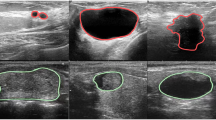
Abstract
Ultrasound guidance images are widely used for high intensity focused ultrasound (HIFU) therapy; however, the speckles, acoustic shadows, and signal attenuation in ultrasound guidance images hinder the observation of the images by radiologists and make segmentation of ultrasound guidance images more difficult. To address these issues, we proposed the superpixel based attention network, a network integrating superpixels and self-attention mechanisms that can automatically segment tumor regions in ultrasound guidance images. The method is implemented based on the framework of region splitting and merging. The ultrasound guidance image is first over-segmented into superpixels, then features within the superpixels are extracted and encoded into superpixel feature matrices with the uniform size. The network takes superpixel feature matrices and their positional information as input, and classifies superpixels using self-attention modules and convolutional layers. Finally, the superpixels are merged based on the classification results to obtain the tumor region, achieving automatic tumor region segmentation. The method was applied to a local dataset consisting of 140 ultrasound guidance images from uterine fibroid HIFU therapy. The performance of the proposed method was quantitatively evaluated by comparing the segmentation results with those of the pixel-wise segmentation networks. The proposed method achieved 75.95% and 7.34% in mean intersection over union (IoU) and mean normalized Hausdorff distance (NormHD). In comparison to the segmentation transformer (SETR), this represents an improvement in performance by 5.52% for IoU and 1.49% for NormHD. Paired t-tests were conducted to evaluate the significant difference in IoU and NormHD between the proposed method and the comparison methods. All p-values of the paired t-tests were found to be less than 0.05. The analysis of evaluation metrics and segmentation results indicates that the proposed method performs better than existing pixel-wise segmentation networks in segmenting the tumor region on ultrasound guidance images.
Similar content being viewed by others
Introduction
High intensity focused ultrasound (HIFU) is a valid image-guided tumor treatment method with non-invasive features that greatly reduces the suffering of patients1,2,3,4. Image guidance plays an important role in HIFU therapy as it assists in integrated therapy planning, real-time control, and evaluation. Currently, HIFU therapy is carried out using either ultrasound or magnetic resonance imaging guidance5. Ultrasound guidance is widely used in HIFU therapy for real-time monitoring of tumor or organ peristalsis because of its flexibility and low cost, and the obtained ultrasound guidance images need to be accurately and rapidly segmented for tumor ablation. During the ablation of a tumor, if the tumor region cannot be accurately located, the tumor tissue may be ablation while surrounding normal tissues may be injured, causing unnecessary harm to the patient6. If the tumor region always needs to be manually segmented and located, the process becomes tedious and greatly reduces treatment efficiency. It is necessary to propose a computer-assisted segmentation method in HIFU therapy to reduce the burden of radiologists and improve the efficiency of treatment.
In recent years, various ultrasound image segmentation methods have emerged, demonstrating excellent performance in segmenting organs and tissues. Traditional ultrasound image segmentation methods can be broadly divided into three categories, edge-based, region-based and those combined with specific theoretical models7,8. The edge-based segmentation method9,10 contains two basic steps: the first step is to identify the edge pixels in the ultrasound image with the image features, and the second step is to connect edge pixels to achieve image segmentation. Edge-based segmentation methods are sensitive to image noise and local features because of the various filters employed in identifying edge pixels. Therefore, edge-based segmentation methods are often used as the initial contours of other automated segmentation algorithms or in combination with other methods. The method of combining specific theoretical models of segmentation is represented by the active contour model11,12, which converges the initial contour to the target contour by minimizing the energy function. The active contour model requires a proper initial contour to ensure performance, and it is usually difficult to converge to the global optimum if the initial contour is far from the true contour13,14. Unlike organs or tissues with fixed shapes, it is challenging to obtain prior shape information for tumors, making it difficult to constrain the segmentation process. Region-based segmentation methods15,16,17,18,19,20 usually over-segment ultrasound images into several sub-regions such as superpixels, combine sub-regions of the same type into a region, and select tumor regions from these regions. A superpixel is a collection of pixels that share similar features and are grouped together, allowing for the extraction of specific features to represent the superpixel. By clustering or classifying the superpixel based on these features, segmentation can be achieved. These traditional methods can achieve satisfactory segmentation of ultrasound guidance images as long as effective features are selected beforehand. Therefore, there is still a need for an automatic feature extraction method with less manual intervention to segment ultrasound guidance images.
Nowadays, some machine learning methods, such as convolutional neural networks (CNN)21,22,23,24,25, have achieved favorable results in medical image segmentation and pioneered new paths for the automatic segmentation of ultrasound images. However, pixel-wise segmentation networks like U-Net26, do not perform as well in the segmentation of ultrasound guidance images from small datasets compared to other medical images, such as computed tomography (CT) scans and magnetic resonance imaging (MRI) scans. This is due to the strong noise interference in ultrasound guidance images, which can introduce randomness in pixel values. The lower quality and signal-to-noise ratio of ultrasound guidance images compared to general ultrasound images are attributed to the mode in which the transducer is mounted during imaging16. Convolution is widely used in common pixel-wise segmentation methods, where features extracted from a fixed-size rectangular box (referred to as the receptive field) centered on the pixel are typically used to determine the pixel’s category26,27,28. Near the boundary, both target and background regions are typically included within the receptive field of a pixel. In low signal-to-noise ratio ultrasound guidance images, this lack of specificity in boundary pixel features is exacerbated by strong noise, further blurring the distinctions between them and resulting in a decline in segmentation performance. This poses a challenge for implementing machine learning-based segmentation on ultrasound guidance images. To enhance segmentation performance on ultrasound guidance images, methods that integrate traditional methods with machine learning have been developed. Ni et al.29 proposed a method combining a deformable contour model and a neural network for segmenting ultrasound guidance images. It exploits deep neural networks to train contrastive loss as a boundary search metric for deformable contours to segment ultrasound image sequences. However, this method relies on a shape prior for the similarity of the lesion region in a sequence of images. The performance and computational efficiency of this method are affected by the initialization of contours, resulting in a computation time that is 2.5 times longer than that of Attention U-Net27.
A superpixel is a homogeneous subregion of an image, composed of a group of pixels with similar features such as position, color, brightness, and texture. Performing feature extraction and analysis with superpixels rather than individual pixels can effectively resist noise interference30,31. By using superpixels as primitives in subsequent steps, the computational cost is reduced and the method’s speed is improved. A well-performed superpixel segmentation method can result in superpixel boundaries that closely approximate the true contours of target regions32. Due to the heterogeneity of the tumor, the complete tumor region will be over segmented into multiple superpixels. After superpixel segmentation, both the tumor region and the background region are over segmented into multiple superpixels. The features extracted from the background superpixels and target superpixels can accurately represent the differences in the characteristics of different regions. It is promising to distinguish superpixels into tumor or background, effectively achieving ultrasound guidance image segmentation.
The traditional image segmentation methods based on superpixel include extracting features from superpixels and implementing feature-based classification or clustering to complete the final target region segmentation30,33,34. Due to the potential presence of diverse background regions in images, such as other organs, clustering methods typically aggregate background superpixels into multiple regions. Additional processing is required to extract the tumor region from these clustered result regions. In contrast, classification methods can be employed to distinguish superpixels as either tumor or background based on the presence or absence of features specific to the tumor region. Artificial neural networks can automatically extract effective features from images and perform classification tasks through machine learning without human intervention. However, superpixels’ shapes and sizes are irregular, posing a challenge on how to enable networks to effectively extract features from these irregular superpixels. To solve this problem, patches rather than superpixels are used as the basic input unit for neural network35,36,37. One method is to select one or more patches from the superpixel and put them into the network for classification36. In this method, if the selected patch is relatively small, the features to describe the superpixel cannot represent the superpixel well; if the selected patch is relatively large, there may be a large number of very small superpixels, which require manual intervention for post-processing if they are discarded; and if they are not discarded and used for classification, a huge number of pixels that do not belong to the superpixel are extracted for features, which may lead to unreliable features. Another method is to divide the image into patches, classify the patches, and then perform boundary correction with superpixels35,37. They only use superpixels as a post-processing method to correct the boundaries, which requires manual intervention. There are few studies that use superpixels as basic input units in neural networks for classification and then merge the superpixels based on these classification labels to achieve medical image segmentation. This is due to the difficulty of extracting information from irregular superpixels and generating feature matrices of uniform size.
Moreover, segmentation through superpixel classification still faces a challenge: classification solely based on features extracted from individual superpixels does not utilize the prior knowledge that tumor regions and background regions are respectively connected, leading to a loss of information from adjacent superpixels. Even with a low error rate, incorrectly classified superpixels may occur in any part of the background or target region, potentially causing serious errors in the final segmentation result. The transformer introduces the self-attention mechanism38 and can use positional information to represent relationships between words. It has achieved great success in natural language processing and has also been successfully applied to computer vision39,40.
To address the problems mentioned above, this study proposes a machine learning network incorporating self-attention based on superpixel classification for the segmentation of ultrasound guidance images. The proposed method addresses the challenge of delivering complete and accurate information with irregularly shaped superpixel to the neural network. Subsequently, a superpixel classification network based on a self-attention mechanism is proposed, which utilizes the mutual aggregation of tumor superpixels to enhance classification accuracy, thereby providing improved segmentation results. The three main contributions of this study are as follows.
-
1.
proposed a method for encoding superpixel information based on texture primitives, which can extract texture features of superpixels with less manual intervention, and designed algorithms to convert superpixel feature matrices with different sizes into the uniform size by dimensionality transformation.
-
2.
proposed a neural network based on self-attention mechanism for the classification of superpixel matrices to achieve tumor segmentation. The network aggregates all superpixel feature matrices for learning and can make full use of the category information of neighboring superpixels in classification to ensure the connectivity of the output tumor region, which solves the problem of random position distribution of tumor labeled superpixels when superpixel is classified individually.
-
3.
The quantitative analysis of the seven metrics was performed to compare the proposed method with several other pixel-wise segmentation methods. The analysis shows that the segmentation results of the method are closer to the ground truth segmentation.
Materials and methods
Datasets
This work includes 140 uterine fibroid ultrasound guidance images of different patients from Model JC200 HIFU systems (Chongqing Haifu Tech Co., Ltd, Chongqing, China) in the First Affiliated Hospital of Chongqing Medical University (Chongqing, China). The guiding transducer (CA430, Esaote, Italy) is fixed to the therapeutic transducer and its center frequency is 3.5 MHz. All data collection complies with the Helsinki Declaration. The study received approval from the Medical Ethics Committee of the First Affiliated Hospital of Chongqing Medical University. Informed consent for the use of all data has been obtained from the patients participating in the study. The original image collected by the HIFU system is 768 × 576 pixels. Because the ultrasound scan area was shaped like a sector rather than a rectangle, the radiologists identified the region of interest (ROI) and corresponding tumor contours using only the imaging data and tumor size without using any of the patient’s personal information. The grayscale of each image is between 0 and 255. The radiologist defined the tumor contour, which was applied as a reference contour in the quantitative evaluation of segmentation results.
There are 140 ultrasound guidance images of uterine fibroids in our dataset, of which 80 images are the training set, 20 are the validation set, and 40 are the test set. To improve the generalization ability and the robustness of the network, data augmentation was applied to the training set and validation set. Specifically, the original images are processed by applying horizontal flipping, vertical flipping, and rotation. These methods expand the dataset to 8 times its original size, providing more samples for the network. Data augmentation techniques41 based on basic image manipulations include geometric transformations, flipping, rotation, translation, color space transformations, noise injection, etc. It has been demonstrated that ultrasound images inevitably feature severe speckle noise, which is neither plain gaussian noise nor multiplicative noise42,43,44, and contain abundant texture features. Therefore, data augmentation is performed with only affine transformation or cropping of the images in order to avoid losing the detailed texture features in the ultrasound guidance images.
Method overview
The flowchart of the method proposed for ultrasound guidance image segmentation is shown in Fig. 1, which is based on superpixel segmentation, classification, and merging. The HIFU image is first over-segmented into superpixels using the iterative multiple region growth (IMRG)17 algorithm. The image is then encoded into a superpixel feature matrix. The steps include extracting the features within the superpixels and converting them into a uniform matrix using dimensionality transformation and then concatenating them into a superpixel feature matrix. The superpixels are then classified using a self-attention-based network. Finally, the superpixels are merged based on the predicted superpixel categories of the classifier and postprocessed to obtain the final segmentation result. The loss function for training the classifier is calculated from the superpixel labels and predicted superpixel classes.
Generating superpixels
A modified algorithm based on IMRG17 is used to generate superpixels on ultrasound guidance images. The simple linear iterative cluster (SLIC)33 algorithm may generate numerous outliers in HIFU ultrasound guidance images with strong noise interference, necessitating forced connections that can result in an uncontrolled number of superpixels. Given that IMRG does not exhibit this issue and demonstrates superior computational efficiency, it is more suitable for generating superpixels in ultrasound guidance images.
The original IMRG algorithm has been optimized in two key aspects. Firstly, we adopted the hash algorithm to index the priority queues to improve the computing speed. Secondly, upon dequeuing and labeling a pixel, an additional step has been introduced to detect whether neighboring pixels possess a cluster label different from the current pixel’s label. When encountering a different label, the algorithm calculates the priority of the pixel and the cluster center of the neighboring pixel. If the calculated priority is lower than the current priority of the pixel, the pixel’s current label is updated to match that of the neighboring pixel. This step enhances algorithmic consistency, particularly at boundaries.
Encoding
Due to the irregular shape and variable size of superpixels, which cannot be directly used as input for a convolutional neural network requiring fixed-size rectangles, an approach to encoding superpixels is proposed to address this issue. The specific flow is shown in Fig. 2. The ultrasound guidance image is initially over-segmented into superpixels. The grayscale values of neighboring regions around each valid pixel within the superpixel are extracted and arranged into a column matrix to represent that pixel. These features from all valid pixels within the superpixel are concatenated to form a superpixel feature matrix describing the entire superpixel. Subsequently, the features extracted from each superpixel are compressed dimensionally and transformed to a uniform size. This approach for extracting superpixel features and encoding for normalization is detailed as follows.
Feature extraction
The grayscale values of pixels within the \(\:l\times\:l\) rectangular window centered at pixel \(\:p\) are sequentially obtained and stored in a column vector of length \(\:{l}^{2}\), which can represent the feature of pixel \(\:p\). This method can reflect the joint probability distribution of the grayscale values of a pixel and its neighboring pixels, considering spatial relationships of this pixel45. Several experiments have validated it as an effective technique for texture feature description46,47. Since each superpixel contains a large number of pixels and represents a homogeneous region, the extracted texture features exhibit better statistical stability due to the increased number of statistical samples. The proposed method does not apply filtering or other preprocessing to the image, which helps preserve the texture information embedded in the speckle noise.
Based on the method of extracting features from pixels, the column feature vectors generated by pixels within each superpixel are concatenated into a second-order structure tensor, forming a superpixel feature matrix of size \(\:{l}^{2}\times\:T\), where \(\:T\) is the number of valid pixels within the superpixel. Due to the irregular shape of superpixels and the rectangular window used for feature extraction, some pixels within the superpixel may have \(\:l\:\times\:\:l\) neighboring pixels that are mostly located outside the same superpixel. This inconsistency in neighboring pixels reduces the coherence of the features. To mitigate this issue, the feature extraction window should be positioned to remain within the boundaries of the same superpixel as much as possible. Nevertheless, if all the pixels near the superpixel boundaries are left unprocessed, it will lead to difficulty in extracting features from small superpixels. Therefore, prior to feature extraction within superpixels, the superpixel masks are dilated to achieve boundary dilation. Using a small kernel for the dilation operation helps maintain internal similarity within the superpixels.
Dimensionality transformation
The size of the superpixel feature matrix obtained from the superpixel is \(\:{l}^{2}\times\:T\). This vector can be further compressed using algorithms to reduce data volume and enhance the efficiency of feature description. In addition, the number of columns \(\:T\) in the superpixel feature matrix is the number of valid pixels in that superpixel. For different superpixels, the numbers of columns in the feature matrices are different, which makes it difficult to input these matrices into a classification neural network. A compression algorithm for the superpixel feature matrix has been designed, which not only enhances the description efficiency of the feature matrix but also converts the feature matrix to a fixed column size.
For the matrix \(\:{A}_{m\times\:n}\), the formula for the singular value decomposition is given in Eq. (1):
where \(\:{\varSigma\:}_{m\times\:n}=diag({\sigma\:}_{1},{\sigma\:}_{2}\dots\:{\sigma\:}_{p}),\:{\sigma\:}_{i}\) is the singular value of \(\:{A}_{m\times\:n}\), \(\:p=\text{m}\text{i}\text{n}(m,n)\), \(\:{\sigma\:}_{1}\ge\:{\sigma\:}_{2}\ge\:{\dots\:\ge\:\sigma\:}_{p}\). In the matrix \(\:{\varSigma\:}_{m\times\:n}\), the singular values \(\:{\sigma\:}_{i}\) are ordered from the largest to the smallest, and decrease rapidly. In many cases, the sum of the first 10% or even 1% of the singular values account for more than 99% of the sum of total singular values. Therefore, the matrix \(\:{A}_{m\times\:n}\) can be approximately restored from the first r nontrivial singular values as shown by Eq. (2), where \(\:{V}_{n\times\:r}\) and \(\:{U}_{m\times\:r}\) both denote matrices composed of the first \(\:r\) column vectors of their original matrix, \(\:{\varSigma\:}_{r\times\:r}\) denotes the diagonal matrix composed of the first \(\:r\) singular values of \(\:{\varSigma\:}_{m\times\:n}\). Projecting \(\:{A}_{m\times\:n}\) into a new eigenspace using Eq. (3) can preserve almost all of its information while transforming the column dimensions.
For simplicity, let \(\:r={l}^{2}\). In this way, the superpixel feature matrix is compressed and transformed to a fixed size of \(\:r\times\:r\).
Inevitably, some superpixels generated in superpixel segmentation are extremely small, resulting in the number of columns in these superpixel matrices being smaller than a reasonable setting of \(\:r\). If such a superpixel matrix appears, it is not possible to apply dimensionality transformation to it. Instead, the superpixel matrix is replaced with an \(\:r\times\:r\) matrix where all elements are zero. After processing, all the superpixels are concatenated in order to produce a matrix of size \(\:r\times\:r\times\:K\), where \(\:K\) is the number of superpixels. An all-1 matrix of \(\:1\times\:K\) is also generated, called the padding mask. The elements representing the small superpixels mentioned previously are set to 0, which means that these superpixels are not focused by the neural network.
Labeling superpixel
Supervised learning for superpixel classification using neural networks involves optimizing the network based on labels assigned to input data. During training and validation, we divided the superpixels into two categories: object (referring to the tumor tissue region) and background (referring to the normal tissue region), with label 0 representing the background and label 1 representing the object. Typically, segmentation results annotated by radiologists are presented as binary images, classified at the pixel-level. Therefore, when assigning labels to superpixels, the same pixel-level perspective is considered. Using the ground truth as the reference, a superpixel is labeled as ‘background’ if the number of pixels belonging to the tumor tissue region within the superpixel is less than 50% of all pixels within the superpixel; otherwise, it is labeled as ‘object ‘. This process results in generating a label \(\:1\times\:K\) matrix. To provide adjacency information of superpixels for enhancing classification performance, the centroid coordinates of each superpixel are stored in a \(\:2\times\:K\) matrix, referred to as ‘SP center coordinate’, which is used by the classifier.
Figure 3 illustrates the steps of superpixel generation, encoding, and labeling for one image, resulting in an \(\:r\times\:r\times\:K\) superpixel feature matrix, a \(\:1\times\:K\) padding mask, and a \(\:2\times\:K\) SP center coordinate, which serve as inputs to the neural network, along with a \(\:1\times\:K\) label for supervised learning.
Network architecture
In the proposed method, the superpixel feature matrix composed of all superpixels is used for classification. This approach utilizes the self-attention mechanism to address the problem of information loss from adjacent superpixels and achieves image segmentation without the need for a decoder.
The specific structure of the proposed network is shown in Fig. 4. First, trainable linear projections are applied to flatten the \(\:r\times\:r\times\:K\) superpixel feature matrix and project it onto a tensor of shape \(\:C\times\:K\). Then, it is reshaped to \(\:H\times\:W\times\:C\), where \(\:H\:\)and \(\:W\:\)are the number of seed points in horizontal and vertical directions of the image, respectively. The SP center coordinate is likewise projected and reshaped into a tensor of shape \(\:H\times\:W\times\:C\), termed as position embedding, to preserve the positional information between each superpixel in an image. The embedded superpixel is composed by adding the projection and position embedding, and is then fed into the subsequent module.
The network is composed of the MSABlock, the SP Merge block, and the prediction head. The details of MSABlock and SP Merge are shown in Fig. 5. The MSABlock consists of a normalization layer and a multi-head attention block, with a residual connect after38. The MSABlock dynamically models feature relationships between embedded superpixels through a dynamic, data-driven mechanism that enables each superpixel to interact with all others in a sequence. Multi-head attention captures diverse semantic or spatial patterns in parallel subspaces. Positional embeddings encode the spatial order of superpixels. The pairwise correlations are computed adaptively by querying the key-value matrix, followed by context-aware feature aggregation. The SP Merge block consists of a convolution layer with a kernel size of 2 and a stride of 2, and a normalization layer. The embedded superpixel is alternately processed by the MSABlock and the SP Merge block, followed by global averaging pooling. Finally, the results are fed into the prediction head, producing a final output of a \(\:1\times\:K\) tensor representing the predicted classes of each superpixel. The activation function in the linear layer is gaussian error linear units (GELU). The activation function used in the output layer of the network is sigmoid.
Merging and postprocess
To obtain the binarized segmentation result, all pixels are set as object in the superpixels identified as object by the classification result, and all pixels are set as background in the superpixels identified as background. For the previously mentioned small superpixels that are not prioritized, their labels will be assigned based on the labels of adjacent superpixels: if the adjacent superpixels are mostly labeled as object (or background), the label of the small superpixel will be set to object (or background). After assigning labels to the superpixels following the above steps and obtaining the binarized segmentation result, the jagged superpixel boundaries are smoothed using the morphological opening operation.
Experiment configuration
During the superpixel generation and encoding, hyperparameters such as the number of superpixels, weighting factor, and kernel size not only affect the size and shape of the superpixels but also relate to the effectiveness of the training and testing of the neural network. Table 1 shows the key hyperparameters utilized in the method. Experiments are performed according to the hyperparameters in the table, and eventually 256 superpixels are obtained for each image. The reason for choosing 256 superpixels is to give the superpixel a reasonable size to be able to extract features from it. After encoding, each superpixel is converted into a matrix of size 25 × 25 and a corresponding label of 0 or 1 is attached to each superpixel.
We implemented the superpixel classification network in PyTorch by training on an NVIDIA RTX 3060 GPU. The AdamW optimizer is applied to optimize our network. The epoch size and batch size of the network are set to 100 and 4, respectively. The base learning rate is set to 0.0001. During training, if the loss of the validation set of 4 epochs remains constant, the learning rate decreases to 1/10 of the previous one. To address the issue of imbalanced positive and negative samples obtained after superpixel generation, we employ a weighted cross-entropy loss function.
Evaluation metrics
The performance of the proposed method is evaluated by comparing the differences between the segmentation results of the method and the clinical technicians. In addition to direct visual observation of images for qualitative evaluation, evaluation metrics were used to quantitatively and objectively assess the quality of automatic segmentation algorithms. The metrics include region-based and distance-based evaluation metrics, both of them were chosen for evaluation in this study. The true positive rate (TPR), false positive rate (FPR) and intersection over union (IoU) belong to the region-based evaluation metrics for evaluating the difference between the automatic segmentation results and the ground truth. The formulas are shown in Eq. (4).
where \(\:{A}_{a}\) represents the predicated foreground region by the method, \(\:{A}_{m}\) represents the foreground region of ground truth, and \(\:{\complement\:}_{U}{A}_{m}\) represents the background region of ground truth.
The distance-based evaluation metrics, which is different from the region-based evaluation metrics, pay more attention to the performance of the segmentation results at the contour. The following Eqs. (5) and (6) are the formulas for two distance-based assessment metrics: Hausdorff distance (HD) and mean absolute distance (MAD).
where \(\:a\) denotes a pixel located on the contour of the predicted mask generated by the method, \(\:{C}_{a}\) represents the contour of predicated mask, and \(\:m\) denotes a pixel located on the contour of the ground truth, \(\:{C}_{m}\) represents the contour of ground truth. The distance from pixel \(\:a\) to contour \(\:{C}_{m}\) is calculated as:
where \(\:\Vert\:a-m\Vert\:\) denotes the two-dimensional Euclidean distance from pixel \(\:a\) to pixel \(\:m\). Since the tumor size in each image varies and HD and MAD are positively correlated with the length of contour, the HD and MAD should be divided by the tumor size factor for normalization when applying HD and MAD to evaluate the segmentation performance. The normalized HD and MAD are defined as Eq. (7).
Results
In this section, to demonstrate the effectiveness of the proposed method for ultrasound guidance image segmentation, the performance of the proposed method is compared with existing pixel-wise segmentation networks. Four networks were selected for the comparison experiments: U-Net26, Attention U-Net27, DeeplabV3plus28, and SETR40. The U-Net has demonstrated high performance on small datasets and has become the mainstream method for medical image segmentation among the numerous CNN-based medical image segmentation algorithms. The encoder-decoder network structure of U-Net has enlightened a huge number of segmentation methods for medical images since it was developed48. DeeplabV3plus also employs the encoder-decoder structure as its main structure and joins the atrous convolution to extend the receptive field while retaining the resolution of the feature map, which improves the performance of the network. Attention mechanisms have played an increasingly prominent role in computer vision over the past decade since they were first proposed49. Oktay et al. introduced the Attention U-Net, which adds attention mechanisms to the U-Net to improve the performance of the network. After self-attention-based transformer was proposed39, it was quickly and widely used in the field of natural language processing with remarkable success, and then the transformer was introduced into the field of computer vision. Sixiao Zeng et al. applied the transformer to semantic segmentation, proposed the SETR. The train set, validation set, and test set used in the comparison networks are consistent with those used in the proposed method, and the loss functions employed are dice loss combined with cross-entropy loss.
The boxplots of the region-based evaluation metrics are shown in Fig. 6. As shown in the figure, the proposed method outperforms the others in terms of TPR, FPR, and IoU. The shorter length of the boxes suggests a more concentrated data distribution for the proposed method. The Table 2 lists the mean values and standard deviations of region-based evaluation metrics for these methods. The proposed method achieved a mean TPR of 89.86% with a standard deviation of 8.84%, a mean FPR of 4.53% with a standard deviation of 4.21%, and the mean IoU of 75.95%, which is an improvement of 5.52% over SETR, with a standard deviation of 11.24%. Furthermore, the paired t-test was employed to demonstrate that the proposed method improves the IoU. The results of the test comparing the proposed method with the other methods are shown in Table 3, with four p-values listed. In addition, in order to quantify the improvement of the proposed method compared to other methods, the Cohen’s d50 for assessing the effect sizes is calculated as shown in Table 3. From the table, it can be inferred that the proposed method outperforms the other comparative models in terms of IoU at a significance level of 0.01. Additionally, a medium improvement in IoU is achieved compared to SETR.
The boxplots of the distance-based evaluation metrics are shown in Fig. 7. As shown in the figure, the proposed method outperforms the others in terms of HD, NormHD, MAD, and NormMAD. The Table 4 lists the mean values and standard deviations of distance-based evaluation metrics for these methods. The proposed method also performs well, achieving an average HD of 27.39 pixels, an average NormHD of 7.34%, an average MAD of 8.03 pixels, and an average NormMAD of 2.17%. The paired t-test was also employed to demonstrate that the proposed method improves the NormHD. The results of the paired t-test comparing the proposed method with the other methods are presented in Table 5. The Cohen’s d is also shown in Table 5. At a significance level of 0.05, the proposed method was found to outperform the other comparison models in NormHD. Moreover, a small improvement in NormHD is achieved by the proposed method compared to SETR.
The segmentation results of these methods for the same image are shown in Fig. 8. As shown in the figure, on images with low noise, all methods give satisfactory segmentation results. However, on images with strong noise and unclear tumor boundaries, the segmentation results of the other methods decrease substantially, whereas the proposed method exhibits a smaller decline and maintains high segmentation accuracy, demonstrating stronger robustness.
Discussion
It is a challenge to achieve the automatic segmentation of ultrasound guidance images under strong noise interference. In this study, we successfully implemented ultrasound guidance image segmentation under strong noise interference based on superpixel classification. This study proposed a superpixel feature extraction method based on texture primitives, and a dimensionality transformation method to convert a coding matrix of variable size into a superpixel feature matrix of fixed size, which solves the problem that superpixels are of variable size and shape and cannot be inputted into the neural network for processing. And a superpixel classification network based on the self-attention mechanism was proposed, which solves the problem of holes in continuous tumor and background regions due to the lack of positional information when individual superpixels are directly classified by introducing positional information and self-attention.
The effectiveness of the proposed method is demonstrated through experimental comparison with other pixel-wise segmentation networks on uterine fibroid ultrasound guidance images. As demonstrated in Table 2; Fig. 6, the proposed method exhibits superior performance in the IoU, TPR and FPR metrics, with improvements of 5.52%, 2.75% and 1.36%, respectively, in comparison to SETR. As shown in Table 4; Fig. 7, the proposed method also performs well with average HD, average NormHD, average MAD and average NormMAD of 27.39 pixels and 7.34%, 8.03 pixels and 2.17%, respectively, which is an improvement of 4.76 pixels, 1.49%, 2.17 pixels and 0.69%, when compared to SETR. The proposed method achieved medium improvement in IoU and NormMAD, and small improvement in NormHD. It demonstrates that the improvement of the proposed method is relatively large at the mean boundary and small at the maximum boundary. This is because misclassified superpixels at the boundary can seriously impact the maximum boundary of the segmentation result. As shown in Fig. 8, the proposed method performs better than other methods on images with strong noise and unclear tumor boundaries. The proposed method does not suffer from the problem of holes in continuous tumor and background regions compared to methods such as U-Net, due to the introduction of the self-attention mechanism and positional information. Compared to pixel-wise segmentation networks, the proposed method can segment the tumor region more efficiently in ultrasound guidance images under strong noise interference, and the mean contour is closer to the contour drawn by the radiologist. In future studies, post-processing methods can be considered to smooth the jagged boundaries to further improve the performance of segmentation. Figure 9 shows examples of input images, labeled images, and attention maps of superpixels. It can be seen that the network focuses more on superpixels at the tumor boundary.
Moreover, the proposed method is not limited to the ultrasound guidance image and is able to be applied to other medical images with strong noise interference. However, the preprocessing step of the method is relatively complex, and the proposed method may not be able to outperform existing pixel-wise segmentation methods for high signal-to-noise ratio medical images or natural images.
Conclusions
We proposed the superpixel based self-attention network, which combines superpixels and self-attention mechanism to automatically segment tumor regions for ultrasound guidance images. Specifically, in this method, the image is first segmented into superpixels, then each superpixel is encoded to the uniform size and concatenate, then fed into a self-attention-based network for classification, and finally, based on the classification result, the superpixels are merged to obtain the segmentation result. Compared with pixel-wise image segmentation networks, the superpixel-based method is more effective for ultrasound guidance images. The effectiveness of the method was further validated by quantitatively evaluating the segmentation results of uterine fibroids ultrasound guidance images from HIFU therapy and comparative experiments with the existing segmentation network.
Data availability
The data analyzed for this study can be accessed from the corresponding author upon reasonable request.
References
Kennedy, J. E., Haar, G. R. & Cranston, D. ter High intensity focused ultrasound: Surgery of the future? Br. J. Radiol. 76, 50 (2003).
Kennedy, J. E. High-intensity focused ultrasound in the treatment of solid tumours. Nat. Rev. Cancer. 5, 321 (2005).
Diana, M. et al. High intensity focused ultrasound (HIFU) applied to hepato-bilio-pancreatic and the digestive system—current state of the Art and future perspectives. Hepatobil Surg. Nutr. 5, 329 (2016).
Li, X. L. et al. High-intensity focused ultrasound in the management of adenomyosis: long-term results from a single center. Int. J. Hyperther. 38, 241 (2021).
Siedek, F. et al. Magnetic Resonance-Guided High-Intensity focused ultrasound (MR-HIFU): technical background and overview of current clinical applications (Part 1). Rofo-Fortschr Rontg. 191, 522 (2019).
Ter Haar, G. & Coussios, C. High intensity focused ultrasound: physical principles and devices. Int. J. Hyperther. 23, 89 (2007).
Xian, M. et al. Automatic breast ultrasound image segmentation: A survey. Pattern Recogn. 79, 340 (2018).
Jiang, J. et al. Segmentation of prostate ultrasound images: the state of the Art and the future directions of segmentation algorithms. Artif. Intell. Rev. 56, 615 (2023).
Yu, Y., Chen, Y. & Chiu, B. Fully automatic prostate segmentation from transrectal ultrasound images based on radial bas-relief initialization and slice-based propagation. Comput. Biol. Med. 74, 74 (2016).
Jaouen, V. et al. Prostate volume segmentation in TRUS using hybrid edge-Bhattacharyya active surfaces. IEEE T Bio-Med Eng. 66, 920 (2018).
Pratondo, A., Chui, C. & Ong, S. Robust edge-stop functions for edge-based active contour models in medical image segmentation. IEEE Signal. Proc. Let. 23, 222 (2015).
Khadidos, A., Sanchez, V. & Li, C. Weighted level set evolution based on local edge features for medical image segmentation. IEEE T Image Process 26 (2017). (1979).
Rodtook, A. & Makhanov, S. S. Multi-feature gradient vector flow snakes for adaptive segmentation of the ultrasound images of breast cancer. J. Vis. Commun. Image R. 24, 1414 (2013).
Huang, Q. et al. A robust graph-based segmentation method for breast tumors in ultrasound images. Ultrasonics 52, 266 (2012).
Xu, M. et al. A Split-and-Merge-Based uterine fibroid ultrasound image segmentation method in HIFU therapy. PLOS ONE 10 (2015).
Zhang, D. et al. Segmentation of tumor ultrasound image in HIFU therapy based on texture and boundary encoding. Phys. Med. Biol.. 60, 1807 (2015).
Zhang, D. et al. A region-based segmentation method for ultrasound images in HIFU therapy. Med. Phys. 43, 2975 (2016).
Daoud, M. I., Atallah, A. A., Awwad, F., Al-Najjar, M. & Alazrai, R. Automatic superpixel-based segmentation method for breast ultrasound images. Expert Syst. Appl. 121, 78 (2019).
Ilesanmi, A. E., Idowu, O. P. & Makhanov, S. S. Multiscale superpixel method for segmentation of breast ultrasound. Comput. Biol. Med. 125, 103879 (2020).
Zhao, Y., Zhang, D., Lin, L. & Yang, Y. A method for eliminating the disturbance of pseudo-textural-direction in ultrasound image feature extraction. Biomed. Signal. Proces. 71, 103176 (2022).
Shin, H. et al. Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE T Med. Imaging. 35, 1285 (2016).
Han, S. et al. A deep learning framework for supporting the classification of breast lesions in ultrasound images. Phys. Med. Biol.. 62, 7714 (2017).
Deepak, S. & Ameer, P. M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 111 (2019).
Leclerc, S. et al. Deep learning for segmentation using an open large-scale dataset in 2D echocardiography. IEEE T Med. Imaging. 38, 2198 (2019).
Roy, S. et al. Deep learning for classification and localization of COVID-19 markers in point-of-care lung ultrasound. IEEE T Med. Imaging. 39, 2676 (2020).
Ronneberger, O., Fischer, P. & Brox, T. U-Net: convolutional networks for biomedical image segmentation. Med. Image Comput. Computer-Assisted Intervention PT III. 9351, 234 (2015).
Oktay, O. et al. Attention u-net: Learning where to look for the pancreas. Preprint at (2018). https://arxiv.org/abs/1804.03999.
Chen, L., Zhu, Y., Papandreou, G., Schroff, F. & Adam, H. Encoder-decoder with atrous separable convolution for semantic image segmentation. In Proceedings of the European conference on computer vision (ECCV), (pp. 801–818) (2018).
Ni, B., Liu, Z., Cai, X., Nappi, M. & Wan, S. Segmentation of ultrasound image sequences by combing a novel deep Siamese network with a deformable contour model. Neural Comput. Appl. 35, 14535 (2023).
Ren & Malik Learning a classification model for segmentation. In Proceedings ninth IEEE international conference on computer vision (pp. 10–17) (2003).
Achanta, R. et al. Superpixels compared to State-of-the-Art superpixel methods. IEEE T Pattern Anal. 34, 2274 (2012).
Stutz, D., Hermans, A. & Leibe, B. Superpixels: an evaluation of the state-of-the-art. Comput. Vis. Image Und 166, 1 (2018).
Ren, Z. & Shakhnarovich, G. Image segmentation by cascaded region agglomeration. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (pp. 2011–2018) (2013).
Ji, S. et al. A new multistage medical segmentation method based on superpixel and fuzzy clustering. Comput. Math. Method M 2014 (2014).
Farag, A. et al. A bottom-up approach for pancreas segmentation using cascaded superpixels and (deep) image patch labeling. IEEE T Image Process. 26, 386 (2016).
Qin, W. et al. Superpixel-based and boundary-sensitive convolutional neural network for automated liver segmentation. Phys. Med. Biol 63 (2018).
Liu, C. & Pang, M. Extracting lungs from CT images via deep convolutional neural network based segmentation and two-pass contour refinement. J. Digit. Imaging. 33, 1465 (2020).
Vaswani, A. et al. Attention is all you need. Adv. Neural Inf. Process. Syst. 30 (2017).
Dosovitskiy, A. et al. An image is worth 16x16 words: Transformers for image recognition at scale. Preprint at (2020). https://arxiv.org/abs/2010.11929
S., Z. et al., Rethinking semantic segmentation from a sequence-to-sequence perspective with transformers. In Proceedings of the IEEE/CVF conference on computer vision and pattern recognition (pp. 6881–6890) (2021).
Shorten, C. & Khoshgoftaar, T. M. A survey on image data augmentation for deep learning. J. Big Data 6 (2019).
Molthen, R. C. et al. Ultrasound echo evaluation by K-distribution. In 1993 Proceedings IEEE Ultrasonics Symposium (pp. 957–960) (1993).
Molthen, R. C., Shankar, P. M. & Reid, J. M. Characterization of ultrasonic B-scans using non-Rayleigh statistics. Ultrasound. Med. Biol. 21, 161 (1995).
Shankar, P. M., Reid, J. M., Ortega, H., Piccoli, C. W. & Goldberg, B. B. Use of non-Rayleigh statistics for the identification of tumors in ultrasonic B-scans of the breast. IEEE T Med. Imaging. 12, 687 (1993).
Varma, M. & Zisserman, A. Texture classification: Are filter banks necessary? In 2003 IEEE computer society conference on computer vision and pattern recognition, vol II, proceedings 691 (2003).
Yang, A. Y., Wright, J., Ma, Y. & Sastry, S. S. Unsupervised segmentation of natural images via lossy data compression. Comput. Vis. Image Und. 110, 212 (2008).
Mobahi, H., Rao, S. R., Yang, A. Y., Sastry, S. S. & Ma, Y. Segmentation of natural images by texture and boundary compression. Int. J. Comput. Vis. 95, 86 (2011).
Yin, X., Sun, L., Fu, Y., Lu, R. & Zhang, Y. U-Net-Based medical image segmentation. J. Healthc. Eng. 2022, 4189781 (2022).
Guo, M. H. et al. Attention mechanisms in computer vision: A survey. Comput. Visual Media. 8, 331 (2022).
Rosenthal, R., Cooper, H., Hedges, L. & Others Parametric measures of effect size. Handb. Res. Synthesis. 621, 231 (1994).
Acknowledgements
This work was supported by the National Basic Research Program of China (Grant no.: 2011CB707900).
Author information
Authors and Affiliations
Contributions
S. W., Y. L., and D. Z. contributed to the conception. S. W. wrote the draft manuscript text. S. W., D. Z., and Y. Y. reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wen, S., Zhang, D., Lei, Y. et al. A superpixel based self-attention network for uterine fibroid segmentation in high intensity focused ultrasound guidance images. Sci Rep 15, 21970 (2025). https://doi.org/10.1038/s41598-025-08711-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-08711-x