects normal and abnormal signals from the sensor. The variables\(\:\:{m}_{0}\:\text{a}\text{n}\text{d}\:{b}_{a}\) are symbolized as normal and abnormal,\(\:\:\gamma\:\) is represented as a prediction. The substitute data is labeled as\(\:\:{u}_{b}\). The sequence error analysis is presented in Fig. 7 based on the correlation process.
The sequence error is less if it reaches below the threshold value, and examines the mapping with the extracted vitals and activities using\(\:\:{t}^{{\prime\:}}\left(\beta\:+{g}_{x}\right)+{d}_{s}\). From this detection phase, identification is carried out to determine whether it is maximum or minimum. In executing this, a disparity detection is performed.\(\:\:{p}^{{\prime\:}}\left({T}_{0}+{y}^{{\prime\:}}\right)*\sum\:_{{e}_{r}}\gamma\:\). This observes the periodic identification of data\(\:\:\left(Ind+\beta\:\right)*\gamma\:-{e}_{r}\left({g}_{x}\right)\). Considering that these error sequences occur due to incomplete data, it is observed that this indicates the assignment of weights for the input (Fig. 7). Following this method, the prediction is followed up with the upcoming algorithm.
Sequence value prediction
The prediction is followed up by mapping with the attained threshold value, where the decision is made accordingly to the variant data analysis. This approach indicates the data sequences and finds the normal and abnormal signals. The algorithm below is used to define the prediction.

Algorithm 4.
The prediction is followed up for the clinical data, where the mapping is followed up by incomplete data observation, represented in the following Equation.
The prediction is done for the existing value from the clinical data observation, which executes the identification of data sequences progressing in the above Equation. Hereafter, the diagnosis coalition between clinical and update is done periodically for the new precision level that has been identified.
The above Equation is derived for the analysis of threshold value generation, generating higher precision values identified with the decision-making approach. This process indicates the periodical observation method for clinical data. It correlates with the existing value, finding whether it is a substitute or incomplete data, and based on this, precision is enhanced. Perceptron Learning is used to perform substitution instances for clinical and previous outcomes. The ensemble perceptron selects the maximum clinical value correlating sensor data to ensure high sequence prediction. The ensembles are updated based on the highest precision-based WS values for diagnosis. This diagnosis-focused coalition between clinical and predicted WS values is updated periodically for the new precision levels identified.
Result analysis and discussion
The strong libraries for machine learning and signal processing, Python 3.8 is used to efficiently develop the suggested ADDT model. Important instruments are pandas/NumPy for data management and scikit-learn for perceptron and ensemble learning. SciPy helps to process signals from wearable sensors. Matplotlib for visualisation and libraries such as PyTorch is included. Sktime improves feature extraction for time-series analysis and MATLAB provides robust signal analysis support using its Neural Network Toolboxes, ML, and Signal Processing. This system allows accurate, real-time health tracking. The existing models, PTL-DTM22, CNN-BiGRU23, and HM-HMS31 are validated using metrics such as accuracy, precision, recall, F1-score, and AUC on various sequence patterns.
Table 2 shows a scenario-based assessment of the ADDT paired with the Multi-Instance Ensemble Perceptron model, stressing its therapeutic relevance and operational efficiency across different patient circumstances. Every scenario mimics real-world situations with missing, incorrect, and replaced wearable sensor data. The assessment criteria are response time, clinical decision support efficacy, disparity detection rate, and sequence prediction accuracy. Results show the model’s robustness in managing data anomalies, preserving high accuracy, and enhancing diagnosis consistency via periodic updates and adaptive learning, confirming its relevance in dynamic healthcare monitoring contexts.
In the discussion section, two metrics\(\:\:\gamma\:\) accuracy and\(\:\:\gamma\:\) precision, are considered for analysis. \(\:Sq\) variants for temperature, heart rate, pulse rate, and respiratory are 12, 48, 24, and 288, respectively. For the varying\(\:\:Sq\), the above metrics are studied for\(\:\:{b}_{a}\) and\(\:\:{m}_{o}\) conditions. If the\(\:\:\beta\:\) illustrated in Fig. 1 is similar to the sensor observed value in any\(\:\:{y}^{{\prime\:}}\) or\(\:\:{e}^{{\prime\:}}\) sequence, then it is\(\:\:{m}_{o}\). The out-of-range values due to missing/erroneous sequences observed result in\(\:\:{b}_{a}\) for which diagnosis, priority is to be given. The dataset observations are jointly used based on time and\(\:\:Sq\) from which appropriate readings/values are obtained. Using these values, the metrics mentioned above are analyzed and represented in Figs. 8 and 9 with a discussion below.
The prediction accuracy is high for varying clinical data and its data sequences. On executing this data sequences are used to provide the desired diagnosis for the incomplete data\(\:\:Ind\left(\beta\:+{d}_{s}\right)+\gamma\:\). This method indicates the existing data where the mapping is followed up with the data storage\(\:\:D{a}^{{\prime\:}}\left(Sq+\beta\:\right)*\gamma\:\). It has progressed under the perceptron learning\(\:\:{t}^{{\prime\:}}\left({w}_{0}+{b}_{i}\right)-{e}_{r}\) which is associated with the activation function to generate a higher prediction among the clinical data (Fig. 8). The precision analysis of\(\:\:\gamma\:\) is discussed in Fig. 9 below.
The prediction precision is high if the input features are associated with the periodic observation.\(\:\:\left(Sq+Da{\prime\:}\right)*\prod\:_{{T}_{0}}{p}^{{\prime\:}}-{e}_{r}\). The time interval is considered for detecting clinical data and performing decision-making. The decision is made using a prediction on perceptron learning, where the correlation is considered.\(\:\:{o}_{r}\left(\gamma\:\right)-\beta\:\). From this observation, the substitution of clinical data is examined for the data sequences\(\:\:\left(\rho\:+Ind\right)*e{\prime\:}\). The activation function is generated where the input features are associated with weight assignment and ensures higher prediction precision (Fig. 9).
Across four signal types listed in Fig. 10 —Temperature, Heart Rate, Pulse Rate, and Respiratory—this Python script visualizes the performance comparison of four models—PTL-DTM, CNN-BiGRU, HM-HMS, and ADDT. Using bar graphs, it assesses three critical measures: Recall, F1-Score, and AUC. Every chart clearly shows Y-axis labels and numerical values atop the bars. The performance of several deep learning models—PTL-DTM, CNN-BiGRU, HM-HMS, and ADDT—on physiological sensor data, including temperature, heart rate, pulse rate, and respiration signals. The study intends to find the most accurate and robust model for sequence-based health monitoring and predictive analysis utilizing examination of recall, F1-score, and AUC across several signal kinds.
Table 3 compares recall, F1-Score, and AUC for four models: PTL-DTM, CNN-BiGRU, HM-HMS, and ADDT, spanning signal kinds and sequence length (Sq) variants. Sq variants (12, 48, 24, and 288) examine Temperature, Heart Rate, Pulse Rate, and Respiratory signals. The ADDT model dominates all signal types with the greatest scores (Recall: 94.1–95.6%, F1-Score: 93.7–95.1%, AUC: 94.8–95.6%). CNN-BiGRU and HM-HMS perform well in the mid-tier, with HM-HMS outperforming. PTL-DTM is a valid baseline despite being behind in all measurements. \(\:Sq\:\)length boosts performance, suggesting the benefit of temporal context. Respiratory signals with the longest sequence perform best across models. These results show that ADDT works and that signal type and temporal granularity matter.
In this research, 10-fold cross-validation has been explicitly used and shows mean ± standard deviation for each metric recall, F1-score, and AUC across folds. This validates the model’s robustness and generalization capability across different data partitions. Table 4 shows the 10-fold cross-validation mean and standard deviation of recall, F1-Score, and AUC. This method evaluates the model on the entire dataset, boosting generalizability and robustness. All measures show that ADDT outperforms baseline models. ADDT had a greater accuracy and lower variance than HM-HMS, with a Recall of 94.2% ± 0.5 compared to 88.6% ± 0.9. The narrow standard deviations show that ADDT is stable over folds and not overfitting to specific data subsets.
The confidence interval (CI) analysis proves that the ADDT model outperforms baseline models as shown in Table 5. Recall, F1-Score, and AUC were measured using 95% CIs from 10-fold cross-validation with bootstrap resampling. ADDT had the highest mean scores and narrow CIs, such as Recall 94.2% (CI: 93.8–94.6). Baseline models like CNN-BiGRU showed lower means and larger CIs (Recall 87.0%, CI: 86.2–87.8). Non-overlapping CIs show ADDT’s statistically significant improvements. It has low performance variance and excellent robustness. CIs provide accurate model performance estimates. Overall, this data supports ADDT’s generalizability and efficacy.
Paired statistical significance testing of ADDT’s improvement over the best baseline model HM-HMS shows t-statistics and p-values for each metric. Table 6 confirms that the ADDT’s performance gains are not due to chance and are statistically significant.
This method evaluates the model on the entire dataset, boosting generalizability and robustness. All measures show that ADDT outperforms baseline models. ADDT had a greater accuracy and lower variance than HM-HMS, with a Recall of 94.2% ± 0.5 compared to 88.6% ± 0.9. The narrow standard deviations show that ADDT is stable over folds and not overfitting to specific data subsets.
Conclusion
This article introduced the novel allied technique to validate the WS data sequence disparity. The missing/ erroneous sequences in the observed intervals are substituted with predicted and clinical data. The data with high precision aiding in diagnosis is thus identified from different regular and irregular sequences. For this purpose, multiple substitution instances from the various sequences ensure perceptron functionalities. The summation and activation functions are responsible for detecting high-precision sensor data inputs. The maximum clinical value correlating input/ sequence is identified using perceptron learning. Therefore, the WS values in the current sequence are updated using the high-precision predicted input. This proposed technique improves the precision of the prediction process by evaluating the clinical data. The data process is carried out periodically, and the sequences are based on the activities. This technique ensures better prediction through correlation with the stored and clinical values.
Though the technique is reliable for predicting erroneous and incomplete WS sensor sequences, the ML-based feature mapping is found to be irresistible for asynchronous intervals. The different operational intervals of the WS cause the problem for which the feature mappings are to be made based on their least possible sequences. Therefore, the proposed technique will add a classification-dependent sequence detection and analysis method.
Strengths and weaknesses
One of ADDT’s main advantages is its flexible merging of clinical and forecast data, which allows the system to maintain continuity in sensor data streams even with dropout, noise, or asynchronous recording periods. An ensemble perceptron learning mechanism enables flexible and precise substitution by using high-precision correlated sequences, hence lowering false alerts and enhancing anomaly detection performance. Moreover, by utilizing periodic updates, the method guarantees that diagnosis-relevant data stays current and accurate.
Even with its encouraging results, the suggested approach has certain drawbacks. Its dependence on machine learning-based feature mapping in asynchronous intervals is one significant difficulty since it could result in poor results if the feature space is not evenly represented. WS data sequence irregularity adds heterogeneity that impedes model generalization across patient profiles and treatment environments.
Future work will focus on the following key directions to address these limitations.
-
Model Generalization: Incorporating transfer learning and domain adaptation strategies to improve the model’s adaptability to different patient populations, sensor types, and clinical conditions.
-
Deployment in clinical settings: Conducting large-scale pilot studies in real-world clinical settings like ICU and patient monitoring platforms to validate the system’s utility, scalability, and integration feasibility.
-
Data Diversity and Augmentation: Enhancing model robustness by including multimodal datasets, covering a range of physiological signals and clinical contexts, while employing advanced data augmentation to simulate rare case scenarios.
Data availability
The data used to support the findings of the study are included within the article.
Abbreviations
- β :
-
Parameter representing a threshold or a scaling factor in data analysis equations; also related to data sequence segmentation.
- S q :
-
Sensor data sequence or specific sensor observation window, representing data collected over time intervals.
- Ind(i,p) :
-
Disparity indicator function; used to detect the presence of data disparity or discrepancies in sequences.
- y′,e′:
-
Predicted or estimated clinical data sequences; reference values for comparison against actual or observed data.
- ba :
-
Out-of-range or erroneous data condition, indicating inaccuracies due to missing, erroneous, or noisy data sequences.
- m′(γ):
-
Function or value related to subscription or mapping parameter, influencing data substitution or prediction modules.
- μ :
-
Threshold parameter ranging from 0 to 1 used in decision-making processes and data analysis, e.g., minimal or maximal thresholds.
- Da′(Sq*y′):
-
Processed or stored dataset incorporating sensor data combined with predicted clinical sequences.
- x′(β):
-
Data variable representing an observed or predicted clinical parameter, influenced by the threshold parameter β.
- ω0 :
-
Weight vector or coefficient in perceptron learning, computed to ensure data meets specified constraints.
- e′,gx :
-
Input variables to the perceptron model, representing constants or variables related to data features or special sequences.
- seq :
-
Sequence instances or combinations of data points used for analysis and prediction.
- Me :
-
Target or output data representing normal/abnormal states or the data used for evaluation in disparity analysis.
- p′:
-
Predicted probability.
References
Liu, Y. et al. Topological data analysis for robust gait biometrics based on wearable sensors. IEEE Trans. Consumer Electron. (2024).
Qin, L., Guo, M., Zhou, K., Chen, X. & Qiu, J. Gait recognition using deep learning with handling defective data from multiple wearable sensors. Digit. Signal Proc. 154, 104665 (2024).
Kim, H., Lee, G., Ahn, H. & Choi, B. Interpretable general thermal comfort model based on physiological data from wearable bio sensors: light gradient boosting machine (LightGBM) and SHapley additive explanations (SHAP). Build. Environ. 266, 112127 (2024).
Van Der Donckt, J. et al. Mitigating data quality challenges in ambulatory wrist-worn wearable monitoring through analytical and practical approaches. Sci. Rep. 14 (1), 17545 (2024).
Kimura, N. et al. Predicting positron emission tomography brain amyloid positivity using interpretable machine learning models with wearable sensor data and lifestyle factors. Alzheimers Res. Ther. 15 (1), 212 (2023).
Kaili, Q. Wearable devices based on wireless sensor network and speech synchronization overlay algorithm application in sports training data simulation. Mob. Netw. Appl. 1–13. (2024).
Kazanskiy, N. L., Khonina, S. N. & Butt, M. A. A review on flexible wearables-Recent developments in non-invasive continuous health monitoring. Sens. Actuat. A Phys. 114993. (2024).
Das, T., Yadav, M. K., Dev, A. & Kar, M. Double perovskite-based wearable ternary nanocomposite piezoelectric nanogenerator for self-charging, human health monitoring and temperature sensor. Chem. Eng. J. 496, 153926 (2024).
Chen, X. et al. Self-powered flexible wearable wireless sensing for outdoor work heatstroke prevention and health monitoring. Chem. Eng. J. 156431 (2024).
Nguyen, T. T. H. et al. Field effect transistor based wearable biosensors for healthcare monitoring. J. Nanobiotechnol. 21 (1), 411 (2023).
Su, M. et al. Wireless wearable devices and recent applications in health monitoring and clinical diagnosis. Biomed. Mater. Dev. 2 (2), 669–694 (2024).
Xue, Z., Gai, Y., Wu, Y., Liu, Z. & Li, Z. Wearable mechanical and electrochemical sensors for real-time health monitoring. Commun. Mater. 5 (1), 211 (2024).
Kim, D., Lee, J., Park, M. K. & Ko, S. H. Recent developments in wearable breath sensors for healthcare monitoring. Commun. Mater. 5 (1), 41 (2024).
Hou, Y. Optical wearable sensor based dance motion detection in health monitoring system using quantum machine learning model. Opt. Quant. Electron. 56 (4), 686 (2024).
Alsadoon, A., Al-Naymat, G. & Jerew, O. D. An architectural framework of elderly healthcare monitoring and tracking through wearable sensor technologies. Multimed. Tools Appl. 1–46. (2024).
Huang, X. et al. Comparison of feature learning methods for non-invasive interstitial glucose prediction using wearable sensors in healthy cohorts: a pilot study. Intell. Med. (2024).
Putra, K. T. et al. A Review on the Application of Internet of Medical Things in Wearable Personal Health Monitoring: A Cloud-Edge Artificial Intelligence Approach. IEEE Access. (2024).
Yang, Y. Application of wearable devices based on artificial intelligence sensors in sports human health monitoring. Meas. Sens. 33, 101086 (2024).
Arslan, M. M. et al. Advancing healthcare monitoring: integrating machine learning with innovative wearable and wireless systems for comprehensive patient care. IEEE Sens. J. (2024).
Shi, H., Hou, Z., Liang, J., Lin, E. & Zhong, Z. Dsfnet: A distributed sensors fusion network for action recognition. IEEE Sens. J. 23 (1), 839–848 (2022).
Gong, W. et al. Embracing Self-Powered wearables for intelligent healthcare data management. IEEE Internet Things J. (2024).
Alruwaili, O., Yousef, A. & Armghan, A. Monitoring the Transmission of Data from Wearable Sensors Using Probabilistic Transfer Learning (IEEE Access, 2024).
Imran, H. A., Riaz, Q., Hussain, M., Tahir, H. & Arshad, R. Smart-wearable sensors and cnn-bigru model: A powerful combination for human activity recognition. IEEE Sens. J. (2023).
Zha, M., Zhu, L., Zhu, Y., Li, J. & Hu, T. A novel SCNN-LSTM model for predicting the SNR confidence interval in wearable wireless sensor network. Intell. Syst. Appl. 22, 200363 (2024).
Tatli, D., Papapanagiotou, V., Liakos, A., Tsapas, A. & Delopoulos, A. Prediabetes detection in unconstrained conditions using wearable sensors. Clin. Nutr. Open Sci. (2024).
Nihar, K., Jain, R. K. & Cheong, S. M. Predicting indoor personalized heat stress using wearable sensors and data-driven models. J. Build. Eng. 97, 110761 (2024).
Matsumura, G. et al. Real-time personal healthcare data analysis using edge computing for multimodal wearable sensors. Device. (2024).
Pini, N. et al. Remote data collection of infant activity and sleep patterns via wearable sensors in the healthy brain and child development study (HBCD). Dev. Cogn. Neurosci. 69, 101446 (2024).
Kim, N., Lee, S., Kim, J., Choi, S. Y. & Park, S. M. Shuffled ECA-Net for stress detection from multimodal wearable sensor data. Comput. Biol. Med. 183, 109217 (2024).
Zadka, A. et al. A wearable sensor and machine learning estimate step length in older adults and patients with neurological disorders. Npj Digit. Med. 7 (1), 142 (2024).
Mahato, K. et al. Hybrid multimodal wearable sensors for comprehensive health monitoring. Nat. Electron. 1–16. (2024).
Gashi, S. et al. Modeling multiple sclerosis using mobile and wearable sensor data. Npj Digit. Med. 7 (1), 64 (2024).
Li, C., Shen, H. & Ge, Z. Optical sensor based athlete player health monitoring and performance enhancement using data analytics based on machine learning model. Opt. Quant. Electron. 56 (1), 109 (2024).
Zhang, W. et al. Wearable sensor-based quantitative gait analysis in parkinson’s disease patients with different motor subtypes. Npj Digit. Med. 7 (1), 169 (2024).
Acknowledgements
This research was supported by the Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R259), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. This work was supported by the Researchers Supporting Project Number (MHIRSP2024005) Almaarefa University, Riyadh, Saudi Arabia. The authors extend their appreciation to the support provided by the University of Business and Technology, Jeddah, 21448, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Mohd Anjum: Conceptualization, Methodology, Software, Writing - Original Draft, Writing - Review & EditingWaseem Ahmad: Methodology, Validation, Formal analysis, Resources, Writing - Review & Editing, VisualizationSana Shahab: Conceptualization, Methodology, Software, Data Curation, Writing - Original Draft, Writing - Review & Editing, Visualization, Funding acquisitionAshit Kumar Dutta: Methodology, Resources, Writing - Review & Editing, Visualization, Funding acquisitionAli Elrashidi: Conceptualization, Methodology, Resources, Writing - Original Draft, Writing - Review & Editing, Funding acquisitionAmr Yousef: Methodology, Validation, Formal analysis, Resources, Data CurationZaffar Ahmed Shaikh: Methodology, Validation, Formal analysis, Data Curation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Anjum, M., Ahmad, W., Shahab, S. et al. Enhancing wearable sensor data analysis for patient health monitoring using allied data disparity technique and multi instance ensemble perceptron learning. Sci Rep 15, 29555 (2025). https://doi.org/10.1038/s41598-025-08051-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-08051-w