Abstract
Colorectal cancer (CRC) treatment remains challenging due to genetic heterogeneity and resistance mechanisms. To address this, we developed a drug discovery pipeline using patient-derived primary CRC cultures with diverse genomic profiles. These cultures closely resemble certain molecular characteristics of primary and metastatic CRC, highlighting their promise as a translational platform for therapeutic evaluation. Importantly, our engineered model and patient-derived cells reflect the complexity and heterogeneity of primary tumors, not observed with standard immortalized cell lines, offering a more clinically relevant system, although further validation is needed. High-throughput screening (HTS) of 4255 compounds identified 33 with selective efficacy against CRC cells, sparing normal, healthy epithelial cells. Among the tested combinations, everolimus (mTOR inhibitor) and uprosertib (AKT inhibitor) demonstrated promising synergy at clinically relevant concentrations, with favorable therapeutic windows confirmed across tested patient-derived cultures. Notably, this synergy, revealed through advanced models, might have been overlooked in traditional immortalized cell lines, highlighting the translational advantage of patient-derived systems. Furthermore, the integration of machine learning into the HTS pipeline significantly improved scalability, cost-efficiency, and predictive accuracy. Our findings underscore the potential of patient-derived materials combined with machine learning-enhanced drug discovery to advance personalized therapies. Specifically, mTOR-AKT inhibition emerges as a promising strategy for CRC treatment, paving the way for more effective and targeted therapeutic approaches.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) is a major public health concern, ranking as the third most prevalent cancer worldwide. In 2020, CRC was responsible for 10% of all cancer diagnoses, amounting to approximately 1,931,590 new cases 1. Interestingly, the incidence of CRC shows a demographic split: it has been decreasing among older adults but is on the rise among younger individuals1,2. This shift emphasizes the importance of understanding CRC across different age groups, especially since early-stage detection can dramatically improve patient outcomes. In the United States, the five-year survival rate for CRC patients stands at about 63% and drops to 13% when diagnosed at an advanced stage3. This statistic highlights the critical role of timely diagnosis, as early detection significantly improves patient outcomes. At the core of CRC’s development is a series of genetic mutations affecting the epithelial tissues of the colon and rectum. These mutations involve key genes such as APC, KRAS, TP53, and SMAD4 and propel the disease along one of three molecular pathways: chromosomal instability, microsatellite instability, or the CpG island methylator phenotype4,5. Each pathway affects crucial cellular processes like DNA mismatch repair, cell cycle regulation, and apoptosis, underlining the complexity of CRC’s molecular basis6,7.
Understanding the mutation burden in CRC has significant implications for treatment. Studies have shown that CRCs with a high mutation load respond better to treatment, particularly due to the increased genetic instability that makes cancer cells more susceptible to chemotherapy and immunotherapy8. This suggests that patients with certain genetic profiles, such as intact APC or mutations in KRAS, BRAF, and TP53, may benefit more from specific therapeutic approaches9. However, despite advancements in targeted therapies, the effectiveness of CRC treatment is heavily dependent on the cancer’s genetic characteristics, necessitating personalized treatment plans.
The management of CRC remains challenging due to the potential for disease recurrence, often with additional oncogenic drivers compared to the primary cancer10. This recurrence can complicate treatment strategies and underscores the need for ongoing research into effective therapies.
Generating primary models representing the early onset of diseases can be highly beneficial for identifying therapeutic targets effective in late-stage cancer. Early-stage models often share common genetic drivers with advanced cancers, as they originate from a common ancestral cell. Studying early models potentially leads to uncovering key pathways and vulnerabilities that persist as the disease progresses11. Consequently, targets identified in early models may provide broader therapeutic value, addressing early and late cancer stages.
This study outlines our approach to high-throughput compound screening, which holds significant potential for identifying novel treatment options for CRC patients. For this purpose, we have developed an intestinal model that employs epithelial stem cells to trace the biological progression of CRC from normal stem cells through sequential genetic alterations, following Vogelstein’s paradigm4. These stem cells are cultured on a monolayer of 3T3 J2 mouse fibroblasts, which effectively emulate the role of stromal cells from the tumor microenvironment—an aspect frequently absent in other research models. This model captures the complex physiology of CRC development while preserving the practicality of immortalized cell lines within the research pipeline, making it an ideal tool for large-scale, high-throughput studies. To address potential discrepancies between genetically engineered models and patient-derived samples, we have extended our research to include cultures derived from CRC patients with diverse genomic profiles. We hypothesize that vulnerabilities common to both the model and patient-derived cultures will reveal the most promising therapeutic targets, with a particular emphasis on those that spare normal cells, thereby advocating their potential as the safest therapeutic options.
Results
Stepwise resistance to niche signaling stimuli drives colorectal cancer progression
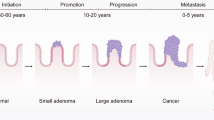
By analyzing the frequency and co-occurrence of mutations in colorectal cancer, we identified key genetic alterations in APC, KRAS, TP53, and SMAD4, which collectively define a large proportion of the CRC mutational landscape. Based on this analysis and data from The Cancer Genome Atlas (TCGA), we engineered a comprehensive in vitro model combining these frequently co-occurring mutations. Remarkably, this model represents over 40% of all CRC tumors (Supplementary Fig. S1a, S1b), demonstrating its relevance in understanding the genetic basis of CRC. To recapitulate a common tumor evolutionary pathway in colorectal cancer, we introduced a truncating mutation in the APC gene (APCtrunc) in normal human intestinal stem cells. This was achieved by delivering Cas9 ribonucleoprotein complex and homology directed repair knock-in template carrying stop codon in exon 15 through the nucleofection process. Damaging the APC gene disrupts the Beta-catenin degradation complex, leading to independence from WNT pathway stimulation. Consequently, we functionally validated mutation-containing clones by withdrawing R-Spondin-1, a WNT pathway agonist essential for wild-type cells, from the culture media (Fig. 1a). As expected, healthy, control cells died after one passage, while cells carrying APC-truncating mutations or APC knockouts grew normally. To mimic the natural progression of tumor evolution, confirmed APC-truncated clones were then selected for further engineering (Supplementary Fig. S2).
Stepwise Resistance to Niche Signaling Stimuli Drives Colorectal Cancer Progression. a Schematic representation of colorectal cancer (CRC) progression from normal intestinal epithelium, following the classic Vogelstein paradigm, which involves the stepwise accumulation of mutations in APC, KRAS, TP53 and SMAD4. Created with BioRender.com. b Growth curves of engineered CRC models (AKT and AKTS) after xenografting into immunocompromised mice, AKT n = 8; AKTS n = 11. c Representative immunostaining of normal intestinal stem cell (WT)-derived colonies used in model development. Intestinal lineage markers (green or red), and nuclei are counterstained with DAPI (blue). Scale bar, 50 μm. d Hematoxylin and eosin staining of sectioned air–liquid interface (ALI)-grown structures derived from successive stages of the engineered CRC model. Scale bar, 100 μm. e Immunofluorescence labeling of intestinal and proliferation markers (green or red) in ALI-derived structures at various stages of CRC progression. Scale bar, 100 μm.
In a subsequent step, we introduced the KRAS G12D mutation, the most common variant in CRC, rendering the cells resistant to the withdrawal of EGF from the culture media and to treatment with the EGFR inhibitor gefitinib. Next, we knocked out the TP53 gene. The modified cells acquired resistance to the nutlin-3 treatment, an MDM2 inhibitor that leads to TP53 upregulation and cell senescence which was used as a selection criterion. Finally, we disrupted the TGF-beta signaling pathway by knocking out the SMAD4 gene and selecting modified cells through TGF-beta treatment, causing differentiation of SMAD4 WT cells. This allowed us to develop the following stages of colorectal cancer evolution: APC-truncated (A), APC-truncated with TP53 knockout (AT), APC-truncated with KRAS G12D substitution (AK), AK with TP53 knockout (AKT), and AKT with SMAD4 knockout (AKTS). Apart from functional validation, all stages were confirmed using Sanger sequencing and Western blotting (Supplementary Fig. S2a and S2b). We also tested the tumorigenicity of the AKT and AKTS models by demonstrating their ability to form tumors in immunocompromised mice (Fig. 1b).
We then performed a series of assays on the developed models to assess the functional consequences of the engineered genotypes. Using immunofluorescence, we characterized intestinal adult stem cells by staining cultured colonies for epithelial markers, including general epithelial cadherin (ECAD) and the intestinal cadherin marker (CDH17). We also confirmed that the stem cells express self-renewal and proliferation markers such as SOX9 and Ki67 but are negative for differentiation markers, including Mucin 2 (MUC2) and Villin 1 (VIL1) (Fig. 1c and Supplementary Table S1). Building on these findings, we next investigated how genetic modifications influence stem cell differentiation into mature intestinal tissue in the Air Liquid Interface (ALI) system. Truncation of APC (A cells) caused minor changes in cell morphology (Fig. 1d) and led to a slight loss of cell polarity. In contrast, constitutive activation of the KRAS protein (AK cells) resulted in significant changes to the spatial organization of the differentiated culture, an increase in Ki67-positive cells, and a loss of MUC2-positive goblet cells (Fig. 1e). The most dramatic changes were observed in AKT and AKTS cells, which exhibited the greatest loss of polarity and the highest number of Ki67-positive cells. Interestingly, the Villin expression was retained in all model variants, even in cells that lost polarity in HE staining.
Engineered model closely resembles early onset of CRC
To gain further insights into the engineered CRC models, we performed RNA sequencing (RNA-seq) analysis and compared their transcriptomes with those of normal wild-type (WT) intestinal stem cells. Our results (Fig. 2a) showed that A and AT cells clustered together with the WT epithelium (Cluster A), while AK, AKT, and AKTS cells formed a separate cluster (Cluster B).
Transcriptomic Characterization of the Engineered Model in the Context of Available CRC References. a Gene expression heatmap displaying the relative expression of 260 differentially expressed genes (DEGs) between normal intestinal epithelial stem cells (WT) and engineered CRC model cells (AKTS). b UMAP embedding of MOBER-derived transcriptomic data integrating engineered CRC models with publicly available CRC datasets. Two primary clusters were identified: Cluster 1, comprising metastatic CRC samples, primary tumors, and most CRC cell lines; and Cluster 2, which includes the engineered CRC models grouped with primary CRC tissues and a subset of metastatic samples. Insets (2A, 2B) highlight the clustering of healthy intestinal stem cells with public healthy tissue samples (2A) and the engineered CRC models with primary CRC tissues (2B). c Differential gene programs identified using ExpiMap overlaid on the clustered data from engineered CRC models and publicly available RNA-seq datasets. Gene programs were derived from GSEA collections, identifying pathways relevant to CRC progression and differentiation.
Upon examining differentially expressed genes (DEGs), we found that Cluster A, which includes WT cells, exhibited upregulated genes associated with digestive system processes, lipid and carotenoid metabolism, and various gastrointestinal tract differentiation markers. Conversely, these genes were downregulated in Cluster B, which contains the more advanced stages of the model (Fig. 2a). Interestingly, Cluster B showed upregulated gene sets related to WNT and TGF-beta signaling, extracellular matrix expression, cell migration, and cancer metastasis, while these gene sets were downregulated in Cluster A. This suggests that AK, AKT and AKTS cells may exhibit preliminary signs of a malignant phenotype.
To contextualize our models within the landscape of patient-derived tissues and transcriptomes published from various other CRC-related studies, we developed a transcriptomic library of approximately 1,500 samples curated from publicly available databases (Supplementary Table S2). A deep learning model – Multi-Origin Batch Effect Remover (MOBER)12, was trained to normalize and cluster these samples, facilitating the identification of differences and similarities between our models and those studied in other projects. The analysis divided all the samples into two distinct clusters (Fig. 2b). Cluster 1 predominantly comprised metastatic CRC samples, primary tumor site samples, and nearly all CRC cell lines. In contrast, Cluster 2 encompassed our engineered CRC models, primarily grouped with CRC tissue samples from primary sites and a smaller subset from metastatic sites, aligning with the stepwise genetic evolution of CRC as it acquires tumor driver mutations. Further analysis revealed that healthy stem cells, used as the origin for CRC model development, clustered with healthy tissue samples from public datasets (Cluster 2A). Notably, patient-derived cells utilized in these studies were identified in both Cluster 1 and Cluster 2, suggesting that our culture system and machine learning-guided analysis lack systemic bias.
To investigate the mechanisms underlying the formation of the two clusters, potentially reflecting the progression from early-stage cancer (Cluster 2) to more advanced, often metastatic tumors (Cluster 1), we employed several analytical approaches. Initially, we applied a consensus molecular subtyping tool, specifically designed for colorectal cancer preclinical models, which leverages gene expression signals13. However, this method failed to assign molecular subtypes to approximately 10% of the samples (Supplementary Fig. S3, Supplementary Fig. S6). To overcome this limitation, we reimplemented and retrained a deep learning-based framework (DeepCC) for cancer molecular subtype classification14. Unlike the CMS caller, which relies solely on gene expression signals, DeepCC utilizes functional spectra to quantify the activities of biological pathways, providing a pathway-centric approach. This shift enabled us to successfully assign consensus molecular subtypes to the vast majority of samples. Additionally, we implemented a pathway-based molecular classification system through the pathway-based molecular classification system (PDS) classifier41. These pathway-focused methods provide deeper insights into the transcriptomic landscape and enhance our understanding of molecular subtypes. When we compared our clusters in the scope of Pathway level subtyping we observe prevalence of PDS2 samples (46% for Cluster 2 and 25% for Cluster 1) enriched for inflammatory and immune signaling pathways, such as interferon-α and interferon-γ response as well as stromal-related epithelial-to-mesenchymal and transforming growth factor β (TGF-β) activation. The trend is opposite for samples classified as PDS1 subtype (8% for Cluster 2, 32% for Cluster 1). Interestingly, we observed that Cluster 2 is enriched with Mixed PDS subtype (32% for Cluster 2 in comparison to 19% for Cluster 1), potentially due to the presence of normal samples. However, none of these colorectal cancer subtyping approaches provided significant insights into the differences between Cluster 1 and 2.
Recognizing the benefits of examining biological pathways rather than focusing solely on individual gene signals, we employed the Explainable Programmable Mapper (Expimap), a machine learning algorithm, to further characterize the transcriptomes. It not only effectively separated all samples in the analysis but also identified statistically significant gene programs distinguishing the clusters (Fig. 2c). While some findings aligned with expectations, such as the involvement of KRAS, EGFR, and TP53 signaling pathways, others were unexpected, including mTORC1 signaling, EZH2 targets, and responses to retinoic acid. These results highlight novel pathways potentially contributing to CRC progression or response to targeted therapies.
Application of CRC model cells in drug discovery research and introduction of machine learning methods for analysis
Following molecular characterization, which confirmed that the transcriptomic profile of our model was consistent with clinical CRC samples, we validated its suitability for high-throughput chemical screenings aimed at identifying novel therapeutic options for CRC patients.
In a pilot study, we evaluated the cytotoxic effects of standard-of-care drugs for CRC including 5-fluorouracil, oxaliplatin, and SN38 (an active metabolite of irinotecan), on healthy cells, engineered CRC models, and patient-derived cells. The results revealed non-selective cytotoxicity of these drugs across all tested cell types. In contrast, a selective G12D inhibitor demonstrated efficacy, specifically targeting cells harboring this driver mutation while sparing healthy cells (Fig. 3a). These results provided initial evidence supporting the capability of the proposed discovery pipeline to potentially identify clinically relevant findings, though larger-scale validation is necessary.
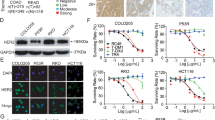
Application of CRC Model Cells in Drug Discovery Research and Machine Learning Methods for Analysis. a Dose–response curves showing the cytotoxic effects of standard-of-care drugs on healthy cells (WT), engineered CRC models (A, AK, AKT, AKTS), and patient-derived cultures (P1–P5). Error bars represent standard deviation, n = 3. b High-throughput screening (HTS) workflow of 4,255 compounds on AKTS cells, n = 3. Screening was performed at a fixed concentration of 1 μM. c Scatter plot showing hits from the primary screening on AKTS cells, with compounds achieving ≥ 70% inhibition selected for further validation. d Scatter plot comparing IC50 values of compound families between WT and AKTS cells. Selective responses in engineered CRC models highlight key drug target families, including mTOR, AKT and EZH2, n = 3. e Heatmap of differential compound responses (Δ AUC) across CRC models and healthy cells, revealing enrichment of inhibitors targeting key pathways, including mTOR, AKT, EZH2, and ALK. f Machine learning pipeline enhancing HTS capabilities: a U-Net-based neural network generates FITC segmentation masks from DAPI-stained images, replacing EdU assays for cell proliferation analysis. g Representative images demonstrating the machine learning model’s input (DAPI), predicted segmentation (FITC), and actual FITC results.
Following a successful pilot study, we expanded the screening to a full-scale experiment using our CRC model. A total of 4255 chemical compounds were screened in triplicate using high-throughput screening (HTS; Fig. 3b and Supplementary Fig. S4a; S4b). The screening assay demonstrated strong technical robustness, with Z′ factor values consistently exceeding 0.5 across plates (Supplementary Fig. S4b), supporting the reproducibility of results. Compounds showing ≥ 70% inhibition in at least two of three replicates were classified as primary hits and selected for further validation. From the primary screening, 304 active compounds were identified (Fig. 3c). To assess the specificity of these compounds for cancerous cells, we conducted dose–response validation assays, comparing their effects on both the CRC model and healthy human intestinal stem cells (Fig. 3d). This step was essential for identifying compounds that selectively target CRC cells while sparing healthy cells, a critical criterion for developing effective and safe therapeutic strategies. As anticipated, inhibitors targeting EGFR were validated as effective in wild-type (WT) cells but showed no efficacy in CRC model cells harboring the KRAS oncogenic mutations. These findings align with established resistance mechanisms in CRC and indicate the potential clinical relevance of our screening method for identifying compounds tailored to specific genetic profiles.
Ultimately, 33 compounds were chosen for further testing on patient-derived CRC samples based on their selective anticancer effects. These compounds were notably enriched in inhibitors representing six distinct drug target families (Fig. 3e). Among these, inhibitors targeting mTOR, EZH2, and retinoic acid pathways aligned with the gene programs identified using the ExpiMap model. Validation in patient-derived samples demonstrated that 10 of the 33 compounds were consistently effective across four tested PDCs and at all stages of engineered models. The effective compounds included everolimus, GW843682X, ensartinib, GSK126, PF-06726304, tazemetostat, uprosertib, miransertib, landiolol, and afuresertib, which target key signaling pathways such as mTOR, PLK, ALK, and EZH2. Specifically, tazemetostat, GSK126, and PF-06726304 target EZH2, while AKT is inhibited by uprosertib, miransertib, and afuresertib. The remaining compounds exhibited variable efficacy, with some being patient-specific or effective only within certain subgroups. These results underscore the promise of our screening approach in advancing personalized therapies for CRC patients, highlighting its strong translational potential.
To explore whether machine learning (ML) could support future high-throughput phenotypic screens, we developed a proof-of-concept pipeline based on a deep vision model. This approach was designed to evaluate whether parts of a labor-intensive and costly assay could be replaced by model predictions trained on a relatively small initial dataset. Specifically, we trained the model to predict the cell proliferation in co-cultures of human CRC and mouse 3T3 cells using only DAPI-stained images, bypassing the need for additional immunofluorescent staining.
To implement this concept, we developed ML-based pipeline that involved generating single-cell segmentation masks for DAPI and FITC images, training a U-Net-based neural network to predict FITC segmentation from DAPI images, and correlating the predicted FITC segmentation with actual cell counts from HTS (Fig. 3f, g). The model demonstrated strong performance achieving high accuracy, with Pearson correlation coefficients of 0.99 for training cell line (AKTS) and 0.97 for unseen WT cell lines (Supplementary Fig. S4c). These results demonstrate the reliability and scalability of our screening process and reinforce the ML model’s value in our drug discovery pipeline. Minimal training data requirements were determined through a learning curve analysis (Supplementary Fig. S4d), showing that control wells from a single plate were sufficient for accurate predictions, while adding additional control data ensured the model’s applicability to WT cells. Once optimal performance was reached, it remained consistent across all subsampled runs, as reflected by the low variance in the learning curve scores—indicating the model’s robustness and low susceptibility to batch effects.
While this ML method was not applied to scale the current primary screen, it serves as a generalizable and accessible framework for reducing assay costs in future screens. By significantly lowering the time, labor, and financial input required for coculture proliferation assays, it enables larger-scale compound screening efforts that may otherwise be constrained by resource limitations. We acknowledge current limitations, including validation restricted to in-distribution data and potential challenges in generalizing to new imaging setups or cell types. Nevertheless, our findings demonstrate the feasibility and broader potential of integrating ML into phenotypic screening workflows, offering a scalable and cost-effective approach for drug discovery.
Inhibition of mTORC1 and AKT shows synergistic effect in CRC cells with KRAS mutational background
Following dose–response validation assays on selected HTS hits, we identified five representatives of enriched drug families targeting specific pathways: uprosertib (AKT), everolimus (mTOR), PF-06726304 (EZH2), givinostat (HDAC), landiolol (adrenergic pathway), and MRTX1133, a specific KRAS G12D inhibitor with the potential to transform the treatment landscape for CRC patients by directly targeting a previously undruggable oncogenic driver mutation. To further evaluate their therapeutic potential, we investigated potential synergistic effects through combination assays using AKTS cells, selecting concentration ranges based on prior dose–response curve (DRC) data.
A strong synergy was observed between everolimus and uprosertib (Fig. 4a, b), which was consistently confirmed by multiple reference models, including Loewe (mean score: 11.87, p = 3.75 × 10⁻4), Bliss (15.73, p = 3.76 × 10⁻5), HSA (13.20, p = 1.93 × 10⁻⁶), and ZIP (18.63, p = 8.56 × 10⁻⁷). This combination was validated across a panel of evolutionary models, CRC patient-derived cultures, and healthy cells. Differential responses were quantified by analyzing the median distributions between normal epithelial stem cells (WTs) and each tested culture (Fig. 4c).
Synergistic Effects of mTORC1 and AKT Inhibition in CRC Models with KRAS Mutational Background. a Loewe synergy analysis of selected drug combinations in AKTS cells, ranked by mean Loewe synergy index, n = 3. Statistical significance is indicated by asterisks: *p < 0.05, **p < 0.01, ***p < 0.001. b Loewe synergy surface plot for the everolimus and uprosertib combination in AKTS cells, highlighting a strong synergistic interaction at clinically relevant concentrations (n = 3). The Loewe model yielded a mean synergy score of 10.5 (p = 1.32 × 10⁻3), indicating a statistically significant effect (**). c Median difference distribution of the everolimus and uprosertib combination across effective concentration ranges in engineered CRC models. The asterisk indicates the selected concentration point used for further visualizations. Generated using Python 3.10 script utilizing the Matplotlib 3.82 library. d Validation of the everolimus and uprosertib combination across evolutionary CRC models (A, AK, AKT, AKTS), patient-derived cultures (P1–5), and healthy control cells (WT). Synergistic responses were quantified using the Loewe Synergy Index (LSI), showing enhanced synergy in KRAS-mutant CRC models. Error bars represent standard deviation, n = 3. e Transcriptomic differences between responders (LSI > 20) and non-responders (LSI < 20) assessed using GSCORE-based pathway enrichment analysis. Dot size and y-axis position reflect pathway-level confidence (–ln[FDR]), while the x-axis (m/n ratio) indicates the proportion of differentially expressed genes within each enriched pathway.
To contextualize these phenotypic differences, we further stratified patient-derived cultures (PDCs) based on their synergy response to the everolimus–uprosertib combination, defining strong responders as those with a Loewe synergy index (LSI) > 20 and weak or non-responders as those with LSI < 20 (Supplementary Fig. 1b). PDC#2 (P2) was excluded from this classification due to its pronounced monotherapy sensitivity to uprosertib, likely reflecting its unique molecular profile (Supplementary Fig. 1b). To investigate mechanisms underlying response heterogeneity, we applied GSCORE, a topology-based pathway enrichment framework that integrates gene expression magnitude with pathway structure, to compare transcriptional programs between responders and non-responders (Fig. 4e). The two groups exhibited distinct pathway-level profiles, with differences observed in KRAS-associated gene signatures, TGF-β signaling, and regulatory activities within the EGFR, mTOR, WNT, AKT, and EZH2 pathways. Notably, non-responders also showed consistent deviation in the "Colorectal Adenoma Downregulated" gene set15, which includes multiple WNT-related transcripts, indicating altered WNT pathway regulation. These preliminary findings, when integrated with mutational profiles, suggest that both genetic and transcriptional features contribute to differential drug sensitivity, supporting a stratified approach to predicting therapeutic response in CRC. However, further investigation—using expanded PDC coverage and complementary analytical approaches—is needed to confirm these trends and elucidate more granular regulatory relationships.
The combination of everolimus and uprosertib demonstrated strong synergy at clinically relevant exposure, showing high efficacy in KRAS-mutant CRC models and patient-derived cultures, with a satisfactory therapeutic window in most cases. The tested concentrations (5–100 nM for everolimus and 0.2–2 µM for uprosertib) were selected based on pharmacokinetic data from clinical trials and fall within the range of plasma exposures achieved in patients (Supplementary Table S7). Notable exceptions included: PDC#2 (P2), which exhibited hypersensitivity to uprosertib alone, with a potent response even at the lowest tested concentrations. As a result, the addition of everolimus provided no further benefit in this model, suggesting a unique dependency on AKT signaling consistent with its PTEN mutation16. In contrast, the APCtrunc model and PDC#5 (P5) showed limited responsiveness to the combination therapy (Fig. 4d and Supplementary Figure S2b;).
Together, these findings highlight the therapeutic potential of dual AKT and mTOR inhibition in KRAS-mutant CRC and underscore the importance of integrating transcriptional and mutational profiling to identify biomarkers of resistance and guide patient stratification for combination therapies.
Interruption of feedback loop between Akt-mTORC1 is detrimental for colorectal cancer
Our investigation into dual inhibition of AKT and mTORC1 elucidates the mechanistic basis underlying their synergistic anti-tumor activity in colorectal cancer (CRC). By examining the interactions within the AKT–mTORC1 signaling, we uncovered pathways that mediate the observed anti-proliferative and pro-apoptotic effects. These insights inform promising strategies for more effective CRC treatment.
Treatment with uprosertib alone led to hyperphosphorylation of AKT at Ser473 and Thr308 (Fig. 5a, b). Paradoxically, this enhanced AKT phosphorylation occurred alongside reduced phosphorylation of downstream AKT targets, including GSK3α/β and the S6 ribosomal protein. This indicates that while uprosertib triggers a feedback-driven increase in AKT activation, it concurrently diminishes AKT-mediated signaling. Notably, uprosertib decreased the phosphorylation of GSK3α at Ser21 and GSK3β at Ser9, effectively restoring GSK3 activity. The reactivation of GSK3 may, in turn, suppress pro-survival pathways and inhibit CRC cell proliferation.
Interruption of Feedback Loop Between AKT–mTORC1 Signaling is Detrimental for Colorectal Cancer. a Immunoblot analysis showing the effects of uprosertib (AKT inhibitor), everolimus (mTORC1 inhibitor), and their combination on AKT–mTORC1 signaling in CRC cells over various time points (0.5, 3, and 48 h). b Densitometric analysis of immunoblots. c Schematic representation of the AKT–mTORC1 signaling pathway and feedback regulation.
Everolimus alone, as expected, inhibited mTORC1 activity, shown by reduced phosphorylation of the S6 ribosomal protein at Ser235/236. However, this suppression triggered a compensatory response, increasing phosphorylation of GSK3α (Ser21) and GSK3β (Ser9) at 48 h post-treatment. Such feedback responses highlight a central challenge: while selective mTORC1 inhibition impedes growth signals, it can also activate alternative survival pathways, ultimately limiting therapeutic efficacy.
The combination of uprosertib and everolimus capitalized on their complementary mechanisms to achieve superior growth inhibition over either agent alone. Although the dual treatment increased AKT phosphorylation at Ser473 and Thr308 relative to everolimus monotherapy, it counteracted the everolimus-induced phosphorylation of GSK3α/β. Restoring GSK3 activity under combination treatment underscores its importance in suppressing proliferative signaling. Additionally, while the combination partially reversed everolimus-induced decreases in S6 Ser235/236 phosphorylation, it also uniquely reduced a mitotic marker—histone H3 Ser10 phosphorylation. This broader impact on chromatin remodeling and transcriptional control further distinguishes the dual therapy’s mechanism of action.
Beyond inhibiting proliferation, the combination therapy markedly enhanced pro-apoptotic effects. Cleavage of PARP and caspase-3—early markers of apoptosis—was significantly increased compared to single-agent treatments. Although uprosertib restored GSK3 activity, we did not observe consistent changes in β-catenin protein levels, suggesting that Wnt/β-catenin signaling is not the primary mediator of apoptosis in this context. While we did not assess differentiation markers directly, our results suggest that the apoptotic response is primarily driven by the disruption of AKT–mTORC1 signaling and not through GSK3-mediated modulation of Wnt pathway output. This enhanced apoptotic response reflects the combination’s ability to disrupt compensatory survival mechanisms within the AKT–mTORC1 axis, ultimately driving CRC cells toward programmed cell death.
In summary, the combined use of uprosertib and everolimus represents a rational therapeutic strategy for CRC, overcoming limitations associated with single-agent interventions by dismantling critical feedback loops and promoting apoptosis. These findings deepen our understanding of AKT–mTORC1 crosstalk and support clinical exploration of this promising dual-targeted approach for improved CRC management.
Discussion
For decades, therapies such as irinotecan, oxaliplatin, and 5-fluorouracil (5-FU) have been the cornerstone of CRC treatment17. While the advent of targeted therapies like EGFR and KRAS inhibitors has introduced more personalized approaches, their efficacy remains limited18, often providing only moderate responses and encountering resistance. These challenges underscore the urgent need to explore novel therapeutic strategies to improve outcomes for CRC patients.
In this study, we developed a drug discovery pipeline that integrates engineered CRC models, patient-derived cultures, HTS, and machine learning to identify and validate novel treatment options (Fig. 6). Using stepwise genetic engineering, we introduced APCtrunc, KRASG12D, TP53KO, and SMAD4KO mutations into normal intestinal stem cells to generate models that closely mimic CRC progression. These models exhibited hallmark CRC features, including altered differentiation, loss of polarity, and increased proliferation19. Transcriptomic analyses further validated these models, revealing that advanced stages (AK, AKT, and AKTS) clustered distinctly from early-stage models (A) and wild-type (WT) cells. These advanced models shared significant transcriptomic similarities with patient-derived CRC samples, particularly those from primary tumor sites, aligning with gene expression profiles associated with malignant phenotypes. This strong concordance underscores their translational relevance for studying CRC progression and therapeutic responses.
The application of an ML-based algorithm, ExpiMap, facilitated the identification of key gene programs and pathways differentiating early and advanced stages of CRC in our models and patient samples. Specifically, the upregulation of pathways related to mTORC1 signaling, EZH2 targets, and retinoic acid responses in advanced models highlighted potential therapeutic targets that are not only critical in CRC progression but also amenable to pharmacological intervention20,21,22. Our high-throughput approach enabled the screening of 4255 compounds, identifying 33 with selective efficacy against CRC cells while sparing normal epithelial cells. These compounds were enriched in drug families targeting mTOR, EZH2, and AKT pathways, aligning with the gene programs identified by ExpiMap. To further dissect response heterogeneity, we stratified PDCs based on their synergy score for everolimus and uprosertib combination into responders (LSI > 20) and non-responders (LSI < 20). GSCORE-based pathway analysis revealed distinct transcriptional landscapes between these groups, including differences in KRAS-associated gene signatures, TGF-β signaling, and regulatory activity across EGFR, mTOR, WNT, AKT, and EZH2 networks. Further studies on a larger cohort of heterogeneous samples are needed to validate the identified mechanisms and assess their utility for patient stratification.
Treatment with mTOR or AKT inhibitors as monotherapies confers only modest anti-tumor effects in KRAS-mutant CRC cells. mTORC1 inhibition reduces S6 phosphorylation but triggers compensatory upregulation of AKT activity, maintaining pro-survival signaling23,24. Conversely, isolated AKT inhibition relieves feedback suppression on mTORC1, preserving growth-promoting pathways25. These adaptive mechanisms highlight the inherent plasticity of the PI3K/AKT/mTOR axis, which undermines the efficacy of single-agent therapies.
Our findings demonstrate that dual inhibition of AKT and mTORC1 effectively circumvents these compensatory feedback loops, resulting in more pronounced anti-tumor response (Fig. 5c). Within 24 h of combined treatment, CRC cells exhibit significant apoptosis, as evident by increased cleavage of caspases and PARP, along with reduced histone H3 and S6 phosphorylation. Notably, these effects were not accompanied by changes in GSK3α/β phosphorylation or β-catenin levels, suggesting that Wnt/β-catenin signaling is not a primary driver of cell death in this context. This dual inhibition approach provides a more sustained suppression of growth and survival pathways, dismantling key feedback loops that otherwise enable resistance. This synergy underscores the critical role of the PI3K/AKT/mTOR axis in CRC progression, as this pathway is frequently dysregulated and drives tumor growth and survival26,27.
The reactivation of GSK3 following AKT inhibition raised the possibility of modulation of β-catenin signaling, given GSK3’s established role in Wnt pathway regulation28. However, our analyses showed no significant changes in β-catenin levels, indicating that canonical Wnt signaling is unlikely to mediate the apoptotic response observed. Although we did not evaluate differentiation markers in this study, the data suggest that GSK3 contributes to the pro-apoptotic effect independently of its classical role in cell fate decisions. This supports the interpretation that the observed synergy arises primarily from feedback disruption within the AKT–mTORC1 axis.
Interestingly, parallels can be drawn to non-small cell lung cancer (NSCLC), where KRAS mutations frequently shift signaling towards the PI3K-AKT-mTOR axis. Recent studies have shown that specific KRAS isoforms, such as KRASG12D, exhibit greater oncogenic potency than the more common KRASG12C isoform, likely due to hyperactive PI3K-AKT-mTOR signaling29. While signaling differences between KRASG12D and KRASG12C diminish during tumor progression, isoform-specific vulnerabilities persist, offering actionable therapeutic targets. For example, KRASG12D-driven tumors rely more heavily on PI3K-AKT-mTOR signaling, while KRASG12C tumors depend on MAPK signaling29. This highlights the importance of tailoring therapeutic strategies to KRAS isoforms and their associated pathway dependencies. Similarly, in CRC, the observed synergy between mTORC1 and AKT inhibitors reflects the translational potential of targeting pathway-specific vulnerabilities in KRAS-mutant cancers.
These findings underscore the functional interdependence of AKT and mTORC1 and highlight the therapeutic advantage of simultaneously disabling both signaling nodes. By dismantling key feedback loops, dual inhibition produces a more sustained suppression of growth and survival pathways than either approach alone. This strategy aligns with emerging evidence supporting multi-node targeting within the PI3K/AKT/mTOR network to enhance therapeutic efficacy, particularly in KRAS-mutant CRC subtypes that display intrinsic resistance to conventional agents30,31,32. Consequently, co-inhibition of AKT and mTORC1 represents a promising framework for a more effective combination therapies development in CRC.
While dual PI3K/AKT pathway inhibition poses potential safety concerns due to the pathway’s role in normal tissues, our platform partially addresses this by including normal intestinal epithelial stem cells as a healthy reference in all assays, allowing direct comparison of drug effects on tumor versus normal cells. Furthermore, our co-culture system incorporates stromal fibroblasts, enabling preliminary assessment of combined treatment impact on the tumor microenvironment components. We acknowledge that comprehensive toxicity evaluation requires more complex models beyond the current scope. However, the integration of AI-driven transcriptomics and CRISPR-based disease modeling within our platform holds promise for future precise patient stratification. With further development and validation, this approach could guide selection of patients most likely to benefit from combination therapies while minimizing adverse effects.
The ML methods integration into our drug discovery pipeline significantly enhanced the efficiency and scalability of HTS. By developing a vision-based ML model to analyze DAPI-stained images, we reduced reagent costs and labor while maintaining high accuracy in predicting cell proliferation. This advancement enables large-scale screenings and accelerates the identification of promising therapeutic candidates.
Despite the strengths of our study, several limitations should be acknowledged. First, while our engineered models capture key genetic alterations in CRC, they may not fully represent the complexity of tumor microenvironments and interactions with immune cells, which play crucial roles in cancer progression and response to therapy. However, unlike many available systems, our approach includes co-culturing cancer cells with supportive stromal cells, thereby creating a more physiologically relevant microenvironment that better recapitulates the complex interplay between tumor and stroma. Second, the patient-derived cultures used in our validation studies, although diverse, represent a limited sample size (n = 5). Larger cohorts would strengthen the generalizability of our findings. Moreover, although the selective KRASG12D inhibitor, MRTX1133, demonstrated potent activity in our engineered model, its effectiveness was diminished in PDCs harboring the G12D mutation. This reduced sensitivity likely stems from drug resistance mechanisms acquired during treatment, which are absent in the background-clean model. In conjunction with combination studies, these findings highlight that our model is particularly effective in identifying universal genetic dependencies, which can be evaluated in patient-specific contexts.
In conclusion, our study demonstrates the power of integrating engineered CRC models, patient-derived cultures, HTS, and ML to identify and validate novel therapeutic strategies. The synergy observed between mTOR and AKT inhibitors offers a promising avenue for treating CRC patients with specific genetic backgrounds. Our findings contribute to the growing body of potential therapies in the CRC.
Materials and methods
Cell culture
This study was conducted in accordance with the Declaration of Helsinki. All analyses involving patient and human samples adhered to the guidelines and procedures of National Institute of Oncology in Warsaw and Gdansk Medical University. The Bioethics Committee of Maria Sklodowska-Curie National Research Institute of Oncology in Warsaw and Bioethics Committee for Scientific Research of Medical University of Gdansk (approval no: 55/2017 and NKBBN/766/2021 respectively), approved protocols in compliance with applicable regulations, requirements, and guidelines, and informed consent was obtained from all participating patients. Stable cultures were established as previously described with slight modifications33. Briefly, tumor tissues were mechanically dissociated into small pieces and incubated with 1 mg/ml collagenase type IV (Gibco) at 37 °C for 45–60 min. The suspension was filtered through 70 µm strainer and washed several times with wash buffer (F12 (Gibco), 5% FBS (Gibco), 1% Penicillin–Streptomycin (Gibco), 1% Gentamicin (Gibco), 0.1% Amphotericin B (Gibco)). Single cell suspension was seeded in culture medium (Supplementary Table S3) onto irradiated 3T3-J2 feeder cells monolayer prepared 24h earlier and cultured at 37 °C in 6% CO2.
Animal studies
All animals were handled in strict accordance with good animal practice as defined by the relevant national and/or local animal welfare bodies. The experiment was approved by the 2nd Local Institutional Animal Care and Use Committee Institute of Pharmacology Polish Academy of Sciences in Cracow (permission no. 342/2022) and performed following ARRIVE guidelines (https://arriveguidelines.org). NSG female mice (Charles River Labs, Animalab), 7–9 weeks old, used for the experiment were housed in SPF conditions. Cells (c.a. 105 per mouse) were suspended in 100 µL of culture medium and Cultrex (R&D Systems, cat. No 3632–005-02) mixture (1:1, v:v) and were injected subcutaneously on the right hind limb. As the tumor growth was progressing, the tumor size was measured twice weekly, and the tumor volume was computed from the formula: TV = (a*a*b)/2 in mm3, where a is the short axis in mm, and b is the long axis in mm. Body weight was measured twice weekly throughout the study. Since day 45 after cells inoculation, selected mice were anaesthetized with isoflurane and euthanized by cervical dislocation to collect tumors for downstream analysis.
Stem cell differentiation methodology (ALI)
The air–liquid interface (ALI) culture of intestinal epithelial cells was conducted using Transwell inserts (Corning, USA) coated with 20% growth factor-reduced Matrigel (BD Biosciences, USA). The inserts were incubated at 37 °C for 30 min to allow the Matrigel to polymerize. Irradiated 3T3-J2 feeder cells were seeded at a density of 200,000 cells per Transwell insert. The seeded inserts were then incubated overnight at 37 °C in a 6% CO2 incubator to establish a feeder layer. Intestinal stem cells were purified by removing the feeder cells using the QuadroMACS Starting Kit (LS) (Miltenyi Biotec, Germany). Purified stem cells were seeded at a density of 200,000–300,000 cells per Transwell insert and cultured in expansion media for approximately 7 days, allowing the formation of a confluent monolayer. At this stage, the medium from the apical compartment of the Transwell insert was carefully removed, and the medium was replaced with expansion media devoid of nicotinamide. The cells were maintained in this differentiation medium for an additional 7–14 days to enable proper differentiation of the stem cells prior to analysis.
Immunofluorescence staining
Normal intestinal epithelial stem cells colonies were fixed with 4% formaldehyde (Thermo Fisher Scientific, USA), permeabilized with 0.3% Triton X-100 for 15 min at room temperature, followed by blocking with 3% BSA + 0.3% Triton™ X-100 in PBS (blocking buffer) for 30 min at room temperature. Primary antibodies (listed in Supplementary Table S4) were diluted in blocking buffer and added for overnight incubation at 4 °C. The next day cells were washed 3 times with wash buffer (0.03% Triton X-100 in PBS). Then secondary antibodies diluted 1:1000 in blocking buffer — Anti-Rabbit IgG F(ab’)2 Fragment conjugated to Alexa Fluor 488 (Cell Signaling Technology Cat. No. 4412S) or Goat anti-Mouse IgG conjugated to Alexa Fluor 546 (Invitrogen Cat. No. A-11003)— were added and incubated for 2 h at room temperature in the darkness. Next, the cells were washed 3 times with PBS and the nuclei were stained with 0.2 µg/mL DAPI dye (Thermo Fisher Scientific, USA).
For immunofluorescence staining of tissues, paraffin-embedded tissues were dewaxed and hydrated, and then the antigen was unmasked with 0.01 mol/L citric acid antigen retrieval solution. These slides were blocked with 1% BSA at room temperature for 1 h, and the subsequent antibody incubation steps were carried out to complete the immunofluorescent experiment. The fluorescence microscope images were captured by Nikon Eclipse Ti2-E inverted fluorescence microscope.
Nucleofection
Normal intestinal epithelial stem cells were edited in a stepwise manner using a ribonucleoprotein (RNP) delivery approach with Lonza 4D-Nucleofector X system. For each nucleofection reaction, 2 × 10^5 primary epithelial stem cells were resuspended in P3 Primary Cell Solution (Lonza) supplemented with 140 pmol sgRNAs (Synthego), 115 pmol Alt-R Cas9 Nuclease V3 and 110 pmol Alt-R Cas9 Electroporation Enhancer (Integrated DNA Technologies Inc.). For homology-directed repair (APC and KRAS mutations), reaction was additionally supplemented with 110 pmol Alt-R HDR donor oligos. Cells were electroporated using the CM-138 program. After nucleofection, cells were immediately transferred to pre-warmed complete culture medium and cultured on feeder cells monolayer in 37 °C, 6% CO2 incubator. Homology-directed repair was encouraged with 1 µM Alt-R HDR Enhancer V2 for 24h. After reaching 70–80% of confluency, cells were passaged into selective culture conditions. Single-cell cloning followed by targeted PCR amplification and Sanger sequencing and/or western blot for specific mutations was performed to identify cells with successful recombination. Details regarding sgRNAs, HDR donor oligos and PCR primer sequences along with selective culture conditions for each nucleofection reactions are available in Supplementary Table S5.
Sanger sequencing
Targeted exons of the selected genes, including APC (exon 15), KRAS (exon 2), TP53 and SMAD4, were first amplified by PCR. Sequencing was performed using BigDye Terminator v3.1 kit Applied Biosystems (Life Technologies). Purified reaction products were separated by electrophoresis on the 3730xl DNA Analyzer according to the manufacturer’s references.
Western blotting
From intestine-feeders cell co-culture, feeders were removed by differential trypsinization, and medium was replaced for fresh media with DMSO or drugs. After indicated time period cells were collected by scrubbing, washed with cold PBS, and total proteins were extracted using RIPA buffer containing protease inhibitors and phosphatase inhibitors. Protein concentration was measured with a DC protein assay (BioRad). For SDS-PAGE, 40 ug of total protein in the Laemmli sample buffer was loaded onto Criterion TGX Stain-Free 4–15% gels (BioRad) and electrophoresed. Proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Trans-Blot Turbo Transfer System). After blocking with 5% non-fat dry milk/TBS-T or 5% BSA/TBS-T, membranes were incubated with specific primary antibodies at 4 °C overnight. After washing in TBS-T, membranes were incubated for 60 min with appropriate secondary peroxidase-conjugated IgG. The immunoreactive proteins were detected using a Clarity Western ECL substrate (BioRad) and visualized with ChemiDoc MP Imaging System. All images were processed with Image Lab version 6.0.0 (BioRad). Densitometry was performed using Image Lab software. The results on the graphs show the ratio of the normalized adjusted volume of target sample to the normalized adjusted volume of Stain Free Gel. The antibodies used in this study were listed on Supplementary Table S6.
Whole exome sequencing data processing
Whole exome sequencing (WES) was performed using the Agilent SureSelect Human All Exon V6 kit for exon capture, and sequencing was conducted by Novogene. Data analysis was carried out with the RYVU in-house pipeline, which was built using Nextflow to ensure scalability, reproducibility, and ease of use. This pipeline integrated several computational tools and algorithms to ensure accurate and comprehensive results.
First, mouse reads were removed using Xengsort. Preprocessing of raw sequencing data was performed with FastP to trim adapters and filter low-quality reads34, and quality control was assessed using FastQC35. Reads were aligned to the human reference genome (hg19) using BWA-MEM. PCR duplicates were marked using the MarkDuplicates tool from the Genome Analysis Toolkit (GATK)36. Hybrid capture performance metrics were evaluated with GATK CollectHsMetrics37, while additional mapping-related metrics were generated using Samtools38. Somatic variant calling was performed by Strelka239 for small variants and short indels, whereas Manta was used to detect structural variants40. Sequencing data used in this study have been deposited in the European Nucleotide Archive with the primary accession code: PRJEB87257.
RNA sequencing and data processing
RNA was extracted using the RNeasy Mini Kit (Qiagen). Total RNA was subjected to library preparation and NGS sequencing at Novogene. To minimize batch effects and ensure homogeneous processing, raw sequencing data from our in-house models, along with external datasets (Supplementary Table S2), excluding TCGA and CCLE data, were processed using the RYVU in-house RNA-Seq pipeline. This pipeline, implemented in Nextflow41 and executed via AWS Batch, enabled efficient cloud-based data processing. Briefly, Xengsort42 was employed to filter out mouse reads in xenograft samples, FASTP34 was used for preprocessing and quality control, STAR43 was utilized for read alignment, and quantification was performed using Slamon44. All necessary references were constructed based on the GRCh38 assembly and gene annotations from GENCODE (v43). Sequencing data used in this study have been deposited in the European Nucleotide Archive with the primary accession code: PRJEB87257.
Transcriptomic-based molecular classification
To classify samples into molecular subtypes, we applied three distinct approaches. First, we utilized the CMS caller13. However, this method failed to assign molecular subtypes to approximately 10% of the samples. To address this limitation, we reimplemented and retrained DeepCC14, a deep learning-based framework for cancer molecular subtype classification. This shift enabled us to assign consensus molecular subtypes to the vast majority of samples. Additionally, we implemented a pathway-based molecular classification system through the PDS classifier45. These pathway-focused methods provide deeper insights into the transcriptomic landscape and enhance our understanding of molecular subtypes. Assigned classes for all 3 classifiers are shown on Supplementary Fig. 3.
MOBER downloading and preprocessing/data harmonization and normalization
The RNA sequencing data from different sources described in Supplementary Table S2 were downloaded and preprocessed in-house by the RNA-seq processing pipeline. To evaluate transcriptomic similarities between RYVU CRC models and the broader spectrum of CRC samples under various conditions, we employed the Multi-Origin Batch Effect Remover (MOBER) model12. MOBER is a deep learning model composed of two types of neural networks: a conditional variational autoencoder, which creates a joint embedding of expression data from different sources, and a source discriminator neural network trained in an adversarial fashion, which accounts for batch effect removal between samples.
The MOBER model was trained on n = 1657 samples using log-transformed Transcripts Per Million (TPM) values. The resulting MOBER-derived embeddings were leveraged for downstream analysis, including sample clustering. Using the Leiden clustering algorithm46 we identified 12 subclusters in the latent space of gene expression data, with RYVU CRC models predominantly grouped in two subclusters (clusters 0 and 12, see Supplementary Fig. S5a, Supplementary Fig. S5b). These subclusters were located close to each other in the UMAP embedding of the MOBER latent space and exhibited similar transcriptomic profiles. For further investigation of transcriptomic differences between RYVU and public CRC models, the two RYVU subclusters were merged into a single cluster. This cluster was contrasted against the remaining CRC models, effectively defining two primary clusters (Supplementary Fig. S5c). The results are visualized in Fig. 2b.
Identification of differential gene programs using ExpiMap model
To investigate transcriptomic differences at the gene set level between the two clusters, we employed the explainable programmable mapper (ExpiMap) model47. ExpiMap is an interpretable conditional variational autoencoder (CVAE) trained on predefined gene programs (GPs)—sets of genes associated with specific pathways, curated from literature, databases, or domain knowledge. The ExpiMap model generates interpretable embeddings while focusing on relevant GPs. It enables the identification of differential gene programs between groups through hypothesis testing in the integrated latent space. Using the hypothesis testing option, the most differential gene programs were selected based on the Bayes Factor, representing the ratio between the two hypothesis probabilities, and visualized on Fig. 2c.
Gene set correlation enrichment analysis
Differential gene expression (DGE) analysis was conducted to identify genes differentially expressed between these two groups. Transcriptomic data preprocessing was performed using the nf-core RNA-seq pipeline48, followed by differential expression analysis with the nf-core differential abundance pipeline [59], both executed using the recommended default parameters. Gene set correlation enrichment analysis was then carried out using GscorePy49 (commit 913ee5e, retrieved from GitHub). The analysis primarily focused on gene sets previously defined by Expimap, which enabled assessment of coordinated expression changes at the pathway level between responder and non-responder groups.
Edu assay/ cell proliferation assay
Proliferation of cells was assessed with Click-iT Plus EdU Cell Proliferation Kit for Imaging, Alexa Fluor 488 dye (Invitrogen) according to manufacturer’s protocol. Briefly, the cells were cultured in 384-well plates in the presence of tested compound or proper control for 6 days. 24 h before the end of experiment, the modified thymidine analogue EdU (5-ethynyl-2′-deoxyuridine) was added to the medium. Then, the cells were fixed, permeabilized and incubated with Click-iT Plus reaction cocktail containing Alexa Fluor 488 picolyl azide for 30 min. Subsequently, the nuclei were stained with 0.2 µg/mL DAPI dye (Thermo Fisher Scientific). High-throughput imaging was performed using a Nikon Eclipse Ti2-E inverted fluorescence microscope equipped with a motorized stage and NIS-Elements AR software (version 5.42.06). Screening plates were imaged using a 4 × air objective with automated perfect focus system. The NIS-Elements software enabled precise multi-well plate scanning with capturing bright-field and DAPI and FITC fluorescence images simultaneously. Bright spot detection on FITC channel was applied for Edu-stained cell count.
Cell proliferation assay data analysis
Dose–response curves were analyzed by importing viability data into GraphPad Prism and fitting it to the four-parameter logistic equation.
High-throughput screening (HTS) data were analyzed using a proprietary, internally developed Python-based tool. The software calculates percent inhibition for single-point assays and employs the four-parameter logistic (4PL) regression model for dose–response experiments. To integrate EC50 and efficacy data derived from the 4PL model, the area under the curve (AUC) was calculated. The AUC was determined using a Python script based on the smallest trapezoid method.
For combination studies, viability data were uploaded to the SynergyFinder Plus web application50, and the values for the Loewe synergy model were used. The best drug combination was selected based on the mean Loewe synergy score for the drug-response matrix (Loewe Synergy Index). Data visualization was conducted using GraphPad Prism, while the selected drug combination and heatmaps were visualized using a Python 3.10 script utilizing the Matplotlib 3.82, Plotly 5.21.0 libraries51,52 and Morpheus application from Broad Institute53.
Machine learning segmentation model
DAPI and FITC images were acquired and segmented using Cellpose54 with pre-trained weights. To predict FITC segmentation from DAPI input alone, we trained a U-Net model55 following a standard architecture as in56. Training input consisted of random 256-pixel crops of DAPI images augmented with random rotations and flips. Pixel intensities were standardized to negative control wells. Predicted masks were binarized using Otsu thresholding (the minimum set to 10% of the intensity range) and top-hat filtered for noise reduction. The model was trained on data from 33 plates (3 replicates of 11 compound sets) of AKTS (KO) cells and evaluated on unseen KO and WT cells. FITC cell counts, obtained using NIS-Elements Nikon software (REF), were correlated with predicted FITC segmentation areas to assess accuracy. A learning curve analysis determined that training on positive and negative control wells from a single plate was sufficient for accurate KO predictions, and adding a small amount of control data preserved performance for WT cells.
Data availability
The datasets generated and/or analyzed during the current study are available in the European Nucleotide Archive(ENA) repository, https://www.ebi.ac.uk/ena/browser/view/PRJEB87257 and with the primary accession PRJEB87257.
References
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics. CA Cancer J. Clin/ 71(2021), 7–33. https://doi.org/10.3322/CAAC.21654 (2021).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/CAAC.21660 (2021).
Hong, Y., Kim, J., Choi, Y. J. & Kang, J. G. Clinical study of colorectal cancer operation: survival analysis. Korean J. Clin. Oncol. 16, 3–8. https://doi.org/10.14216/KJCO.20002 (2020).
Vogelstein, B. et al. Cancer genome landscapes. Science 340(2013), 1546–1558. https://doi.org/10.1126/SCIENCE.1235122/SUPPL_FILE/VOGELSTEIN.SM.COVER.PAGE.PDF (1979).
Guinney, J. et al. The consensus molecular subtypes of colorectal cancer. Nat. Med. 21(11), 1350–1356. https://doi.org/10.1038/nm.3967 (2015).
Grady, W. M. & Carethers, J. M. Genomic and epigenetic instability in colorectal cancer pathogenesis. Gastroenterology 135, 1079–1099. https://doi.org/10.1053/J.GASTRO.2008.07.076 (2008).
Li, Q. et al. Signaling pathways involved in colorectal cancer: pathogenesis and targeted therapy. Signal Transduct. Target. Ther. 9(1), 1–48. https://doi.org/10.1038/s41392-024-01953-7 (2024).
Le, D. T. et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357(2017), 409–413. https://doi.org/10.1126/SCIENCE.AAN6733/SUPPL_FILE/AAN6733_TABLES-S8-S9-S10.XLSX (1979).
Wang, C. et al. Wild-type APC is associated with poor survival in metastatic microsatellite stable colorectal cancer. Oncologist 26, 208–214. https://doi.org/10.1002/ONCO.13607 (2021).
Arnold, D. et al. Prognostic and predictive value of primary tumour side in patients with RAS wild-type metastatic colorectal cancer treated with chemotherapy and EGFR directed antibodies in six randomized trials. Ann. Oncol. 28, 1713–1729. https://doi.org/10.1093/ANNONC/MDX175 (2017).
Drost, J. & Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 18(7), 407–418. https://doi.org/10.1038/s41568-018-0007-6 (2018).
S. Dimitrieva, R. Janssens, G. Li, A. Szalata, R. Gopal, C. Parmar, A. Kauffmann, E.Y. Durand, Biologically relevant integration of transcriptomics profiles from cancer cell lines, patient-derived xenografts and clinical tumors using deep learning, BioRxiv (2022) 2022.09.07.506964. https://doi.org/10.1101/2022.09.07.506964.
Eide, P. W., Bruun, J., Lothe, R. A. & Sveen, A. CMScaller: an R package for consensus molecular subtyping of colorectal cancer pre-clinical models. Sci. Rep. 17(1), 1–8. https://doi.org/10.1038/s41598-017-16747-x (2017).
Gao, F. et al. DeepCC: a novel deep learning-based framework for cancer molecular subtype classification. Oncogenesis 8(9), 1–12. https://doi.org/10.1038/s41389-019-0157-8 (2019).
Sabates-Bellver, J. et al. Transcriptome profile of human colorectal adenomas. Mol. Cancer Res. 5, 1263–1275. https://doi.org/10.1158/1541-7786.MCR-07-0267 (2007).
Kingston, B. et al. Exceptional response to AKT inhibition in patients with breast cancer and germline PTEN mutations. JCO Precis Oncol https://doi.org/10.1200/PO.19.00130 (2019).
Nielsen, D. L., Palshof, J. A., Larsen, F. O., Jensen, B. V. & Pfeiffer, P. A systematic review of salvage therapy to patients with metastatic colorectal cancer previously treated with fluorouracil, oxaliplatin and irinotecan +/− targeted therapy. Cancer Treat Rev 40, 701–715. https://doi.org/10.1016/J.CTRV.2014.02.006 (2014).
Amodio, V. et al. Egfr blockade reverts resistance to krasg12c inhibition in colorectal cancer. Cancer Discov 10, 1129–1139. https://doi.org/10.1158/2159-8290.CD-20-0187/333482/AM/EGFR-BLOCKADE-REVERTS-RESISTANCE-TO-KRAS-G12C (2020).
Brabletz, T. et al. Variable β-catenin expression in colorectal cancers indicates tumor progression driven by the tumor environment. Proc Natl Acad Sci U S A 98, 10356–10361. https://doi.org/10.1073/PNAS.171610498/ASSET/01EAA5E9-0913-4AF7-942E-10A5B771806F/ASSETS/GRAPHIC/PQ1716104005.JPEG (2001).
O’Brien, E. et al. Pharmacological EZH2 inhibition combined with retinoic acid treatment promotes differentiation and apoptosis in rhabdomyosarcoma cells, Clin. Clin. Epigenet 15, 1–16. https://doi.org/10.1186/S13148-023-01583-W/FIGURES/5 (2023).
Sabour-Takanlou, M., Sabour-Takanlou, L. & Biray-Avci, C. EZH2-associated tumor malignancy: a prominent target for cancer treatment. Clin. Genet. 106, 377–385. https://doi.org/10.1111/CGE.14576 (2024).
Fricke, S. L. et al. MTORC1/2 inhibition as a therapeutic strategy for PIK3CA mutant cancers. Mol Cancer Ther. 18, 346–355. https://doi.org/10.1158/1535-7163.MCT-18-0510/87725/AM/MTORC1-2-INHIBITION-AS-A-THERAPEUTIC-STRATEGY-FOR (2019).
Tamburini, J. et al. Mammalian target of rapamycin (mTOR) inhibition activates phosphatidylinositol 3-kinase/Akt by up-regulating insulin-like growth factor-1 receptor signaling in acute myeloid leukemia: rationale for therapeutic inhibition of both pathways. Blood 111, 379–382. https://doi.org/10.1182/BLOOD-2007-03-080796 (2008).
Mateo, J. et al. A first in man, dose-finding study of the mTORC1/mTORC2 inhibitor OSI-027 in patients with advanced solid malignancies. Br. J. Cancer 114(8), 889–896. https://doi.org/10.1038/bjc.2016.59 (2016).
Chandarlapaty, S. et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell 19, 58–71. https://doi.org/10.1016/J.CCR.2010.10.031 (2011).
Bahrami, A. et al. Therapeutic potential of targeting PI3K/AKT pathway in treatment of colorectal cancer: rational and progress. J Cell Biochem 119, 2460–2469. https://doi.org/10.1002/JCB.25950 (2018).
Soleimani, A. et al. Role of regulatory oncogenic or tumor suppressor miRNAs of PI3K/AKT signaling axis in the pathogenesis of colorectal cancer. Curr Pharm Des 24, 4605–4610. https://doi.org/10.2174/1381612825666190110151957 (2019).
Clevers, H. & Nusse, R. Wnt/β-catenin signaling and disease. Cell 149, 1192–1205. https://doi.org/10.1016/J.CELL.2012.05.012 (2012).
McDaid, W. J. et al. The PI3K-AKT-mTOR axis persists as a therapeutic dependency in KRASG12D-driven non-small cell lung cancer. Mol. Cancer 23, 1–18. https://doi.org/10.1186/S12943-024-02157-X (2024).
Shimizu, T. et al. The clinical effect of the dual-targeting strategy involving PI3K/AKT/mTOR and RAS/MEK/ERK pathways in patients with advanced cancer. Clin. Cancer Res. 18, 2316–2325. https://doi.org/10.1158/1078-0432.CCR-11-2381/84708/AM/THE-CLINICAL-EFFECT-OF-THE-DUAL-TARGETING-STRATEGY (2012).
Belmont, P. J. et al. Resistance to dual blockade of the kinases PI3K and mTOR in KRAS-mutant colorectal cancer models results in combined sensitivity to inhibition of the receptor tyrosine kinase EGFR. Sci. Signal https://doi.org/10.1126/SCISIGNAL.2005516/SUPPL_FILE/7_RA107_TABLE_S1.ZIP (2014).
Burgenske, D. et al. Abstract B50: targeting KRASG12D colorectal cancer with combined inhibition of PI3K/mTOR and MAPK signaling. Mol. Cancer Ther. 14, B50–B50. https://doi.org/10.1158/1538-8514.PI3K14-B50 (2015).
Zhao, Y. et al. Colorectal cancer patient-derived 2D and 3D models efficiently recapitulate inter- and intratumoral heterogeneity. Adv. Sci. 9, 2201539. https://doi.org/10.1002/ADVS.202201539 (2022).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. https://doi.org/10.1093/BIOINFORMATICS/BTY560 (2018).
Babraham Bioinformatics - FastQC a quality control tool for high throughput sequence data, (n.d.). https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed December 16 2024.
MarkDuplicates (Picard) – GATK, (n.d.). https://gatk.broadinstitute.org/hc/en-us/articles/360037052812-MarkDuplicates-Picard. Accessed December 16 2024.
CollectHsMetrics (Picard) – GATK, (n.d.). https://gatk.broadinstitute.org/hc/en-us/articles/360036856051-CollectHsMetrics-Picard. Accessed December 16, 2024.
Samtools, (n.d.). https://www.htslib.org/. Accessed December 16 2024.
Kim, S. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods 15(8), 591–594. https://doi.org/10.1038/s41592-018-0051-x (2018).
Chen, X. et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics 32, 1220–1222. https://doi.org/10.1093/BIOINFORMATICS/BTV710 (2016).
di Tommaso, P. et al. Nextflow enables reproducible computational workflows. Nat. Biotechnol. 35(4), 316–319. https://doi.org/10.1038/nbt.3820 (2017).
Zentgraf, J. & Rahmann, S. Fast lightweight accurate xenograft sorting. Algorithms Mol. Biol. 16, 1–16. https://doi.org/10.1186/S13015-021-00181-W/TABLES/6 (2021).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/BIOINFORMATICS/BTS635 (2013).
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A. & Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14(4), 417–419. https://doi.org/10.1038/nmeth.4197 (2017).
Malla, S. B. et al. Pathway level subtyping identifies a slow-cycling biological phenotype associated with poor clinical outcomes in colorectal cancer. Nat. Genet. 56(3), 458–472. https://doi.org/10.1038/s41588-024-01654-5 (2024).
Traag, V. A., Waltman, L. & van Eck, N. J. From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep. 9(1), 1–12. https://doi.org/10.1038/s41598-019-41695-z (2019).
Lotfollahi, M. et al. Biologically informed deep learning to query gene programs in single-cell atlases. Nat. Cell Biol. https://doi.org/10.1038/s41556-022-01072-x (2023).
Ewels, P. A. et al. The nf-core framework for community-curated bioinformatics pipelines. Nat. Biotechnol. 38, 276–278. https://doi.org/10.1038/S41587-020-0439-X (2020).
Chang, L. Y. et al. Gene set correlation enrichment analysis for interpreting and annotating gene expression profiles. Nucleic. Acids Res. 52, e17. https://doi.org/10.1093/NAR/GKAD1187 (2024).
Zheng, S. et al. SynergyFinder Plus: toward better interpretation and annotation of drug combination screening datasets. Genom. Proteom. Bioinform. 20, 587–596. https://doi.org/10.1016/J.GPB.2022.01.004 (2022).
Data Apps for Production | Plotly, (n.d.). https://plotly.com/. (accessed December 6, 2024).
Matplotlib — Visualization with Python, (n.d.). https://matplotlib.org/. Accessed May 28 2025.
Morpheus, (n.d.). https://software.broadinstitute.org/morpheus/#. Accessed December 6 2024.
Stringer, C., Wang, T., Michaelos, M. & Pachitariu, M. Cellpose: a generalist algorithm for cellular segmentation. Nat. Methods 18(1), 100–106. https://doi.org/10.1038/s41592-020-01018-x (2020).
Weng, W. & Zhu, X. U-Net: convolutional networks for biomedical image segmentation. IEEE Access 9, 16591–16603. https://doi.org/10.1109/ACCESS.2021.3053408 (2015).
GitHub - PARMAGroup/UNet-Instance-Cell-Segmentation: [Pytorch] This project aims to perform well at instance segmentation on the BBBC006 cells dataset. We tested UNet over several configurations including the loss function, evaluation function and the datasets., (n.d.). https://github.com/PARMAGroup/UNet-Instance-Cell-Segmentation/tree/master. Accessed December 19 2024.
Acknowledgements
The results from CRC data modelling with MOBER and ExpiMap models shown here are partially based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. Project is co-financed by the European Union under the Operational Programme European Funds for Modern Economy 2021-2027. Project title: "ONCO Prime: new possibilities for personalised anti-cancer therapy based on patient-derived primary cell cultures, omics characterisation, and functional assays". Grant Agreement no: FENG.01.01-IP.02-0095/23.
Author information
Authors and Affiliations
Contributions
MD, AM, and EZ designed the study and wrote the manuscript with input from all authors. MS, JSz, MC, IW, BL, AS, AK, KS, JSzk, JD, ASW, KP and OD planned and executed the experiments. Data analysis was performed by KBa, OB and JK. Machine learning work was conducted by MCh and KW, with support from MOand KR. MM, PF, and RD assisted with the collection of clinical samples from hospitals. KB provided HTS figures. RD, TR, and AT contributed to the study design. KB, MD, and AM supervised the project.
Corresponding authors
Ethics declarations
Competing interests
MD, EZ, JS, MS, JS, OD, AK, IW, MC, BL, KS, JK, OB, ASW, KP, JD, KB, TR, AT, KB, AM are/were employees of Ryvu Therapeutics and some of the authors are shareholders in Ryvu Therapeutics. The other authors have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Duleba, M., Zimoląg, E., Szuszkiewicz, J. et al. Integrated transcriptomic and functional modeling reveals AKT and mTOR synergy in colorectal cancer. Sci Rep 15, 26643 (2025). https://doi.org/10.1038/s41598-025-08649-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-08649-0