Abstract
Long non-coding RNA (lncRNA) exhibits a crucial role in multiple human malignancies. The expression of lncRNA MIR155HG, reportedly, is aberrantly up-regulated in several types of tumors. In this research, we studied the role and mechanism of MIR155HG in the progression of cervical cancer (CC). We analyzed the expression pattern of MIR155HG in CC patients using public database TCGA, and overexpressed its expression in HeLa cells to explore its cellular functions. Cellular phenotype experiments were conducted to assess the influence of MIR155HG on HeLa cells, including proliferation, cell cycle, migration, invasion, and cisplatin sensitivity. Then we evaluated the impact on epithelial-mesenchymal transition (EMT) of MIR155HG and identified the underlying regulatory mechanism. We found that MIR155HG was highly expressed and positively correlated with overall survival (OS) prognosis results in CC patients. Overexpression of MIR155HG (MIR155HG-OE) demonstrated higher proliferation and migration levels in HeLa cells, while MIR155HG inhibited the apoptosis rate of HeLa cells. Meanwhile, we found MIR155HG can reduce the effect of cisplatin sensitivity on HeLa cells. Besides this, MIR155HG-OE significantly changed the expression pattern of EMT biomarkers, including ZEB1, TWIST1, Vimentin, N-cadherin, and E-cadherin. To explore the underlying mechanism, we found MIR155HG can promote ZEB1 expression by sponging miR-409-3p, forming a competitive endogenous RNA system to inhibit cisplatin sensitivity. In this study, we deeply explored the molecular and cellular functions of MIR155HG in CC cells. Our results highlight the important role and potential targeting value of MIR155HG in CC pathogenesis and development.
Similar content being viewed by others
Introduction
Cervical cancer (CC) is the most common gynecological malignancy among women worldwide, which poses a serious threat to female health, with approximately 760,000 CC newly diagnosed cases and more than 410,000 predicted deaths by 20301,2. Besides the human papillomavirus (HPV) infection3, the pathogenesis of CC has been extensively investigated by identifying multiple risk factors. The cervical microbiota4, genomic variations5, protein-coding genes6, micro RNAs (miRNAs)7, and long noncoding RNAs (lncRNAs)8 are risk factors, while their potential functions and underlying mechanisms are largely unknown. It is imperative to further clarify the molecular mechanism and biomarkers underlying CC tumorigenesis, including targets associated with tumor immune microenvironment and chemodynamic therapy, so as to improve the treatment of CC9,10,11.
It is well documented that lncRNAs are important regulators of the malignant phenotypes of cancer cells12,13. For example, the significant augmentation of FOXP4-AS1 expression in esophageal squamous cell carcinoma (ESCC) tissues and cell lines is reported, and its high expression markedly promotes the proliferation, invasion and migration ability of ESCC cells by enriching MLL2 and H3K4me3 in the FOXP4 promoter through a molecular scaffold14. Deletion of LINC00839 suppresses nasopharyngeal carcinoma cells rapid growth, invasive capacity and epithelial-mesenchymal transition (EMT)15. Moreover, MIR155HG exhibits cancer-promoting effects in multiple types of cancers. For example, down-regulate the expression of MIR155HG impairs the proliferative, migratory, and invasive activity in melanoma cells via the miR-485-3p/PSIP1 axis, and levels of MIR155HG are linked to decreased overall survival (OS) in melanoma patients16. LncRNA MIR155HG promotes temozolomide resistance by activating the Wnt/beta-Catenin pathway via binding to PTBP1 in glioma17. LncRNA MIR155HG knockdown can also suppress cell proliferation, migration and invasion in non-small cell lung cancer by upregulating TP53INP1 directly targeted by miR-155-3p and miR-155-5p18. Our previous research showed that dysregulated MIR155HG act as competitive endogenous RNAs and are associated with cervical cancer development in UYGHUR Women19, while the underlying regulatory mechanism is unknown.
Previous studies report that miRNAs play crucial roles in cancer progression. MiRNAs are capable of binding to 3’ untranslated region (3’-UTR) of mRNA, further inducing target mRNA degradation or translational inhibition and hence regulating gene expression at the post-transcriptional level20. MicroRNA-133b plays essential role in cervical cancer development by modulating the ERK and AKT1 pathways21. MicroRNA-301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer22. The functions of miRNAs in tumor development were also reported in hepatocellular carcinoma23 and gastric cancer24. In CC, hsa-miR-409-3p was detected with differential expression in liquid and tissue biopsies and can regulate the expression levels of HPV16/18-E6 mRNA25,26. For mechanisms, hsa-miR-409-3p can be sponged by circular RNA circEPSTI1, which has the ability to accelerates cervical cancer progression27. MiR-497-5p, a well-known tumor suppressor in cancer biology, induces cell cycle arrest and represses cancer cell proliferation by targeting CBX428. Nonetheless, the molecular mechanism of miR-497-5p dysfunction in CC requires further delineation.
In this study, we aimed to further explore the functions and cellular influences of MIR155HG in CC cells, as well as the underlying mechanisms. To achieve this goal, we overexpressed and silenced the expression of MIR155HG in HeLa cells, and further assessed its influences on cellular phenotypes. At the same time, we also performed experiments to test whether MIR155HG can modulate the treatment effect of cisplatin. Finally, the molecular targets of MIR155HG and the regulatory axis was confirmed by dual luciferase reporter assay. In summary, our study made a comprehensive exploration on the biological functions and molecular targets of MIR155HG in CC cells, and highlighted its potential regulatory role in CC development, suggesting that MIR155HG can be served as a potential biomarker and therapeutic targets for CC in future.
Materials and methods
Cell cultivation and transfection
The human HeLa cells were derived from Procell Biotech (CL-0101, Wuhan, China), and the cell culture conditions were MEM medium + 10% FBS + 1% PS, 37℃, 5% CO2, and saturated humidity. The HeLa cells have been identified by STR profiling for validation, and tested negative for mycoplasma.
MIR155HG overexpression experiment
We used Lentivirus method to overexpress MIR155HG in this study. We prepared 2 mL of cell suspension with a density of 5 × 104 cells/mL using complete culture medium. Then we took 100 µL/well and added it to a 96 well plate for a total of 12 wells, with 3 wells serving as the control group, which were then incubated at 37 ℃ for 24 h until the cell confluence is around 30%. Took out the negative control virus from the refrigerator, slowly melt it on ice, and dilute the virus with serum-free medium to titers of 1 × 108 TU/mL, 1 × 107 TU/mL, 1 × 106 TU/mL, each 50 µL. Remove the supernatant from each well, replace the culture medium, add virus and corresponding infection enhancement solution, mix well, and continue cultivation. After 12 h of infection, switch back to normal culture medium. After about 48 h of infection, observe under a microscope when the fluorescence expression abundance is high. The infection efficiency is around 80%, and the infection conditions and MOI corresponding to the group with good cell growth can be used as the basis for subsequent infection experiments.
Cell counting kit-8 (CCK-8) assay
The HeLa cells (2 × 103/well) were seeded into a 96-well plate and cultured overnight. Subsequently, the cells in each well were incubated with 10 µL of CCK-8 reagent (Dojindo, Kumamoto, Japan) for 2 h, and then a microplate reader was used to detect the optical density (OD) values of the cells at 450 nm wavelength. With the same method, the absorbance values were detected for three consecutive days.
Transwell assay
The cell suspensions (2 × 105 cells/mL) of HeLa cells were prepared using serum-free medium. The transwell system (Nest, Wuxi, China) was used for this experiment, with a total of 0.2 mL cell suspension in the upper compartment, and medium containing 10% FBS in the lower compartment. After 12 h incubation of the cells in the system, the migrated and invaded cells were fixed by 4% paraformaldehyde and stained with 0.5% crystal violet solution. Finally, the stained cells were screened and selected in each group to count the cell number by the microscope. The invasion assay was performed with the bottom of the Transwell system coated with a layer of Matrigel (30 µg/well; BD, San Jose, CA, USA) before the inoculation of the cells.
Western blot
The extraction of total protein from cells was performed employing RIPA lysis buffer (Sigma-Aldrich, St. Louis, MO, USA). After the samples were mixed with 5×loading buffer and denatured in boiling water for 5 min, the separation of the proteins was completed via SDS-PAGE, followed by subsequent transfer of separated proteins onto polyvinylidene difluoride (PVDF) membranes (Beyotime, Shanghai, China). After that, the membranes were blocked in 5% skimmed milk for 2 h at room temperature and then incubated with primary antibodies against ZEB1 (1: 1000; ab203829; Abcam, Shanghai, China), TWIST (1: 1000; ab50581; Abcam, Shanghai, China), E Cadherin (1: 1000; ab40772; Abcam, Shanghai, China), Vimentin (1: 1000; ab92547; Abcam, Shanghai, China), SNAIL (1: 1000; ab216347; Abcam, Shanghai, China), N Cadherin (1: 1000; ab76011; Abcam, Shanghai, China), and β-actin (1: 2000; ab5694; Abcam, Shanghai, China) overnight at 4 °C. Next, the membranes were incubated with horseradish peroxidase-labeled secondary antibodies (1:1000, Beyotime, Shanghai, China) at room temperature for 2 h. The development of protein bands was conducted employing an enhanced chemiluminescent kit (Biozym, Hessisch Oldendorf, Germany).
Reverse transcription and quantitative polymerase chain reaction (RT-qPCR)
The U6 and GAPDH were used as the internal control genes for lncRNA and mRNA in RT-qPCR experiment, respectively. The total RNA was extracted from HeLa cells by TRIZOL (15596026, Ambion). The cDNA was synthetized by standard procedures and RT-qPCR was conducted by the PCR system (MyCycler Thermal Cycler, Bio-Rad) and real time PCR instrument (ABI 7500 Fast, ABI) according to the steps following published studies29,30. All data were expressed as mean ± standard deviation, and statistical analysis was performed on the gene content in each group using SPSS 19.0 software. ANOVA analysis method was used for statistical analysis: P < 0.05 indicates significant differences. The primer sequences were presented in Table S1.
Dual-luciferase reporter gene assay
The C-33 A cells (CL-0045, Procell Biotech, Wuhan, China) were used for dual-luciferase reporter gene assay to further confirm the regulatory axis in another cell line. The amplification of wild-type (WT) MIR155HG sequence and miR-409-3p sequence were completed by PCR, followed by the selective mutation of the sequences. Next, mutant (MUT) MIR155HG sequence at the interacting site of has-miR-409-3p, has-miR-409-3p NC sequence, MIR155HG-WT sequence and WT miR-409-3p sequence were inserted into pmirGLO luciferase reporter vectors (Promega, Madison, WI, USA). Subsequently, pmirCHECK2-MIR155HG-MUT and pmirCHECK2-MIR155HG-MUT were transfected into C-33 A cells, respectively, together with the transfection of has-miR-409-3p mimics and has-miR-409-3p NC. After 24 h, the Dual-Luciferase Reporter Assay System (Promega, Madison, WI, USA) with a GloMax 20/20 Luminometer (Promega, Madison, WI, USA) were used to detect the luciferase activity of the cells in each group in line with manufacturer’s protocol.
Statistical analysis
All experiments were performed in triplicate, and all experimental data expressed by mean ± standard deviation were analyzed using GraphPad Prism 8.0 (GraphPad Software Inc., La Jolla, CA, USA). Kaplan-Meier method31 with log-rank test was performed to evaluate the relationship between MIR155HG expression and progression-free survival (PFS) and overall survival (OS) of CC patients. The comparison of data was performed using student’s t-test (between two groups) and one-way ANOVA (among multiple groups), with P < 0.05 statistically indicating a significant difference.
Results
MIR155HG was highly expressed in CC tissues and cells
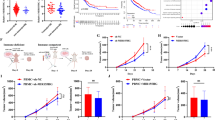
Our previous study has demonstrated that MIR155HG was dysregulated in cervical cancer tissues and is probably associated with the development of CC19. Then we analyzed its expression difference in TCGA database, and found significant increase (p-value < 0.01) in tumor samples (Fig. 1A), although the normal sample size (n = 3) was very low. The promoter region of MIR155HG was lower methylated in primary tumor samples (Fig. 1B), showing consistency with the evaluated expression level. However, we also found that CESC patients with higher expression level of MIR155HG showed better overall survival (OS) prognosis result compared with patients with lower expression level (Fig. 1C). Prognosis analysis for the mature has-miR-155 showed similar result with the host gene MIR155HG (Fig. 1D), indicating that MIR155HG may have unexpected functions in CC and need to be further investigated. To predict the potential functions of MIR155HG in cervical cancer cells, we assessed its expression level and difference in nucleus and cytoplasm. The results demonstrated that MIR155HG was dominantly expressed in nucleus but not in cytoplasm (Fig. 1E).
MIR155HG was dysregulated in CESC patients and associated with the prognosis results of CESC patients. (A) Box plot showing the expression level of MIR155HG in normal and primary tumor samples of CESC patients. Student’s t-test, P-value < 0.01. (B) Box plot showing the methylation level at the promoter region of MIR155HG in normal and primary tumor samples of CESC patients. Student’s t-test, P-value < 0.01. (C) Line plot showing the different OS probability of two CESC patient groups divided by MIR155HG expression levels. (D) The same as (C) but for the OS analysis result by GEPIA2 software. (E) Bar plot showing the cellular location of MIR155HG in HeLa cells by RT-qPCR experiment. N = 5; Student’s t-test, *** p-value < 0.001.
MIR155HG enhanced proliferation, migration and invasion of CC HeLa cells
To further examine the cellular functions of MIR155HG in CC, we overexpressed its expression in HeLa cells. Compared with the blank and negative control (NC) samples, MIR155HG showed significant increase in MIR155HG-OE samples (Fig. 2A). We then performed cellular phenotype experiments to explore the functions of MIR155HG in HeLa cells. Apoptosis assay by flow cytometry demonstrated the decreased apoptosis level in MIR155HG-OE samples (Fig. 2B). Consistent with the apoptosis result, proliferation assay by CCK8 assay showed that MIR155HG-OE significantly increased the proliferation level of HeLa cells (Fig. 2C). To further confirm the regulation on cell proliferation by MIR155HG, we performed cell cycle experiment and found that MIR155HG-OE obviously altered the G0/G1 proportion of cells compared with NC, with increased and decreased percentages in S and G2/M stages but not significant, respectively (Fig. 2D). In summary, these results illustrated that MIR155HG can promote the growth of HeLa cells, implying its cancer promoting functions in CC. We then explored the potential functions in cancer metastasis of MIR15HG, and found that MIR155HG-OE also significantly increased the number of migrated and invaded cells by performing cellular migration and invasion assays (Fig. 2E).
The effects of MIR155HG on proliferation,migration and invasion of CC cells. (A) Bar plot showing the successful overexpression of MIR155HG in CC cells by RT-qPCR experiment; N = 6. (B) Cell flow cytometry and bar plot showing the decreased apoptosis level of CC cells by MIR155HG-OE; N = 3. (C) Bar plot showing the increased proliferation level of CC cells by MIR155HG-OE; N = 3. (D) Cell flow cytometry and bar plot showing the percentage of cells at different cell cycle states; N = 3. Two-way ANOVA test, * P < 0.05. (E) Cell culture and bar plot showing the increased migration and invasion levels of CC cells by MIR155HG-OE; N = 9. Two-way ANOVA test, * P < 0.05; ** P < 0.01; *** P < 0.001; and **** P < 0.0001.
MIR155HG significantly regulated the expression levels of EMT biomarkers
To further explore the influence of MIR155HG on cancer metastasis, we checked the expression levels of epithelial-mesenchymal transition (EMT) biomarkers. The results demonstrated that MIR155HG increased the expression levels of TWIST1, Vimentin, Snail, and N-cadherin, while decreased the expression of E-cadherin by RT-qPCR experiment (Fig. 3A). Then we tested the expression levels of these EMT biomarkers using western blot experiment. The protein levels of N-cadherin, Snail, TWIST1, and Vimentin were increased in MIR155HG-OE samples, while the expression level of E-cadherin was decreased (Fig. 3B, Supplementary Figure S1). To further quantify the expression levels of these EMT biomarkers, we calculated the ratio between EMT biomarkers and β-actin for their band intensities, and found that all of them showed significant changes in MIR155HG-OE group compared with other two groups (Fig. 3C). In summary, these results demonstrated that MIR155HG can significantly regulate the expression levels of EMT biomarkers in CC cells and probably modulate EMT process of CC patients.
MIR155HG significantly regulated expression levels of EMT biomarkers. (A) Bar plot showing the quantitative results for RT-qPCR results. N = 5; Student’s t-test, *** p-value < 0.001, **** p-value < 0.0001. (B) Western blot result showing the expression levels of EMT biomarkers. (C) Quantitative results for the WB experiment in (B). N = 3; Student’s t-test, ** p-value < 0.01.
MIR155HG can reduce the effect of cisplatin sensitivity on cervical cancer cells
Then we investigate whether MIR155HG can influence the sensitivity of cisplatin treatment on HeLa cells. By treating the CC HeLa cells with different doses of cisplatin according to previous study32, we found the proliferation level of CC HeLa cells was significantly inhibited with a dose dependent manner (Fig. 4A). Then we transfected the MIR155HG-OE plasmid into HeLa cells. There was a significant difference of inhibition level of cellular proliferation in MIR155HG-OE group compared with the other two groups when the cisplatin doses were 2 and 5µM (Fig. 4B). We next explored how MIR155HG influenced the phenotypes of HeLa cells under cisplatin treatment. The number of cell clones was significantly increased in MIR155HG-OE + Cisplatin group compared with Cisplatin and NC + Cisplatin groups (Fig. 4C, p-value < 0.01). At the same time, the apoptosis level in MIR155HG-OE + Cisplatin group was significantly inhibited compared with the Cisplatin and NC + Cisplatin groups (Fig. 4D, p-value < 0.01). Then we assessed the proliferation levels of HeLa cells under MIR155HG-OE and cisplatin treatment, and found that the proliferation level was increased by MIR155HG-OE after 48, 72, and 96 h after cisplatin treatment (Fig. 4E). In summary, these results demonstrated that MIR155HG has the ability to reduce the sensitivity of cisplatin treatment in HeLa cells.
Effect of MIR155HG on cisplatin sensitivity of cervical cancer cells. (A) Growth inhibition of Hela cells was observed at different concentrations of cisplatin by CCK-8 method (mean ± STD, n = 5). (B) Growth inhibition of Hela cells was reduced after overexpression of MIR155HG at different concentrations of cisplatin. (C) Plate clonality assays was used to measure the impact of cisplatin to cell clonality in Hela cells after overexpression of MIR155HG. (D) Flow cytometry was adopted to examine apoptotic rate in Hela cells with cisplatin after overexpression of MIR155HG. (E). Effect of cisplatin on proliferation of Hela cells after overexpression of MIR155HG at different time. N = 3 for C-D, and N = 5 for A-B and E; One-way ANOVA test, * p-value < 0.05, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001; ns, non-significant.
MIR155HG was directly targeted by miR-409-3p
To explore the underlying regulatory mechanism of MIR155HG in cervical cancer, we analyzed its potential as a compete endogenous RNA (ceRNA). By predicting miRNA binding sites on MIR155HG using LncTar software33 with default parameters, we found that miR-409-3p can potentially interact with MIR155HG (Fig. 5A). We then performed dual luciferase reporter assay to validate the interaction between miR-409-3p and MIR155HG. The results demonstrated that a significant difference of luciferase signal was detected between MIR155HG-WT + miR-409-3p and other groups, indicating their tight interaction (Fig. 5B). We then performed RT-qPCR experiment to check the expression pattern of miR-409-3p after MIR155HG-OE, and found that MIR155HG-OE can significantly decrease the expression level of miR-409-3p (Fig. 5C), indicating that MIR155HG can influence the expression level of miR-409-3p in CC cells. At the same time, we also predicted that.
MIR155HG interacted with miR-409-3p and repressed its expression in HeLa cells. (A) Base pair presentation for MIR155HG and has-miR-409-3p that was predicted by LncTar. (B) Bar plot showing the quantitative results for dual luciferase reporter assay. N = 3; Student’s t-test, ** p-value < 0.01, **** p-value < 0.0001. (C) Bar plot showing the expression level for miR-409-3p in three groups. N = 3; Student’s t-test, **** p-value < 0.0001.
MIR155HG can rescue the expression of ZEB1 by sponging miR-409-3p in CC cells
Based on the interaction between MIR155HG and miR-409-3p, we propose that MIR155HG can rescue the expression of genes that were targeted by miR-409-3p. Thus, we predicted the potential targets of miR-409-5p and found that ZEB1 was one of the targets of miR-409-5p. Then we analyzed whether the expression level of ZEB1 can be regulated by MIR155HG. The results demonstrated that ZEB1 mRNA level was significantly increased in MIR155HG-OE samples (Fig. 6A). The protein level of ZEB1 was also increased by MIR155HG-OE in HeLa cells with β-actin as internal control (Fig. 6B). To further validate if ZEB1 was regulated by MIR155HG/miR-409-3p axis, we predicted the targets of miR-409-3p by TargetScan34 and found ZEB1 was among them (Fig. 6C). Then we compared the expression levels of MIR155HG, ZEB1, and miR-409-3p under cisplatin treatment. The results demonstrated that MIR155HG and ZEB1 were significantly repressed, while miR-409-3p was significantly increased under cisplatin treatment (Fig. 6D).
MIR155HG regulated the expression of ZEB1 in HeLa cells. (A) Bar plot showing the expression level changes of ZEB1 in three groups. (B) Western blot showing the expression pattern of ZEB1 in HeLa cells. Left panel was the gel result, and right panel was the quantitative result. (C) Base pair presentation for ZEB1 and has-miR-409-3p that was predicted by TargetScan. (D) Bar plot showing the expression levels of MIR155HG and its target genes after cisplatin intervention. N = 5; Student’s t-test, ** p-value < 0.01, *** p-value < 0.001, **** p-value < 0.0001.
Discussion
Currently, surgery, chemotherapy, radiotherapy, targeted therapy, and immunotherapy are the main treatment strategies for cancers35. For instance, circulating biomarkers such as cytokines and soluble PD-L1, have a role in personalized treatments for melanoma patients36. Meanwhile, next-generation CAR T-cell strategies that adapt to altered tumor metabolism are being explored to overcome the metabolic barriers of solid tumors37. Finally, interrupting the crosstalk between tumor cells and tumor-associated macrophages mediated by small extracellular vesicles offers a novel avenue to reprogram immunosuppressive macrophages and bolster anti-tumor immunity38. However, these methods still couldn’t cure most of the human malignancies, especially the metastatic diseases. In recent years, non-coding RNA therapeutics have attracted a lot of attention, and are promising solutions to improve the prognosis of the patients39. LncRNAs possess multiple functions including serving as a host gene of miRNA, preventing RNA and protein from binding to targets, or acting as a molecular scaffold to guide proteins to their targets40. In this study, we systematically explored the functions of MIR155HG in cervical cancer cells, and confirmed its regulatory mechanism to facilitate the progression of HeLa cells by acting as a sponge for miR-409-3p and increasing the expression of ZEB1. Our study clearly showed the important role of MIR155HG in HeLa cells and illustrated its potential therapeutic value in the treatment of cervical cancer.
Previous study has demonstrated the functions and clinical significance of MIR155HG in tumor-infiltrating macrophages of lung adenocarcinoma41. Our study provides novel insights into the role of MIR155HG in cervical cancer (CC). We demonstrated that MIR155HG is aberrantly upregulated while it is negatively correlated with poor prognosis in CC patients. This finding is consistent with previous studies showing that MIR155HG is often overexpressed in various human malignancies, including breast cancer, colorectal cancer, and hepatocellular carcinoma. It is also suggested that MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers42. Recent studies have highlighted advancements in liquid biopsies for cancer diagnostics and monitoring, such as circulating tumor DNA (ctDNA) and DNA methylation signatures by next-generation sequencing43,44,45. We propose that the expression or methylation level of MR155HG can also be detected in CC using such methods. Thus, the upregulation of MIR155HG in CC tissues suggests that it may serve as a potential biomarker for disease progression and prognosis.
In this study, our experimental data revealed that overexpression of MIR155HG (MIR155HG-OE) significantly enhanced the proliferation and migration capabilities of HeLa cells, while simultaneously inhibiting apoptosis. Another study demonstrated that MIR155HG can promote the expression of PD-L1 by functioning as a ceRNA to modulate the miR-223/STAT1 axis in hepatocellular carcinoma HepG2 cells, suggesting the important role of MIR155HG in cancer immune escape46. MIR155HG can also promote the progression and is associated with prognosis of pancreatic cancer patients through negative regulation of miR-80247. We also observed that miR155HG functions as a competing endogenous RNA for miR-185. Moreover, miR-185 directly targets and inhibits ANXA2, which exhibits oncogenic functions in glioblastoma multiforme48. It has been reported that MIR155HG expression in cancer-associated fibroblasts was associated with overall survival among ovarian cancer patients, and was also associated with higher infiltrates of immune cell subsets49. These results highly demonstrated the MIR155HG has profound influence on cellular phenotypes by acting as a regulatory RNA. Prior studies have demonstrated that lncRNAs can act as critical modulators of inflammation and cell proliferation via ceRNA networks, such as KCNQ1OT1 in Parkinson’s disease50, FOXD2-AS1 in oral squamous-cell carcinoma51, supporting our conclusion that MIR155HG may contribute to the aggressive behavior of cervical cancer cells by promoting cell survival and metastasis. This is particularly relevant given that metastasis is a major cause of treatment failure and mortality in CC patients. By enhancing cell viability and migration, MIR155HG likely facilitates the spread of cancer cells to distant sites, thereby exacerbating disease progression. Moreover, the interaction between coding genes and lncRNAs is increasingly being studied by gene–lncRNA network analyses through large-scale bioinformatics approaches52,53, indicating that the function of MIR155HG can be further predicted by bioinformatics approaches54. Thus, it would be valuable to deeper explore the function and interaction network of MIR155HG by using multi-omics datasets and analysis tools in future studies.
The epithelial-mesenchymal transition (EMT) is a critical process in cancer metastasis, characterized by the loss of epithelial markers (E-cadherin) and the gain of mesenchymal markers (such as Vimentin and N-cadherin)55. Our results showed that MIR155HG-OE significantly altered the expression pattern of key EMT biomarkers, including upregulation of ZEB1, TWIST1, Vimentin, and N-cadherin, as well as the downregulation of E-cadherin. This suggests that MIR155HG may drive the EMT process in CC cells, thereby enhancing their migratory and invasive capabilities. The induction of EMT is a hallmark of metastatic cancers, and our findings highlight the potential role of MIR155HG in promoting this process in cervical cancer. In clear cell renal cell carcinoma (ccRCC), blocking lncRNA MIR155HG/miR-155-5p/-3p inhibits proliferation, invasion and migration of cancer cells, suggesting that MIR155HG is a potential target for early diagnosis and precise treatment of ccRCC56. Besides, MIR155HG can also be served as prognostic lncRNAs in kidney renal clear cell carcinoma57. Based on the above discovery, we propose that MIR155HG can probably regulate the metastasis state of CC by modulating the expression patterns of EMT marker proteins. Meanwhile, this hypothesis should be further validated using in vivo experiments, such as metastatic mouse model, in future studies.
To elucidate the molecular mechanism underlying the effects of MIR155HG, we identified a regulatory axis involving MIR155HG, miR-409-3p, and ZEB1. Specifically, we found that MIR155HG can sponge miR-409-3p, thereby relieving its inhibitory effect on ZEB1 expression. This interaction forms a competitive endogenous RNA (ceRNA) system, where MIR155HG competes with ZEB1 mRNA for binding to miR-409-3p, ultimately leading to increased ZEB1 expression. ZEB1 is a well-known transcription factor that promotes EMT and metastasis in various cancers58. By upregulating ZEB1, MIR155HG indirectly enhances the EMT process, contributing to the malignant phenotype of CC cells. In laryngeal squamous cell carcinoma (LSCC), TGF‑β can induce the expression of MIR155HG, which can further promote the progression and EMT of LSCC by regulating the miR-155-5p/SOX10 axis59. This ceRNA mechanism provides a novel perspective on how lncRNAs can regulate gene expression and influence cancer progression. Therefore, MIR155HG/miR-409-3p/ZEB1 is a potential regulatory axis for the development of CC. Meanwhile, further experiments such as luciferase and RNA pull-down should be conduct to confirm this regulatory axis.
We also observed that MIR155HG can significantly reduce the sensitivity of HeLa cells to cisplatin treatment, which is a cornerstone of chemotherapy for cervical cancer but is rendered ineffective owing to drug resistance. Thus, resistance to cisplatin is a significant clinical challenge60. This finding suggests that MIR155HG may contribute to chemoresistance in cervical cancer, potentially by enhancing cell survival pathways and inhibiting apoptosis. It has been reported that knockdown of MIR155HG can suppress M2 macrophage polarization, and proliferation, migration, invasion, as well as oxaliplatin resistance of CRC cells61. Another lncRNA, linc00958, can also inhibit the cellular resistance to cisplatin by modulating the miR-185-5p/RSF-1 axis in SiHa/DDP cells of cervical cancer62, indicating that lncRNAs are important players in drug resistance of cervical cancer therapy. Targeting MIR155HG or its downstream effectors, such as antisense oligos or CRISPR screening63, may therefore represent a promising strategy to overcome cisplatin resistance and improve therapeutic outcomes in CC patients, which need to be further validated using animal model and clinical experiments in future.
Our study highlights the multifaceted role of MIR155HG in cervical cancer progression, from promoting cell proliferation and migration to driving EMT and contributing to chemoresistance. These findings underscore the potential of MIR155HG as a therapeutic target in CC. Future research should focus on validating these findings in larger cohorts of CC patients and exploring the therapeutic potential of targeting the MIR155HG/miR-409-3p/ZEB1 axis. Additionally, further investigation into the downstream effectors of MIR155HG and its interactions with other signaling pathways may reveal additional mechanisms by which this lncRNA contributes to cervical cancer pathogenesis. Meanwhile, in vivo experiments, such as patient-derived xenograft (PDX) models64 and the rescue experiments at the ZEB1 protein level, should be performed to further validate the functions and clinical applicability of MIR155HG in CC.
Conclusions
In conclusion, our study demonstrates that MIR155HG can regulate the phenotypes and functions of cervical cancer cells, and extends the molecular regulatory mechanism of MIR155HG, illustrating the regulatory axis of MIR155HG/miR-409-3p/ZEB1 as potential therapeutic pathway of cervical cancer. Further studies with animal models and new technologies65, will deeper decipher the functions and regulatory mechanism of MIR155HG in future. The findings highlight the potential therapeutic value of targeting MIR155HG and its associated pathways, offering new avenues for improving the management of cervical cancer.
Data availability
The datasets used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
Siegel, R. L., Giaquinto, A. N. & Jemal, A. Cancer statistics. CA Cancer J. Clin. 74(1), 12–49 (2024).
Li, Z. et al. Global landscape of cervical cancer incidence and mortality in 2022 and predictions to 2030: the urgent need to address inequalities in cervical cancer. Int. J. Cancer. 157 (2), 288–297 (2025).
Choi, S. et al. HPV and cervical cancer: A review of epidemiology and screening uptake in the UK. Pathogens 12(2), 1–16 (2023).
Wu, S. et al. The feature of cervical microbiota associated with the progression of cervical cancer among reproductive females. Gynecol. Oncol. 163 (2), 348–357 (2021).
Ramachandran, D. & Dork, T. Genomic risk factors for cervical cancer. Cancers (Basel) 13(20) (2021).
Revathidevi, S. et al. APOBEC: A molecular driver in cervical cancer pathogenesis. Cancer Lett. 496, 104–116 (2021).
Doghish, A. S. et al. MiRNAs role in cervical cancer pathogenesis and targeted therapy: signaling pathways interplay. Pathol. Res. Pract. 244, 154386 (2023).
Modabber, N. et al. Evaluation of long Non-coding RNA (LncRNA) in the pathogenesis of chemotherapy resistance in cervical cancer: diagnostic and prognostic approach. Mol. Biotechnol. 66 (10), 2751–2768 (2024).
Sonkin, D., Thomas, A. & Teicher, B. A. Cancer treatments: Past, present, and future. Cancer Genet. 286–287, 18–24 (2024).
Liu, H. & Dilger, J. P. Different strategies for cancer treatment: targeting cancer cells or their neighbors? Chin. J. Cancer Res. 37 (2), 289–292 (2025).
Joshi, R. M. et al. Overview of perspectives on cancer, newer therapies, and future directions. Oncol. Translational Med. 10 (3), 105–109 (2024).
Renganathan, A. & Felley-Bosco, E. Long noncoding RNAs in Cancer and therapeutic potential. Adv. Exp. Med. Biol. 1008, 199–222 (2017).
Puvvula, P. K. LncRNAs regulatory networks in cellular senescence. Int. J. Mol. Sci. 20(11) (2019).
Niu, Y. et al. LncRNA FOXP4-AS1 promotes the progression of esophageal squamous cell carcinoma by interacting with MLL2/H3K4me3 to upregulate FOXP4. Front. Oncol. 11, 773864 (2021).
Feng, Y. et al. LINC00839 knockdown restrains the metastatic behavior of nasopharyngeal carcinoma by sponging miR-454-3p. Ageing 13(24), 26022–26033 (2021).
Huo, J. et al. The LncRNA MIR155HG is upregulated by SP1 in melanoma cells and drives melanoma progression via modulating the MiR-485-3p/PSIP1 Axis. Anticancer Agents Med. Chem. 22 (1), 152–159 (2022).
He, X. et al. LncRNA MIR155HG promotes Temozolomide resistance by activating the Wnt/beta-Catenin pathway via binding to PTBP1 in glioma. Cell. Mol. Neurobiol. 41 (6), 1271–1284 (2021).
Ren, X. Y., Han, Y. D. & Lin, Q. Long non-coding RNA MIR155HG knockdown suppresses cell proliferation, migration and invasion in NSCLC by upregulating TP53INP1 directly targeted by miR-155-3p and miR-155-5p. Eur. Rev. Med. Pharmacol. Sci. 24 (9), 4822–4835 (2020).
Chen, Y. et al. Dysregulated LncRNAs act as competitive endogenous RNAs and are associated with cervical Cancer development in UYGHUR women. Technol. Cancer Res. Treat. 20, 1533033821989711 (2021).
Bartel, D. P. Metazoan MicroRNAs. Cell 173 (1), 20–51 (2018).
Qin, W. et al. MicroRNA-133b is a key promoter of cervical carcinoma development through the activation of the ERK and AKT1 pathways. Oncogene 31 (36), 4067–4075 (2012).
Zhang, W. et al. MicroRNA-301a promotes migration and invasion by targeting TGFBR2 in human colorectal cancer. J. Exp. Clin. Cancer Res. 33 (1), 113 (2014).
Li, T. T. et al. MicroRNA-138-1-3p sensitizes Sorafenib to hepatocellular carcinoma by targeting PAK5 mediated beta-catenin/ABCB1 signaling pathway. J. Biomed. Sci. 28 (1), 56 (2021).
Zhou, T., Chen, S. & Mao, X. miR-145-5p affects the differentiation of gastric cancer by targeting KLF5 directly. J. Cell. Physiol. 234 (5), 7634–7644 (2019).
Shukla, V. et al. Enumeration of deregulated MiRNAs in liquid and tissue biopsies of cervical cancer. Gynecol. Oncol. 155 (1), 135–143 (2019).
Sommerova, L. et al. The role of miR-409-3p in regulation of HPV16/18-E6 mRNA in human cervical high-grade squamous intraepithelial lesions. Antiviral Res. 163, 185–192 (2019).
Wu, P. et al. Circular RNA circEPSTI1 accelerates cervical cancer progression via miR-375/409-3P/515-5p-SLC7A11 axis. Aging (Albany NY). 13 (3), 4663–4673 (2021).
Chen, Y. et al. MicroRNA-497-5p induces cell cycle arrest of cervical Cancer cells in S phase by targeting CBX4. Onco Targets Ther. 12, 10535–10545 (2019).
Wu, Z. et al. Icaritin induces MC3T3-E1 subclone14 cell differentiation through Estrogen receptor-mediated ERK1/2 and p38 signaling activation. Biomed. Pharmacother. 94, 1–9 (2017).
Peng, C. et al. Syzygium aromaticum enhances innate immunity by triggering macrophage M1 polarization and alleviates Helicobacter pylori-induced inflammation. J. Funct. Foods 107 (2023).
Lanczky, A. & Gyorffy, B. Web-Based survival analysis tool tailored for medical research (KMplot): development and implementation. J. Med. Internet Res. 23 (7), e27633 (2021).
Xu, Y. et al. Inhibition of autophagy enhances cisplatin cytotoxicity through Endoplasmic reticulum stress in human cervical cancer cells. Cancer Lett. 314 (2), 232–243 (2012).
Li, J. et al. LncTar: a tool for predicting the RNA targets of long noncoding RNAs. Brief. Bioinform. 16 (5), 806–812 (2015).
Agarwal, V. et al. Predicting effective MicroRNA target sites in mammalian mRNAs. Elife 4 (2015).
Zhou, Y. et al. Current standards in the management of early and locally advanced cervical cancer: Update on the benefit of neoadjuvant/adjuvant strategies. Cancers (Basel) 14(10) (2022).
Splendiani, E. et al. Immunotherapy in melanoma: can we predict response to treatment with Circulating biomarkers? Pharmacol. Ther. 256, 108613 (2024).
Ramapriyan, R. et al. Altered cancer metabolism and implications for next-generation CAR T-cell therapies. Pharmacol. Ther. 259, 108667 (2024).
Niu, L. et al. Small extracellular vesicles-mediated cellular interactions between tumor cells and tumor-associated macrophages: implication for immunotherapy. Biochim. Biophys. Acta Mol. Basis Dis. 1870 (2), 166917 (2024).
Winkle, M. et al. Noncoding RNA therapeutics - challenges and potential solutions. Nat. Rev. Drug Discov. 20 (8), 629–651 (2021).
Tim, R. & Dinger, M. M. Long non-coding rnas: insights into functions. Nat. Rev. Genet. 10 (3), 155–159 (2009).
Guo, Y. et al. A LncRNA signature of tumor-infiltrating macrophages is associated with prognosis and tumor immunity in lung adenocarcinoma. Comput. Biol. Med. 148, 105655 (2022).
Peng, L. et al. MIR155HG is a prognostic biomarker and associated with immune infiltration and immune checkpoint molecules expression in multiple cancers. Cancer Med. 8 (17), 7161–7173 (2019).
Jahangiri, L. Updates on liquid biopsies in neuroblastoma for treatment response, relapse and recurrence assessment. Cancer Genet. 288-289, 32–39 (2024).
Ohyama, H. et al. Development of a molecular barcode detection system for pancreaticobiliary malignancies and comparison with next-generation sequencing. Cancer Genet. 280-281, 6–12 (2024).
Gonzalez, T. et al. Methylation signatures as biomarkers for non-invasive early detection of breast cancer: A systematic review of the literature. Cancer Genet. 282-283, 1–8 (2024).
Peng, L. et al. Lipopolysaccharide facilitates immune escape of hepatocellular carcinoma cells via m6A modification of LncRNA MIR155HG to upregulate PD-L1 expression. Cell. Biol. Toxicol. 38 (6), 1159–1173 (2022).
Qin, Y. et al. Long noncoding RNA MIR155HG facilitates pancreatic cancer progression through negative regulation of miR-802. J. Cell. Biochem. 120 (10), 17926–17934 (2019).
Wu, W. et al. The miR155HG/miR-185/ANXA2 loop contributes to glioblastoma growth and progression. J. Exp. Clin. Cancer Res. 38 (1), 133 (2019).
Colvin, E. K. et al. Expression of long noncoding RNAs in cancer-associated fibroblasts linked to patient survival in ovarian cancer. Cancer Sci. 111 (5), 1805–1817 (2020).
Li, M. M. et al. LncRNA KCNQ1OT1 promotes NLRP3 inflammasome activation in parkinson’s disease by regulating pri-miR-186/mature miR-186/NLRP3 axis. Biochim. Biophys. Acta Mol. Basis Dis. 1870 (8), 167454 (2024).
Liu, J. et al. LncRNA FOXD2-AS1 promotes the growth, invasion and migration of OSCC cells by regulating the MiR-185-5p/PLOD1/Akt/mTOR pathway. Cancer Genet. 284-285, 48–57 (2024).
Liu, H. & Tang, T. Pan-cancer genetic analysis of disulfidptosis-related gene set. Cancer Genet. 278, 91–103 (2023).
Liu, H. & Tang, T. Pan-cancer genetic analysis of Cuproptosis and copper metabolism-related gene set. Front. Oncol. 12, 952290 (2022).
Duan, L. et al. Bioinformatics analysis of the association between miR-942-5p–induced downregulation of PIEZO-type mechanosensitive ion channel component 1 and poor prognosis in non–small cell lung cancer mediated by the mitogen-activated protein kinase pathway signaling pathway. Oncol. Translational Med. 10 (6), 272–280 (2024).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell. Biol. 15 (3), 178–196 (2014).
Tao, M. et al. Blocking LncRNA MIR155HG/miR-155-5p/-3p inhibits proliferation, invasion and migration of clear cell renal cell carcinoma. Pathol. Res. Pract. 216 (2), 152803 (2020).
Song, J. et al. Identification and validation of two novel prognostic LncRNAs in kidney renal clear cell carcinoma. Cell. Physiol. Biochem. 48 (6), 2549–2562 (2018).
Caramel, J., Ligier, M. & Puisieux, A. Pleiotropic roles for ZEB1 in Cancer. Cancer Res. 78 (1), 30–35 (2018).
Cui, W. et al. TGF-beta-induced long non-coding RNA MIR155HG promotes the progression and EMT of laryngeal squamous cell carcinoma by regulating the miR-155-5p/SOX10 axis. Int. J. Oncol. 54 (6), 2005–2018 (2019).
Bhattacharjee, R. et al. Cellular landscaping of cisplatin resistance in cervical cancer. Biomed. Pharmacother. 153, 113345 (2022).
Zhou, L. et al. LncRNA MIR155HG induces M2 macrophage polarization and drug resistance of colorectal cancer cells by regulating ANXA2. Cancer Immunol. Immunother. 71 (5), 1075–1091 (2022).
Tian, J. et al. linc00958/miR-185-5p/RSF-1 modulates cisplatin resistance and angiogenesis through AKT1/GSK3beta/VEGFA pathway in cervical cancer. Reprod. Biol. Endocrinol. 20 (1), 132 (2022).
Liu, H. & Wang, P. CRISPR screening and cell line IC50 data reveal novel key genes for Trametinib resistance. Clin. Exp. Med. 25 (1), 21 (2024).
Lee, S. J. et al. hsa-miR-CHA2, a novel microrna, exhibits anticancer effects by suppressing Cyclin E1 in human non-small cell lung cancer cells. Biochim. Biophys. Acta Mol. Basis Dis. 1870 (6), 167250 (2024).
Chen, Z. & Er Saw, P. Integration in biomedical science 2024: emerging trends in the Post-Pandemic era. BIO Integr. 5 (1), 998 (2024).
Acknowledgements
We are thankful for the assistance and discussion from colleagues of Wuhan Nissi Biotech.
Funding
This study was supported by projects of state key laboratory of pathogenesis, prevention and treatment of high incidence diseases in central Asia (SKL-HIDCA-2020-16), and the general project of natural science foundation of Xinjiang Uyghur Autonomous Region (2020D01C238). This study was also supported by national natural science foundation of China (82260613).
Author information
Authors and Affiliations
Contributions
YC and CM conceptualized, designed and supervised the study. YC and JW performed experiments, interpreted the data, and drafted the manuscript. YH, YG, and KD collected the samples. CM revised and approved the manuscript. All authors read and approved the final manuscript. All authors contributed to editorial changes in the manuscript. All authors have participated sufficiently in the work and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, Y., Wang, J., Heng, Y. et al. MIR155HG promotes metastasis and cisplatin resistance of cervical cancer cells by regulating the miR-409-3p and ZEB1 axis. Sci Rep 15, 20541 (2025). https://doi.org/10.1038/s41598-025-08727-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-08727-3