Abstract
Accurate estimation of plant disease severity is pivotal for effective management and decision-making. Field experiments were conducted to understand the correlation and predict the yellow mosaic disease severity in yard-long beans using visible image indices. A total of 45 visible / Red Green Blue (RGB) indices were derived from the RGB images and correlated with disease severity, and also used as inputs for predicting disease severity using nine machine learning (ML) models. Out of 143 genotypes screened based on final disease severity 3, 18, 18, 17, 34 and 53 genotypes were grouped in immune, resistant, moderately resistant, moderately susceptible, susceptible and highly susceptible categories, respectively. Model performances was evaluated using R2, d-index, mean bias error, and normalized Root Mean Square Error (n-RMSE) metrics. Results revealed that 34 indices exhibited significant correlations (p < 0.01) with YMD severity, with 23 positively and 12 negatively correlated. Among these, Red Color Composite (RCC) and Excessive red (ExR) demonstrated the highest and equal positive correlations (0.87), while Green red difference (GRD) exhibited the largest negative correlation (-0.88) with disease severity. The ML models achieved commendable performance, attaining R2 and d-index values exceeding 0.92 and 0.98, respectively, in calibration, and 0.88 and 0.96 in validation, underscoring their effectiveness in predicting YMD severity using RGB images only. Random Forest (RF), Cubist, XGBoost (XGB), K-Nearest Neighbors (KNN), and Gradient Boosting Machine (GBM) emerged as the five top-performing models for predicting YMD severity using visible indices in yard-long beans. These findings hold practical implications for timely disease management strategies, expediting breeding programs, and aiding policy planners and farmers in making well-informed decisions.
Similar content being viewed by others
Introduction
The world’s food supply is greatly dependent on plants. Plant diseases caused by a various biotic and abiotic factor leads to significant yield losses. According United Nations, Department of Economic and Social Affairs, Population Division, there will be nine billion people on the planet by the year 2050 (United Nations, Department of Economic and Social Affairs, Population Division, 2017)1. which will increase demand for agricultural products by fifty percent as compared to 20132. Climate change leading to rising temperatures and shift in weather patterns often lower the crop yields due heat waves, drought and other adverse conditions3. The cowpea, Vigna unguiculata (L.) Walp., is well-known for its ability to withstand high temperatures and dry conditions4. There are roughly 150 species in the genus Vigna, and it has been proposed that the wild cowpea originated in southern Africa4. According to FAOSTAT5, cowpeas are cultivated worldwide, covering an estimated 15.1 million hectares of land with a total annual production of 9.7 million metric tons. It stands out as the sole arid legume widely distributed across continents, thriving particularly well in arid and semi-arid regions due to its extensive adaptability6. It is a vital legume crop crucial for ensuring food and nutritional security in tropical and subtropical regions across the globe7,8. The short cowpea [V. unguiculata subsp. cylindrica (L.) Estt.], the common cowpea [V. unguiculata subsp. unguiculata (L.) Walp.], and the yardlong bean [Vigna unguiculata subsp. sesquipedalis (L.) Verdc.] are the three cultivated subspecies of cowpea. In India, this crop has a remarkable regenerative capability and can be cultivated during the kharif (rainy season), summer, and winter seasons. Despite its versatility, the average grain yield of this crop remains comparatively low, currently averaging around only 350 kg/ha9. The production of this crop is often challenged by several biotic stressors, among which viral infections are major limitations causing yield losses ranging from 10-100%10. Disease symptoms first appear as small chlorotic spots or yellow flecks in young leaves. Late it progresses from initial chlorotic spots to a characteristic mosaic pattern on leaves leading to complete yellowing in susceptible genotypes. As the infection progresses, the symptoms become more pronounced. Size of leave is generally not considerably affected. A few, smaller and malformed pods are produced by the affected plants11. Plants affected by Yellow Mosaic Disease (YMD) exhibit reduced flower and pod production, with malformed pods and the yield loss can potentially reach 100%12.
The traditional method of manual monitoring of plant diseases in field is time-consuming and has a limitation of investigator’s subjectivity, resulting in excessive use of resources. Remote sensing provides an efficient, non-destructive, and real-time alternative which is cost-effective and practical. Visible (RGB) imaging has shown strong potential for detecting plant stress. This is because the changes in plant pigments and their spectral responses due to stresses can be effectively captured using visible sensors. Moreover, visible imaging is particularly important because it is low-cost and is widely available, even in farmers’ smartphones, making it a more accessible solution for monitoring plant disease. In the new Artificial Intelligence (AI) era, use of visible images, with or without Machine Learning (ML) models is becoming well-known for agricultural disease detection, severity assessment, high throughput phenotyping and disease forecasting/prediction13,14,15,16,17. Precisely estimating crop disease severity using imaging and AI techniques is a potentially useful approach for tracking and effectively managing crop health. This helps to reduce the overabundance of harmful chemicals in agriculture ecosystems, speeds up preventive tactics and can save nearly 30% of crop yield18. The ML based approaches are reported to be more effective at detecting plant disease in the early phases of growth stages19. Several works have been conducted by researchers for detection and prediction of plant diseases using combination of imaging techniques and ML. Schor et al.20 developed a robotic disease detection system with a sensory apparatus (a visible camera and a laser sensor) to identify powdery mildew and tomato spotted wilt virus of bell pepper in greenhouses using PCA-based methods with accuracy levels of above 80%. Es-saady et al.21 utilized 284 photos shot against a black backdrop with Support Vector Machine (SVM) classifiers to identify three pests (leaf miners, Thrips, and Tuta absoluta) and three crop diseases (early blight, late blight, and powdery mildew) of tomato with recognitions accuracies above of 87.8%. Similarly, Sabrol and Satish22 categorized tomato late blight, bacterial spot, Septoria spot, bacterial canker, and tomato leaf curl using the decision tree method with an accuracy of 97.3%. Several researchers attempted to classify and predict plant diseases using imaging and ML approaches in different crops like cucumber23, potato24, tomato25, soybean26, and several other plant diseases27. However, only few similar studies have been conducted on any vegetable disease15,23,28,29. While these studies primarily concentrate on classification problems, determining the extent of disease severity becomes the subsequent crucial step, which is pivotal for effectively managing the disease while minimizing resource utilization and environmental impact. Also, most of the previous studies where carried out under controlled or greenhouse conditions, which do not capture the complex and variable interactions present in natural field settings. Unlike the controlled environments, field experiments captures real-world variability in weather, soil, and pathogen interactions and enhances the practical relevance and robustness of the predictions. Recently, few studies were conducted for yellow rust of wheat30 and chickpea wilt31 to estimate the disease severity under field conditions using combination of thermal and visible imaging with ML models. Feng et al. (2022) quantified powdery mildew severity in wheat using hyperspectral data and ML models achieving an R2 value of 0.85. Similarly, Mandal et al.32 utilized hyperspectral data to predict disease blast disease severity in rice using ML models achieving an R2 value of 0.99. However, to best of our knowledge, none of the studies were conducted to assess the disease severity of the yellow mosaic of yard-long under field conditions been using image analysis and ML. Also, most of the studies on disease severity assessment are conducted under controlled environments conditions which do not fully capture the complexity and variability of real field conditions. To address these gaps, the present study was conducted with the objective to quantify yellow mosaic disease (YMD) severity in yard long bean vegetable using visible imaging coupled with ML models under field conditions making the outcomes more robust and applicable to practical crop management.
Material and method
Study site and screening of germplasm
Field experiments were conducted at ICAR-Research Complex for Eastern Region (ICAR-RCER), Patna experimental farm (25.59° N latitude, 85.13° E longitude, and an altitude of 53 m above mean sea level), India. The climate of Patna is humid subtropical with hot summer and dry winter. The soil type of the experimental location is slightly alkaline and non-calcareous in nature which is classified under Indo-Gangetic alluvium. Genetically diverse one hundred forty three genotypes of yardlong bean were collected from ICAR-National Bureau of Plant Genetic Resources, New Delhi and different location of Bihar and Odisha. These genotypes were grown during April to August at ICAR-RCER, Patna with recommended package of practices on raised beds in plot size of 2 × 2 m2 area in augmented block design. Crops were grown with 90 cm row to row spacing and 30 cm plant to plant spacing. One hundred forty three genotypes of yardlong bean were grown during April to August at ICAR-RCER, Patna with recommended package of practices on raised beds. Plant symptoms, viz., yellowing and necrosis, were seen visually in the field at vegetative to post flowering stage and per cent disease severity was recorded. The genotypes were then classified using a scale of 0–9 depending on the proportion of infected leaves, or impacted foliage area (Table 1)33. Germplasms were categorized as Immune (I), Resistant (R), Moderately Resistant (MR), Moderately Susceptible (MS), Susceptible (S), and Highly Susceptible (HS) based on severity of yellow mosaic. Observation for disease severity was taken at early vegetative stage, late vegetative stage and flowering stage (reproductive stage). However, as the disease is transmitted by vector (insect), at early vegetative stage severity of disease was very less with few genotypes showing yellow mosaic symptoms. As the crop growth stage advanced, the disease severity also progressed and at the time of flowering disease severity was maximum. So, disease data and images from flowering stage were considered in the present study.
Image acquisition for estimating yellow mosaic severity
Visible images were obtained using iPhone 13 rear camera, which comes with dual 12MP camera setup. (Main camera: Sony IMX603,12.2 MP, Aperture f/1.65, focal length 26 mm, pixel size 1.7 µm. Ultra-wide camera: Sony IMX 372, 12.2 MP, Aperture f/2.4, focal length 13 mm, pixel size 1.0 µm). Out of a total of 143 genotypes grown, images were acquired from 100 genotypes during the post-flowering stage. These genotypes were carefully selected to represent the range of phenotypic variability and disease response observed in the field, ensuring sufficient coverage of field heterogeneity. At roughly 13.30–14.30 IST, when the crop plants were experiencing the maximum day temperature and less windy situations, images were captured from the nadir view angle. Images of individual plant were taken from three different locations (where disease severity was maximum) for each genotypes from a height of 0.5 m above the canopy using a reference stick marked at 0.5 m. For each genotype, the data collected from three locations was averaged to obtain a representative value for further analysis.
Visible indices and their correlation
The RGB tristimulus was extracted using R version 4.1.2 and RStudio (version 2021.9.1.372), and visible indices were computed using this data. We also used normalized RGB values (as indicated by r,g,b) to minimize the impact of illumination and colour variation sensitivity in some indices34. The whole set comprised of 45 visible indices generated using RGB images (Table 2). Pearson’s correlation analysis was conducted to assess the relationship between image-derived vegetation indices and Yellow Mosaic Disease (YMD) severity, recorded as percentage of affected area. A two-tailed t-test was conducted to determine the statistical significance of the results at a 5% significance level35.
Machine learning (ML) models
Dataset generated were used to develop 9 ML models, which include elastic net (ELNET), random forest (RF), multivariate adaptive regression spline (MARS), partial least square regression (PLS), L2 Regularized Support Vector Regression (dual) with Linear Kernel (SVM), Stochastic Gradient Boosting (GBM), k-nearest neighbours (KNN), extreme gradient boosting (XGB), and Cubist. The ELNET stands as a frequently employed penalized regression technique, aiming to conduct both variable selection and regularization concurrently. This method incorporates both lasso (L1) and ridge (L2) regression to eliminate superfluous coefficients. RF represents a machine learning model based on decision trees, where each tree generates a unique prediction, and the collective average of these predictions forms the output of the regression. Meanwhile, MARS offers another adaptive non-parametric strategy for tackling intricate regression challenges. It leverages ensembles of simple linear model to deliver optimal predictions in multivariate non-linear regression scenarios. PLS models the correlation between observed values and latent variables. Its significance lies in its capability to concurrently conduct dimensionality reduction and model construction. Support Vector Regression (SVM) employs a linear kernel function for regression tasks, with a minor variation in the objective function optimized compared to its classification counterpart. Its fundamental principle involves identifying the points within the decision boundary and establishing a hyperplane that maximizes the number of points, thereby defining the best-fit line. KNN operation involves computing the Euclidean distance of K neighboring points from the target value and forecasting the output as the mean of these k-nearest neighbors. GBM and XGB are ensemble models based on decision trees, utilizing the gradient boosting algorithm to mitigate overfitting in predictions. Cubist operates as a rule-based regression model, employing linear regression models at the terminal nodes for prediction. These machine learning models were developed using the “caret” package using the R 4.1.2 and RStudio (version 2021.9.1.372) software.
Evaluation of machine learning models
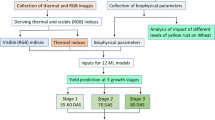
We took image data of 100 genotypes, 70% were allocated for training the ML models, while the remaining 30% were reserved for testing. The hyperparameters of the ML models were optimized using tenfold cross-validation with 5 repetitions, facilitated by the ‘caret’ package in R software36. The performance of the models were assessed using multiple statistical criteria (R2, d-index, MBE and n-RMSE) of validation and the models were finally ranked based on standardized Ranking Performance Index (sRPI) using formulas suggested by Aschonitis et al.37. The overall methodology is depicted in Fig. 1.
Results
Disease progress in yard long bean and screening of germplasm
The progression of disease severity in one hundred forty three yardlong genotypes under research recorded up to 95% disease severity based on the genotypes. Six groups—Immune (I), Resistant (R), Moderately resistant (MR), Moderately susceptible (MS), Susceptible (S), and Highly susceptible (HS) were distinguished based on the severity of the disease (Fig. 2). For training and testing the ML models, all one hundred forty three of the yard-long bean genotypes that were taken into consideration in this study provided varying degrees of disease severity. No visible symptoms were observed in genotypes IC-626170, IC-630412, IC-20514, IC-20298 and IC-586954. Genotypes IC-622570, IC-622574, IC-626139, IC166140, IC-626143, IC-630388, IC-630390, IC-630391, IC-626163, IC-630413, IC-398083, IC-202893, IC-361502 and IC-471938 were found to be resistant. Rest of the genotypes were categorized in moderately resistant, moderately susceptible, susceptible and highly susceptible categories (Table 3).
Correlation analysis between indices and disease severity
Table 4 presents an analysis of the Pearson’s correlation coefficient (PCC) between 45 visible indices and the YMD severity. Of the visible image indices, the correlation coefficients were significant for 34 indices (p < 0.01), of which 23 had a positive association with the YMD severity and the remaining 12 had a negative correlation. The image indices RCC and ExR (r = 0.87) showed the highest positive correlations, while the GRD (r = −0.88) showed the largest negative correlation. Out of 34 significant correlations, 19 indices have very strong correlations with YMD disease severity above ± 0.7.
Performance of ML models
For estimating the visible indices based disease severity of YMD, all the ML models were assessed using R2, d-index, MBE and n-RMSE. The values of these statistics for all models during the calibration and validation stages are reported in Table 5 and illustrated in Figs. 3 and 4 respectively using a 1:1 plot. The sRPI values shown in Table 6 were used to rank the models’ performances.
All of the ML models performed satisfactorily during calibration, as evidenced by R2 values above 0.92, d-index values above 0.98, and least error values (nRMSE and MBE) (Fig. 3). R2 (0.99) and d-index (1.00) values were highest for the Cubist and XGB model, while R2 (0.92) was lowest for the PLS and SVM. The PLS, SVM, KNN, and ELNET, on the other hand, all performed similarly with the lowest d-index value of 0.98. While SVM displayed the lowest MBE value (−0.46), the KNN model indicated the highest MBE value (2.19). The n-RMSE was also lowest for the Cubist (3.70) model and highest for the SVM (9.30) model (Table 5). All other models with positive MBE values, with the exception of SVM, somewhat overestimated during the calibration phase; however, the model-generated results for YMD severity were found to be excellent, with a n-RMSE of less than 10%.
During validation, R2 and d-index were determined to be marginally lower than the calibration values, with ceiling values of 0.95 and 0.98, respectively. The results indicated that the performance of XGB, Cubist, and RF was identical with the highest R2, and that the performance of XGB, Cubist, KNN, RF, and GBM was also identical with the highest d-index value (Fig. 4). Conversely, MARS was determined to have the lowest performance in terms of both statistical parameters (R2 of 0.87 and d-index of 0.96). Nearly all of the models provided slightly overstated but still acceptable results (n-RMSE < 15%) during validation with + ve values of MBE as well. The MARS model had the highest (12.8%), whereas RF had the lowest n-RMSE (7.80) (Table 5). The top five models throughout the validation phase were the RF, GBM, KNN, XGB, and Cubist (R2 > 0.91, d-index > 0.97, and n-RMSE < 10).
In the validation phase, sRPI values were used to rank all nine ML models from high to low performing (Table 6). As per sRPI values RF was the best performing models, while MARS is the poorest performing model to predict the disease severity. According to this validation sRPI-based ranking, the best models for predicting YMD severity in yard-long beans using visible indices were RF, followed by Cubist, XGB, and KNN.
Discussion
The utilization of imaging technologies in conjunction with ML models for identifying diseases in crops has gained considerable attention recently38. However, the majority of the studies were limited to the detection and classification of different diseases. Only a handful of studies had been conducted to estimate the extent or severity of crop diseases in field. While reviewing the literature it was discovered that, this is the first study to employ visible image-based indices to estimate YMD severity in any vegetable crop and first of its kind under Indian conditions. The use of visible image derived data along with ML models to estimate YMD in the yard long bean field has not been tested so far. Our results revealed significant correlations visible image derived indices with YMD severity in different yard long bean genotypes and the potential of using these indices to estimate the YMD severity by training the ML models.
The RGB indices in this study were well correlated with r values between −0.88 to 0.87. Out of 45 indices, 34 indices had significant correlations at 1% significance level. Also, the correlations with 19 indices are very strong with YMD disease severity. Recently, researchers reported weak to medium correlation coefficients of the RGB indices in chickpea wilt31 and wheat yellow rust30 under field conditions, which might be due to the different dates of observations in their both work. The strong correlations in the current study were majorly attributed to collection of all images on same date at same crop growth stages with dominant disease symptoms, which have kept the environmental factor constant in all images. Alves et al.39 reported correlations up to 0.99 between the RGB indices like GLI, NGRDI, BI, HI, SI, VARI, BGI and gray with wheat leaf blast, potato late blight, soybean rust, Calonectria blight of eucalypts, which supports higher correlation values of our work. Similarly, the work of Bhandari et al.40 on stripe rust reported GI, GLI and NDI values correlations ranging from 0.63–0.68 and 0.72–0.79, which is also comparable to the present work.
In our research, the ML models effectively predicted disease severity based on the RGB indices. Given the absence of prior studies on assessing YMD severity, we benchmarked our results against analogous investigations conducted by various researchers in different agricultural contexts. Our observed values of R2, d-index, n-RMSE, and MBE align closely with those reported in similar studies32,41,42,43. The patterns observed in the statistical parameters employed for evaluating the ML models during validation lack consistency. With nine models evaluated across four statistical parameters (d-index, r-square, MBE, and n-RMSE), determining the optimal model became more complex. The utilization of sRPI37 in this investigation, streamlined this process, offering a robust metric for straightforward ranking of the machine learning models based on multiple statistics during validation. Using similar approach44,45) also ranked several ML models under different disease severity level, while predicting disease severity and crop yield under field conditions.
The models in our study attained an R2and d-index above 0.92 and 0.98, respectively, in calibration, and 0.88 and 0.96 in validation. This demonstrates the ML models’ capability to predict YMD severity effectively. The MBE of all the models were slightly higher side during validation as compared to calibration indicating over-prediction of YMD severity by the ML models. The n-RMSE of the ML models during validation remained below 10%, indicating “excellent” performance during Calibration. At validation stage, except ELNET, MARS and PLS all other ML models have again n-RMSE < 10%, which indicates “excellent” prediction of the disease. This is also in line with the final sRPI rankings of the ML models as the best three models ranked using the sRPI, including the RF, Cubist and XGB has < 10% of n-RMSE in both calibration and validation stage31,44.) also reported Cubist, XGB and RF models as best predictors of disease severity under field condition for yellow rust and chickpea wilt, respectively. Duarte-Carvajalino et al.46 assessed the severity of late blight in potatoes by employing multispectral imagery from UAVs and analyzed the data using RF and SVR algorithms. They found that only the RF algorithm achieved satisfactory results, with an R2 of 0.75 and an n-RMSE of 16%, which is less accurate then our findings. Chemura et al.42 detected coffee leaf rust (CLR) using a greenhouse-based spectroradiometer with PLS regression and reported R2 = 0.92, and RMSE = 6.1%. Feng et al.47 found R2 values ranging from 0.54 to 0.68 when forecasting powdery mildew in wheat through PLSR, SVR, and RF models. They indicated RF as the superior model compared to PLSR and SVR in predicting powdery mildew, aligning with our observations supported by sRPI rankings and n-RMSE. Kang et al.48 reported an R2 value of 0.91–0.93 and RMSE between 5.77% and 6.35% using a CNN approach for detecting verticillium wilt in cotton through a combination of field-collected images and spectral data, results that are comparable to our findings. Similarly, Martinez-Martinez et al.49 achieved an R2 of 0.87 in assessing angular leaf spot in beans using spectral measurements ranging from 440 to 850 nm with an ANN model, which is slightly lower than the outcomes observed in our study. Liu et al.50 assessed the severity of apple mosaic using ELNET, GPR, KNN, and SVR algorithms applied to hyperspectral data, reporting prediction accuracies and model rankings that closely align with our findings. Mandal et al.32 effectively demonstrated the use of hyperspectral remote sensing combined with machine learning models for rapid assessment of rice blast disease severity. They reported high predictive accuracy and with calibration and validation R2 values of 0.99 and 0.94, respectively.
Overall, our study demonstrates the potential of machine learning models, optimized with random parameter selection, for accurately estimating YMD disease severity in field conditions. This study provides a robust framework for non-destructive, scalable disease monitoring that aligns with real-world agricultural practices and supports early disease detection and timely management decisions, contributing to yield stability and resource-efficient farming. The findings also support the development of early warning systems for timely disease management and integrating such predictive tools into crop insurance schemes can also aid in risk assessment and premium planning. Future work could benefit from the integration of grid search optimization and advanced techniques like deep learning and transfer learning. Also, by expanding the dataset through additional field experiments of multiple seasons and geographical location could further refine the predictive accuracy and robustness of the models. Integrating this approach into mobile or UAV-based platforms may also enable real-time, high-throughput field phenotyping and disease surveillance. Such advancements would not only improve the management of YMD but also contribute to the broader goal of developing reliable, data-driven solutions for plant disease monitoring and crop management in diverse agricultural settings.
Conclusions
In this study, visible imaging was used with ML models for YMD severity estimation at the canopy level in yard-long beans. The ML models could predict the yellow mosaic disease severity using visible (RGB) images, offering a quick and easy way to measure the severity of YMD in yard-long bean fields. RF model was found to be the best for predicting YMD severity in yard long beans under field settings. Based on the sRPI-based model ranking RF, Cubist, XGB, KNN, and GBM were the top five models for predicting YMD severity in yard-long beans using visible imaging, while MARS was the poorest performing model. The results of this study point to the potential of visible imaging and machine learning (ML) models as a quick and accurate way to measure the severity of plant diseases under field conditions. The findings of this study can serve as a valuable resource for real-time monitoring of crop diseases, aiding stakeholders in making informed decisions.
Data availability
The datasets used for this research study are available from the corresponding author on reasonable request.
References
United Nations Department of economic and social affairs PD world population prospects 2017 Data booklet (ST/ESA/SER.A/401) cover (2017).
FAO.. The future of food and agriculture Trends and challenges. Food and agriculture organization Rome Italy 180 p https://openknowledge.fao.org/server/api/core/bitstreams/2e90c833-8e84-46f2-a675-ea2d7afa4e24/content (Accessed 08 February 2024) (2017).
Kerr, B. R., Hasegawa, T., Lasco, R., Bhatt, I., Deryng, D., Farrell, A., Gurney-Smith, H., Ju H., Lluch-Cota, S., Meza, F., Nelson, G., Neufeldt H. & Thornton P. Chapter 5: Food fibre and other ecosystem products. In climate change 2022: Impacts, Adaptation and vulnerability. Contribution of working group II to the sixth assessment report of the intergovernmental panel on climate change [Portner, H.O., Roberts, D.C., Tignor, M., Poloczanska, E.S., Mintenbeck K., Alegria, A., Craig, M., Langsdorf, S., Loschke, S., Moller, V., Okem, A., Rama, B. (eds.)]. (Cambridge University Press Cambridge UK and New York NY USA 2022)
Smykal, P. et al. Legume crops phylogeny and genetic diversity for science and breeding. Crit. Rev. Plant Sci. 34, 43–103. https://doi.org/10.1080/07352689.2014.897904 (2015).
FAOSTAT. Data. food and agriculture organization of the United Nations. Rome Italy. https://www.fao.org/faostat/en/#data/QCL (Accessed 05 February, 2024) (2022)
Carvalho, M. D. & Halecki, W. Modeling of cowpea (Vigna unguiculata) yield and control insecticide exposure in a semi-arid region. Plants 10(6), 1074. https://doi.org/10.3390/plants10061074 (2021).
Kebede, E. & Bekeko, Z. Expounding the production and importance of cowpea (Vigna unguiculata (L.) Walp.) in Ethiopia. Cogent Food & Agri. 6(1), 1769805. https://doi.org/10.1080/23311932.2020.1769805 (2020).
Mekonnen, T. W., Gerrano, A. S., Mbuma, N. W. & Labuschagne, M. T. Breeding of vegetable cowpea for nutrition and climate resilience in Sub-Saharan Africa: Progress, opportunities, and challenges. Plants 11(12), 1583. https://doi.org/10.3390/plants11121583 (2022).
IIPR. (2024). Cowpea. ICAR-Indian Institute of Pulses Research, Kanpur, India. https://iipr.icar.gov.in/cowpea/ (Accessed 10 February 2024)
Kareem, K. T. & Taiwo, M. A. Interactions of viruses in cowpea: Effects on growth and yield parameters. Virol. J. 4, 1–7. https://doi.org/10.1186/1743-422X-4-15 (2007).
Akram, M. & Singh, N. P. Yellow mosaic of mungbean and urdbean: current status and future strategies. J. Food Legum. 29, 77–93 (2016).
Karthikeyan, A. et al. Mungbean yellow mosaic virus (MYMV): A threat to green gram (Vigna radiata) production in Asia. Int. Journal of Pest Manag. 60(4), 314–324. https://doi.org/10.1080/09670874.2014.982230 (2014).
Bock, C. H. & Nutter, F. W. Jr. Detection and measurement of plant disease symptoms using visible-wavelength photography and image analysis. CABI Rev. 2011, 1–15. https://doi.org/10.1079/PAVSNNR20116027 (2011).
Domingues, T., Brandão, T. & Ferreira, J. C. Machine learning for detection and prediction of crop diseases and pests: A comprehensive survey. Agriculture 12(9), 1350. https://doi.org/10.3390/agriculture12091350 (2022).
Kim, W. S., Lee, D. H. & Kim, Y. J. Machine vision-based automatic disease symptom detection of onion downy mildew. Comput. Electron. Agric. 168, 105099. https://doi.org/10.1016/j.compag.2019.105099 (2020).
Singh, K. K. An artificial intelligence and cloud based collaborative platform for plant disease identification, tracking and forecasting for farmers. 2018 IEEE International Conference on Cloud Computing in Emerging Markets (CCEM) 49–56 https://doi.org/10.1109/CCEM.2018.00016. (2018)
Ullah, M. R., Dola, N. A., Sattar, A. & Hasnat, A. Plant diseases recognition using machine learning. 2019 8th International Conference System Modeling and Advancement in Research Trends (SMART) 67–73. https://doi.org/10.1109/SMART46866.2019.9117284. (2019)
Muppala, C. & Guruviah, V. Machine vision detection of pests, diseases and weeds: A review. J. Phytol. 12, 9–19. https://doi.org/10.25081/jp.2020.v12.6145 (2020).
Backhaus, A., Bollenbeck, F. & Seiffert, U. Robust classification of the nutrition state in crop plants by hyperspectral imaging and artificial neural networks. Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS) https://doi.org/10.1109/WHISPERS.2011.6080898 (2011).
Schor, N. et al. Robotic disease detection in greenhouses: Combined detection of powdery mildew and tomato spotted wilt virus. IEEE Robot. & Autom. Lett. 1(1), 354–360. https://doi.org/10.1109/LRA.2016.2518214 (2016).
Es-saady, Y., El Massi, I., El Yassa, M., Mammass, D. & Benazoun, A. Automatic recognition of plant leaves diseases based on serial combination of two SVM classifiers. International Conference on Electrical and Information Technologies (ICEIT) 561–566 https://doi.org/10.1109/EITech.2016.7519661. (2016)
Sabrol, H. & Satish, K. Tomato plant disease classification in digital images using classification tree. 2016 International Conference on Communication and Signal Processing (ICCSP) 1242–1246. https://doi.org/10.1109/ICCSP.2016.7754351. (2016)
Krithika, P. & Veni, S. Leaf disease detection on cucumber leaves using multiclass support vector machine. 2017 International Conference on Wireless Communications, Signal Processing and Networking (WiSPNET) 1276–1281. https://doi.org/10.1109/WiSPNET.2017.8299969. (2017)
Islam, M., Dinh, A., Wahid, K. & Bhowmik, P. Detection of potato diseases using image segmentation and multiclass support vector machine. IEEE 30th Canadian Conference on Electrical and Computer Engineering (CCECE) 1–4 https://doi.org/10.1109/CCECE.2017.7946594. (2017)
Lu, J., Zhou, M., Gao, Y. & Jiang, H. Using hyperspectral imaging to discriminate yellow leaf curl disease in tomato leaves. Precision Agric. 19, 379–394. https://doi.org/10.1007/s11119-017-9524-7 (2018).
Selvaraj, M. G. et al. AI-powered banana diseases and pest detection. Plant Methods 15, 1–11. https://doi.org/10.1186/s13007-019-0475-z (2019).
Goel, L. & Nagpal, J. A systematic review of recent machine learning techniques for plant disease identification and classification. IETE Tech. Rev. 40(3), 423–439. https://doi.org/10.4018/IJSI.315657 (2023).
Asif, M. K. R., Rahman, M. A. & Hena, M. H. CNN based disease detection approach on potato leaves. 2020 3rd International Conference on Intelligent Sustainable Systems (ICISS) 428–432 https://doi.org/10.1109/ICISS49785.2020.9316021. (2020).
Mim, T. T., Sheikh, M. H., Shampa, R. A., Reza, M. S. & Islam, M. S. Leaves diseases detection of tomato using image processing. 2019 8th International Conference System Modeling and Advancement in Research Trends (SMART), 244–249. https://doi.org/10.1109/SMART46866.2019.9117437. (2019).
Singh, R., Krishnan, P., Bharadwaj, C. & Das, B. Improving prediction of chickpea wilt severity using machine learning coupled with model combination techniques under field conditions. Eco. Inform. 73, 101933. https://doi.org/10.1016/j.ecoinf.2022.101933 (2023).
Singh, R. N., Krishnan, P., Singh, V. & Das, B. Estimation of yellow rust severity in wheat using visible and thermal imaging coupled with machine learning algorithms. Geocarto Int. https://doi.org/10.1080/10106049.2022.2160831 (2023).
Mandal, N. et al. Spectral characterization and severity assessment of rice blast disease using univariate and multivariate models. Front. Plant Sci. 14, 1067189. https://doi.org/10.3389/fpls.2023.1067189 (2023).
Mayee, C. D. & Datar, V. V. Phytopathometry; Technical Bulletin-1 (Special Bulletin-3) 72 (Marathwada Agricultural University, 1986).
Cheng, H. D., Jiang, X. H., Sun, Y. & Wang, J. Color image segmentation: advances and prospects. Patt. Recogn. 34(12), 2259–2281. https://doi.org/10.1016/S0031-3203(00)00149-7 (2001).
Rafiq, M. et al. Exploration of resistance level against black scurf caused by Rhizoctonia solani in different cultivars of potato. Plant Stress 12, 100476 (2024).
Kuhn, M. et al. Package ‘caret’. R J. 223, 7 (2020).
Aschonitis, V. G. et al. A ranking system for comparing models’ performance combining multiple statistical criteria and scenarios: The case of reference evapotranspiration models. Environ. Model. Softw. 114, 98–111. https://doi.org/10.1016/j.envsoft.2019.01.005 (2019).
Chadha, S., Sharma, M. & Sayyed, A. Advances in sensing plant diseases by imaging and machine learning methods for precision crop protection. In Microbial management of plant stresses 157–183 Elsevier. https://doi.org/10.1016/B978-0-323-85193-0.00012-7 (2021).
Alves, K. S. et al. RGB-based phenotyping of foliar disease severity under controlled conditions. Trop. Plant Pathol. https://doi.org/10.1007/s40858-021-00448-y (2021).
Bhandari, M. et al. Assessing winter wheat foliage disease severity using aerial imagery acquired from small Unmanned Aerial Vehicle (UAV). Comput. Electron. Agric. 176, 105665. https://doi.org/10.1016/j.compag.2020.105665 (2020).
Ashourloo, D., Aghighi, H., Matkan, A. A., Mobasheri, M. R. & Rad, A. M. An investigation into machine learning regression techniques for the leaf rust disease detection using hyperspectral measurement. IEEE J. Sel. Topics Appl. Earth Obs. & Remote Sens. 9(9), 4344–4351. https://doi.org/10.1109/JSTARS.2016.2575360 (2016).
Chemura, A., Mutanga, O., Sibanda, M. & Chidoko, P. Machine learning prediction of coffee rust severity on leaves using spectroradiometer data. Tropical Plant Pathol. 43(2), 117–127. https://doi.org/10.1007/s40858-017-0187-8 (2018).
Osuna-Caballero, S., Olivoto, T., Jiménez-Vaquero, M. A., Rubiales, D. & Rispail, N. RGB image-based method for phenotyping rust disease progress in pea leaves using R. Plant Methods 19(1), 86. https://doi.org/10.1186/s13007-023-01069-z (2023).
Singh, R. N., Krishnan, P., Singh, V. K. & Banerjee, K. Application of thermal and visible imaging to estimate stripe rust disease in wheat using supervised image classification methods. Ecol. Inf. https://doi.org/10.1016/j.ecoinf.2022.101774 (2022).
Singh, R., Krishnan, P., Singh, V. K., Sah, S. & Das, B. Combining biophysical parameters with thermal and RGB indices using machine learning models for predicting yield in yellow rust affected wheat crop. Sci. Rep. 13(1), 18814. https://doi.org/10.1038/s41598-023-45682-3 (2023).
Duarte-Carvajalino, J. et al. Evaluating late blight severity in potato crops using unmanned aerial vehicles and machine learning algorithms. Remote Sens. 10(10), 1513. https://doi.org/10.3390/rs10101513 (2018).
Feng, Z. et al. Monitoring wheat powdery mildew based on hyperspectral, thermal infrared, and RGB image data fusion. Sensors 22(1), 31. https://doi.org/10.3390/s22010031 (2021).
Kang, X. et al. Assessing the severity of cotton Verticillium wilt disease from in situ canopy images and spectra using convolutional neural networks. Crop J. 11(3), 933–940. https://doi.org/10.1016/j.cj.2022.12.002 (2023).
Martinez-Martinez, V., Gomez-Gil, J., Machado, M. L. & Pinto, F. A. C. Leaf and canopy reflectance spectrometry applied to the estimation of angular leaf spot disease severity of common bean crops. PLoS ONE 13(4), e0196072. https://doi.org/10.1371/journal.pone.0196072 (2018).
Liu, Y., Zhang, Y., Jiang, D., Zhang, Z. & Chang, Q. Quantitative assessment of apple mosaic disease severity based on hyperspectral images and chlorophyll content. Remote Sens. 15(8), 2202. https://doi.org/10.3390/rs15082202 (2023).
Acknowledgements
The authors gratefully acknowledge ICAR-National Bureau of Plant Genetic Resources (NBPGR), New Delhi, for providing the planting material used in this study.
Author information
Authors and Affiliations
Contributions
A. K. D.: Conceptualization, Data curation, Formal analysis, Methodology, Writing – original draft; P. J.: Formal analysis, Writing – review & editing; K. S.: Data curation, Writing – review & editing; R. N.S.: Conceptualization, Formal analysis, Methodology, Writing – original draft; M.T.: Conceptualization, Formal analysis, Writing – original draft, Writing – review & editing; S.S.:Writing – review & editing;S. K.: Writing – review & editing; S. K.: Supervision, Writing – review & editing; R. K.: Writing – review & editing; K.S.: Writing – review & editing; R. K.: Writing – review & editing; A. D.: Supervision, Writing – review & editing; P. V. V. P.: Writing – review & editing; A. K. C.: Supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Dubey, A.K., Jha, P.K., Shubha, K. et al. Predicting yellow mosaic disease severity in yardlong bean using visible imaging coupled with machine learning model. Sci Rep 15, 24886 (2025). https://doi.org/10.1038/s41598-025-09176-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09176-8