Abstract
Current evidence for predictive models of post-stroke depression (PSD) risk based on machine learning (ML) remains limited. The aim of this study is to develop a superior predictive model based on ML algorithms for PSD in the Chinese population. We retrospectively gathered data from 507 patients who were hospitalized for ischemic stroke between June 2022 and August 2023 at a medical facility in China. The patients were separated into a training group (70%) and a validation group (30%). After selecting the core predictors of PSD using LASSO regression dimension reduction, six machine learning (ML) algorithms were used to statistically model the risk prediction of PSD. Based on five-fold cross-validation, we evaluated the predictive performance of the developed model. Shape Additive Explanation (SHAP) analysis was then used to interpret the best-performing model. PSD developed in 158 (31.16%) of the 507 eligible patients. Frontal lobe lesion, number of lesions, ALB, NIHSS, PSQI, and MMSE were considered significant predictors of PSD. The AUC(0.941), accuracy (0.876), sensitivity (0.822), specificity (0.899) , F1 score (0.802), and average precision (AP) value (0.858) of XGBoost model were superior to other ML models. The XGBoost model offers an interpretable PSD prediction tool using key clinical indicators, though external validation is needed to confirm generalizability.
Similar content being viewed by others
Introduction
Post-stroke depression (PSD), the most common psychiatric complication after stroke, is becoming a serious social and public health problem that seriously threatens patients’ functional recovery, rehabilitation response, and quality of life1. Stroke is one of the top three causes of disease burden2, and about 55% of stroke patients develop depression at some point after the vascular event3. Currently, PSD has a current prevalence of between 20 and 60%4, but low clinical detection rates and delayed diagnosis5, as well as significant underdiagnosis and treatment. Moreover, the effects of depression and stroke interact in a cyclical manner, as depression itself is identified as a substantial risk factor for stroke development6. Furthermore, the occurrence of PSD is associated with an increased risk of mortality and poor rehabilitation outcomes among stroke survivors7, further emphasizing the importance of post-stroke depression events.
Despite recent efforts to study predictors and risk factors for PSD, the evidence remains limited, and there are still no validated and reliable tools available for its screening and diagnosis8. Currently, the identification of PSD predominantly relies on the Diagnostic and Statistical Manual of Mental Disorders (DSM) guidelines and the integration of multiple depression scales9, yet the prediction of PSD continues to pose a significant challenge. PSD is a multifaceted disease that encompasses psychological, social, and biological aspects and their synergies. There are numerous risk factors for PSD, typically including genetic factors, age, sex, a history of depression, stroke severity, lesion location, levels of education, marriage, social support, and others9. Risk prediction models can be utilized to stratify patients by high or low risk and may also be integrated with multiple predictive factors to more accurately assess the potential for future disease events10. Despite the fact that some traditional predictive models have been developed recently for assessing PSD risk based on risk predictors1,11,12, research on the use of these models in combination with machine learning (ML) techniques is still scarce13. Building ML models based on risk factors introduces a novel paradigm for disease prediction. ML offers substantial advantages in data processing, induction, and synthesis. Its application value in risk factor assessment and disease outcome prediction is significant14. In addition, ML techniques often outperform traditional rule model algorithms, enhancing the performance and generalization capabilities of predictive models15. As a result, additional evidence is critical for further development of predictive PSD ML models to assist healthcare professionals in making earlier and more accurate diagnosis and treatment decisions.
The aim of this study was to identify high-risk factors affecting PSD and to develop a predictive ML model with superior performance to assist screening and decision-making in the clinical setting of PSD.
Methods
Study design
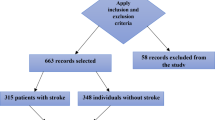
This retrospective study protocol for this study was approved by the Ethics Committee of the Affiliated Hospital of Shandong University of Traditional Chinese Medicine. All participants received and signed informed consent forms and were recruited to the Department of Neurology, Affiliated Hospital of Shandong University of Traditional Chinese Medicine between June 2022 and August 2023. Inclusion criteria were met by: (1) patients aged 18–80 years old; (2) patients diagnosed with ischemic stroke; (3) diagnosis confirmed by magnetic resonance imaging (MRI) or computed tomography (CT); and (4) patients with complete clinical data. Exclusion criteria were met by: (1) patients with severe dementia, previous psychosis, and depression, combined with neurodegenerative diseases; (2) patients who had recently received hormones, sedatives, antipsychotics or antidepressants; (3) patients with severe heart, liver, and kidney dysfunction; (4) patients with severe impairment of consciousness, hearing, and speech; (5) patients with epilepsy, infection, malignant tumor, infectious disease, blood disease, thyroid disease, heart failure, and heart valve disease; (6) patients who were pregnant or lactating; and (7) patients in the postoperative rehabilitation period. PSD was diagnosed by psychiatrists through a detailed clinical interview, in which they evaluated depressive symptoms in line with the criteria outlined in the fifth edition of the Diagnostic and Statistical Manual of Mental Disorders (DSM-5), and patients were identified by Hamilton Depression Scale-24 score ≥ 8 points16. In this study, we assessed the incidence of depression in patients during the 3–6 month follow-up period after stroke onset. After the strict implementation of inclusion and exclusion criteria, a total of 14 samples with missing data, 17 samples with abnormal data, and 10 patients with severe cognitive impairment (MMSE score < 18 points) were excluded. Finally, a total of 507 patients were included in the study, including 158 patients with PSD and 349 patients without PSD. The specific sample selection and modeling procedures are illustrated in Fig. 1.
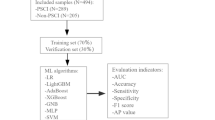
The flowchart of the sample determination and modeling process. The figure illustrates the flowchart of sample selection and ML modeling pipeline. Abbreviations: PSD: Post-stroke depression; MMSE: Mini-mental state examination; AUC: Area under the curve; XGBoost: eXtreme gradient boosting; LR: Logistic regression; LightGBM: Light gradient boosting machine; SVM: Support vector machines; GNB: Gaussian naive bayes.
Sample size calculation
According to previous research17, the average depression incidence within 5 years post-stroke in China was 29%. Using α = 0.05, β = 0.10, δ = 0.05, Zβ = 1.282, Zα = 1.960, and n = [(Zβ + Zα)/δ]2 × π0 × (1 − π0). A two-tailed test was conducted. It was calculated that the minimum sample size should be 317 patients according to the estimation formula. Considering cases lost to follow-up or shedding, the final number of patients included in the study was 507.
Cohort definition and screening of variables
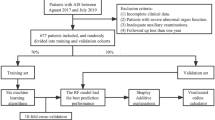
The study data was partitioned into a 7:3 ratio for training and validation purposes. The training set served to screen and develop the prediction model, while the validation set was utilized to confirm the outcomes of the training process. We selected general information, imaging test parameters, laboratory test parameters, scale evaluation parameters, and medication information for use as easily collected clinical indicators. These indicators were then used to enhance the development of the ML model in this study. Specifically, we gathered socio-demographic, disease history, family history, medication status, and lifestyle information for each patient through the use of questionnaires and related scales. Blood pressure was measured using three consecutive 2-min intervals, and the calculated average of these measurements was recorded as the patient’s blood pressure18. The imaging examination was conducted and evaluated by the hospital’s imaging department, with input from the neurologist. The imaging test parameters included the specific location, number of lesions, and side on which the lesions were located. The laboratory examination data, including blood routine and blood biochemistry, was meticulously analyzed by the professional laboratory physicians, providing reliable results. Furthermore, we incorporated the utilization of several relevant assessment scales to gather crucial patient information such as the National Institute of Health Stroke Scale (NIHSS), Mini-Mental State Examination (MMSE), Pittsburgh Sleep Quality Index (PSQI), and family function assessment, including adaptation, partnership, growth, affection, and resolve (APGAR).
Data balancing and preprocessing pipeline
To address class imbalance (349 non-PSD vs 158 PSD cases), we employed the BalanceCascade ensemble undersampling algorithm.In addition, to ensure comparability across features, we standardized key continuous variables (e.g., white blood cells, NIHSS, MMSE) to zero mean and unit variance.
Dataset partitioning and cross-validation
The complete dataset (N = 507) was rigorously partitioned into: ① Training set (70%): Used for n-fold cross-validation during model development Hold-out; ② Validation set (30%): Reserved exclusively for final performance evaluation We implemented stratified fivefold cross-validation on the training set, ensuring each fold maintained identical class distribution. The optimal model identified through cross-validation was retrained on the full training set, with all final performance metrics calculated strictly using the untouched validation set.
Statistical analyses
All statistical analyses were performed using SPSS Statistics 27.0, GraphPad Prism 9.0.0, R 3.6.3, and Python 3.7. Quantitative data were expressed as the mean ± standard deviation (SD) or the median (quartile)[M(P25, P75)] for better illustration. The differences between the groups were compared using either the independent test or Mann–Whitney U test for robust statistical analysis. Frequency (%) was used for ordinal data, and Chi-square or Fisher’s test was used to compare differences between groups. Dimensionality reduction methods such as univariate regression and LASSO regression were utilized to screen the most appropriate predictors for the analysis. Multiple prediction models were developed, including Logistic Regression (LR), LightGBM, AdaBoost, XGBoost, Gauss Naive Bayes (GNB), and support vector machines (SVM), to select the most accurate prediction model for PSD. We performed systematic hyperparameter tuning via Grid Search for all models, optimizing key parameters to maximize performance. Below are the optimal configurations: XGBoost: reg_lambda = 1, min_child_weight = 2, max_depth = 8, learning_rate = 0.3; LR: C = 0.1, penalty = l2, tol = 1e-6, max_iter = 100; LightGBM: num_leaves = 100, n_estimators = 5, max_depth = 20, learning_rate = 2; AdaBoost: n_estimators = 50, learning_rate = 1.0; SVM: C = 1.0, kernel = rbf’, tol = 0.001; GNB: var_smoothing = 1e-7. Five-fold cross-validation was utilized to identify the model with the best predictive accuracy, with the following performance indicators: AUC, accuracy, sensitivity, specificity, average precision (AP) value, and F1 score. Additionally, a correction curve was used to assess the model’s prediction ability. SHAP analysis was employed to interpret the prediction model’s results, while the clinical utility was evaluated through decision curve analysis (DCA). Furthermore, a nomogram was used to facilitate visualization of the ML prediction model. Two-tailed test determined that P < 0.05 was statistically significant.
Results
Comparative analysis of demographic and clinical characteristics between populations with and without PSD
After rigorous screening, 507 ischemic stroke patients aged 30 to 80 years were selected for this study. Of these patients, 158 had PSD and 349 did not have PSD, leading to a detection rate of 31.16% for PSD. The baseline data comparison between the two groups revealed notable variations in the following characteristics: gender, educational levels, marital status, smoking habits, diabetes, hyperlipidemia, atrial fibrillation, frontal lobe lesion, temporal lobe lesion, basal ganglia lesion, lesion count, white blood cell (WBC) count, red blood cell distribution width (RDW), albumin (ALB), platelet count (PAB), serum creatinine (SCr), fasting blood glucose (FBG), apolipoprotein A1 (APOA1), NIHSS, PSQI, and MMSE. The details were provided in Table 1.
Exploring PSD-associated phenomena through univariate regression
As shown in Table 2, we selected 22 significant indicators from Table 1 for logistic regression analysis. The results demonstrated that gender (women), marital status (unmarried, divorced or widowed), diabetes, hyperlipidemia, atrial fibrillation, smoking, frontal lobe lesion, temporal lobe lesion, basal ganglia lesion, number of lesions (multiple), WBC, RDW, ALB, PAB, FBG, APOA1, NIHSS, PSQI, and MMSE were significant influencing factors of PSD.
The LASSO regression model identified significant modeling predictors
To determine the most influential factors from the logistic regression model, we utilized LASSO regression to reduce the feature dimensions. To mitigate potential collinearity issues, the five-fold cross-validation technique was utilized, with a threshold of 0.042 for the standard error of the smallest distance. Ultimately, the model identified a subset consisting of six key predictors of PSD—frontal lobe lesion, the number of lesions, NIHSS, PSQI, MMSE. As illustrated in Fig. 2, these variables were determined to be most suitable for our predictive model.
Comparison and optimal model of different ML algorithms
Figures 3, 4 and Table 3 summarize the PSD prediction model’s design and performance. As can be seen in Fig. 3A and Table 3, six ML models have been trained and tested based on the provided training dataset. The results show that XGBoost, among the other models, demonstrates superior performance with an AUC of 0.992 (0.986–0.998), an accuracy of 0.964(0.948–0.979), a sensitivity of 0.970(0.962–0.977), a specificity of 0.961(0.937–0.984), and a F1 score of 0.944(0.921–0.966). The proposed second model is the AdaBoost model, which provides an AUC of 0.965 (0.950–0.981), accuracy of 0.899 (95%CI: 0.883–0.915), sensitivity of 0.926 (95%CI: 0.901–0.951), specificity of 0.887 (95%CI: 0.859–0.915), and F1 score of 0.851 (95%CI: 0.833–0.870). The third recommended model is SVM, which presents an AUC of 0.932 (0.907–0.957), an accuracy of 0.865(95%CI: 0.860–0.871), a sensitivity of 0.880 (95%CI: 0.855–0.904), a specificity of 0.859 (95%CI: 0.840–0.878), and an F1 score of 0.803 (95%CI: 0.799–0.806). Among the four suggested models, LR obtains an AUC of 0.898 (0.867–0.929), an accuracy of 0.821 (95%CI: 0.806–0.837), a sensitivity of 0.873 (95%CI: 0.843–0.904), a specificity of 0.798(0.763–0.833), and an F1 score of 0.753 (95%CI: 0.743–0.764). The fifth recommended model is GNB, which has an AUC of 0.870 (0.835–0.906), accuracy of 0.810 (95%CI: 0.791–0.828), sensitivity of 0.851(95%CI: 0.820–0.882), specificity of 0.791(95%CI: 0.751–0.830), and an F1 score of 0.737 (95%CI: 0.724–0.749). The LightGBM model has demonstrated subpar performance, with an AUC of 0.766 (0.710–0.822), accuracy of 0.800(0.759–0.840), sensitivity of 0.759(0.649–0.870), specificity of 0.818(0.756–0.880), and F1 score of 0.700(0.634–0.767), when compared with other ML models. As illustrated in Fig. 3B and Table 3, the performance of the different ML models trained and evaluated using the validation set data has been presented. The XGBoost model exhibited an outstanding predictive performance, achieving an AUC of 0.941 (0.895–0.987), an accuracy of 0.876(0.854–0.897), a sensitivity of 0.822(0.717–0.926), a specificity of 0.899(0.848–0.950), and an F1 score of 0.802(0.762–0.843). Furthermore, the PR curve analysis indicates that the AP values of the XGBoost model developed from the training set and the validation set are 0.962(95%CI:0.953–0.971, Fig. 4A) and 0.858(95%CI: 0.831–0.885, Fig. 4B), respectively. Furthermore, as shown in Fig. 5, the prediction model demonstrates a superior degree of calibration (P > 0.05), indicating a positive correlation between the predicted probability of PSD and the actual probability of PSD.
Plots of PR curve for different ML models. Larger AP values represent better model performance. Abbreviations: PR: Precision Recall; AP: Average precision; XGBoost: eXtreme gradient boosting; LR: Logistic regression; LightGBM: Light gradient boosting machine; SVM: Support vector machines; GNB: Gaussian naive bayes.
DCA curve of the best performing ML model
Figure 6 presents the decision curve analysis (DCA) of the XGBoost model, demonstrating its clinical applicability. The results show that the model provides substantial net benefit across multiple threshold probabilities, indicating strong clinical utility.
Nomogram of PSD prediction model
Figure 7 depicts a nomogram designed to estimate the risk of PSD in the presence of ALB, NIHSS, PSQI, MMSE, frontal lobe lesion, and number of lesions. The final score was a combination of individual score calculation, and the majority of the study’s patients had a total risk point for PSD that was situated between 0 and 240. Thus, we can predict a patient’s likelihood of developing PSD based on the risk value corresponding to their total score.
Nomogram model to predict the PSD. The nomogram model’s application protocol is as follows: First, a single node value is assigned to each variable axis, subsequently the corresponding score is derived under each variable state, and finally the scores of all variables are aggregated to generate the patient’s total score, which then corresponds to the lowest risk value. Abbreviations: ALB: Albumin, PSQI: Pittsburgh sleep quality index, MMSE: Mini-mental state examination, NIHSS: National institutes of health stroke scale.
SHAP results of the model
As illustrated in Fig. 8, personalized prediction profiles demonstrate variable feature importance through SHAP value analysis. Each predictor’s SHAP value reflects its directional impact on PSD risk prediction, where positive values correspond to increased risk probability. The current study results indicate that the PSQI is the most significant predictor of predicting PSD. Elevated PSQI scores showed positive associations with both Shapley values and PSD risk. Furthermore, elevated NIHSS scores, frontal lobe lesions, and the presence of multiple lesions (indicated in pink) demonstrated positive correlations with Shapley values, representing significant risk factors for post-stroke depression. Conversely, reduced MMSE scores and lower albumin levels (denoted in blue) showed inverse relationships with Shapley values, serving as protective indicators against post-stroke depression (Fig. 8A and B).
Feature importance ranking and correlation direction of predictors based on SHAP analysis.The vertical axis displays feature information and the horizontal axis displays SHAP data. The correlation between the predictor and PSD risk was shown using two distinct colors, where pink indicated a positive correlation and blue indicated a negative correlation.
Figure 9 shows several randomly selected cases using SHAP force plots to predict the risk of PSD and Non-PSD. Figure 9A illustrates the SHAP force plot that correctly predicts PSD; the SHAP value indicates that a lower ALB and higher PSQI support the prediction model, with a prediction probability of 0.830. Figure 9B demonstrates the SHAP force plot that correctly predicts Non-PSD; the SHAP value indicates that a lower ALB and MMSE, higher PSQI support the prediction model, with a prediction probability of 0.930.
à
Discussion
This study used an excellent ML model to develop and validate a model based on frontal lobe lesion, the number of lesions, ALB, NIHSS, PSQI, and MMSE for predicting PSD in Chinese populations. The XGBoost model, with its superior predictive performance, has an AUC of 0.941, an accuracy of 0.876, a sensitivity of 0.822, a specificity of 0.899, and an F1 score of 0.802. With an up-to-date PSD prediction tool, clinicians can gain a more comprehensive understanding and thus improve their management of the risk associated with PSD.
It is critical in this study with its many indicators and small sample size to implement scientific methods to accurately and judiciously select key predictor variables. To avoid problems such as overfitting, generalization failure, and feature selection bias due to limited sample sizes, this study employs a set of strategies such as dimensionality reduction, cross-validation, and alternative methods for identifying the most relevant predictors for building effective prediction models. LASSO regression analysis is advantageous in its ability to precisely select characteristic variables by reducing noise variables to zero19, especially in scenarios involving small samples, multiple variables, and high-dimensional characteristic datasets. In recent years, LASSO regression analysis has gained tremendous popularity in the process of predicting risk factors of diseases, which involves building predictive models20,21,22. The traditional significance analysis of the current study is more susceptible to the effect of the number of samples and features, since the initial number of variables is large. Consequently, it is more likely that the selection of core variables may be hindered by the limitations of statistical analysis results. In this study, we used an LR model to identify key independent factors and a LASSO regression analysis to reduce dimensionality. By carefully selecting the most critical and vital factors, we have developed different models for both training and evaluation. This method can effectively identify the core factors that influence the development of PSD prediction models.
The findings of this research highlight the importance of patients diagnosed with ischemic stroke closely monitoring their frontal lobe lesions, the number of lesions, ALB, NIHSS, sleep patterns, and cognitive function. Further SHAP analysis revealed that poor sleep quality stands out as the first feature to be prioritized in the ischemic stroke population. Emerging evidence suggests that sleep disorders, especially PSD, are significantly associated with the occurrence and progression of ischemic stroke. In addition, vitamin D deficiency may contribute to this association23. The PSQI, an important indicator of subjective sleep quality, effectively reflects the patient’s normal sleep state. Patients with higher scores on the PSQI scale generally have worse sleep quality, and the components of this scale, such as subjective sleep quality and sleep disturbance, are found to be correlated with PSD24. Furthermore, another research indicates that inadequate sleep before a stroke could potentially be a unique factor contributing to the development of PSD25. Sleep disorders increase the risk of depression by affecting neuroendocrine functions and disrupting circadian rhythm26. Research has shown that the development of PSD in high-risk individuals may be tied to neurobiological mechanisms such as inflammation, hypoperfusion, and neurosuppression27. Long-term insomnia can lead to frequent recurrence of depression symptoms in patients with post-stroke depression, which can reduce the body’s immunity, thereby increasing the risk of other diseases to some extent, ultimately exacerbating the prognosis of PSD. In addition, patients with PSD have poor self-regulation and damage neurological function, which will further exacerbate sleep disorders. SHAP analysis suggests that the second feature to focus on is cognitive function. Cognitive impairment is a common complication after stroke and is also thought to be closely related to the occurrence of PSD. It has been reported that cognitive impairment is considered an independent predictor of major depression after stroke28. In fact, depression and cognitive decline in post-stroke patients can interact. On the one hand, patients with impaired cognitive function often suffer from depression or aggravation of original depressive symptoms due to decreased learning ability and decreased self-emotional regulation ability, and the mechanism of its effect may be related to changes in the internal structure of the brain29. On the other hand, PSD patients are often affected by reduced subjective initiative in treatment, thus affecting the recovery of neurological and cognitive functions, which may be related to changes in the levels of some neurotransmitters and brain-derived neurotrophic factor30. The third feature that needs close attention in the stroke population is serum ALB. A previous observational cohort study determined that lower serum ALB is associated with long-term depressive symptoms in the elderly after stroke31. Although both malnutrition and immune/inflammatory responses have been reported to be associated with the pathogenesis or exacerbation of PSD32,33, previous evidence suggests that the mechanism by which low ALB are associated with depression is more likely to be attributed to an immune/inflammatory response than to poor nutritional status34,35,36. NIHSS is the fourth predictor highlighted, which has been considered to be an effective indicator of neurological impairment and is considered to have positive predictive ability for PSD37. The higher the NIHSS score, the more serious the neurological function impairment after stroke, the worse the recovery of the patient’s physical function, aggravating the patient’s psychological burden and generating negative emotions, thus increasing the risk and severity of PSD38. In general, neurological impairment indicates pathological changes in brain tissue, which affect neurotransmitters and depression-related parts. Coupled with changes in psychological pressure during rehabilitation, it is easy to lead to depression. In addition, current results also suggest that lesion location is an important predictor of PSD. The occurrence of PSD is mostly based on the presence of infarcts in frontal lobe, temporal lobe and hippocampus, which regulate the emotional function of human beings39,40. Multiple studies have shown that the left frontal lobe and basal ganglia lesions are more likely to cause depression after stroke41,42,43. Depression has been found to be associated with frontal lobe lesions, while apathy is more often associated with basal ganglia lesions44. When acute brain injury occurs, the inhibition of enzymes during ischemia leads to a decrease in the synthesis of monoamine neurotransmitters in the brain. The transmitters reach the frontal cortex through the basal ganglia and thalamus behind the deep cortex, forming a frontal subcortical neural circuit, which may be related to the regulation of emotional, sleep and cognitive functions45. Once there are lesions in the frontal lobe and basal ganglia, it reflects the damage of the circuit, and the loss of emotional, sleep and cognitive regulation, which leads to the occurrence of depression.
ML, as the core of artificial intelligence, surpasses traditional rule model algorithms in terms of enhancing prediction model performance and generalization ability. It also provides a new paradigm for modeling epidemiological risk scenarios. Studies on the use of ML predictive models in depression have been conducted in recent years, but similar research with individuals with ischemic stroke is lacking. An analysis using data from the 2020 Korea National Health and Nutrition Examination developed a predictive model for depression using three machine learning algorithms. It was found that the Logistic Regression model exhibited superior performance, achieving an AUC of 0.812 (95% CI: 0.787–0.837)46. In another previous study, researchers built a ML predictive model for PSD using blood metal elements. The study found that the XGBoost model was an effective algorithm for diagnosing depression, achieving an AUC of 0.88 and an accuracy of 0.8947. Abdulla et al.48 developed an Extreme Gradient Boosting model based on inflammation Markers for patients with depression, and its AUC and accuracy were 0.95 and 0.89, respectively, and it is suggested that MCP-1 and IL-1βare significant predictors of depression. Liu et al.49 developed a postpartum depression risk model, showing that the XGBoost model had the best identification, with an AUC of 0.789 (95% CI 0.742–0.836). In addition, research from multiple medical institutions in Taiwan using data revealed that the four ML models achieved average sensitivity and specificity values of 0.30–0.48 and 0.83–0.91, respectively. Additionally, various predictors of PSD at various time points were identified, such as age, height, weight, DBP, etc.13 Gong et al.12 built five ML model for the risk with PSD based on six liver function test indices, the results show that the gradient boosting decision tree (GBDT) model obtain the best prediction performance (AUC = 0.761, F1 = 0.498). Zhang et al.50 used LASSO and random forest analysis to identify two characteristic genes, SDHD and FERMT3, which proved to have good reliability and validity, with AUCs of SDHD and FERMT3 being 0.896 and 0.964. Our current ML model appears to be simpler and more economical than previously reported, because we take into account common clinical indicators that are easily accessible in clinical practice. More importantly, our SHAP analysis provides interpretability to the predictive model, helping to discern the significant predictors and the implications of their positive and negative effects. In addition, we evaluated the performance of the XGBoost model using five-fold cross-validated ROC curves, DCA analysis, and calibration curves. This revealed the model’s reliability and applicability, providing further evidence of its effectiveness.
However, there are several limitations to this study that should be considered. First, the study faced challenges in terms of time, human resources and other resources. This has resulted in a limited number of samples collected by a single centre, which may still lead to bias and imbalance in sample selection, despite the necessary data pre-processing and sample balancing measures we have taken. Second, this current study has only performed internal validation of the prediction model, which has limited its ability to accurately assess the model’s generalization capabilities due to the lack of multicenter external validation. Third, despite our efforts to systematically select predictors and algorithms, some critical indicators (e.g., socioeconomic status) and advanced machine learning methods (e.g., ensemble models) were excluded due to data availability and computational constraints. To further enhance the prediction performance and generalization ability of our PSD prediction model, we plan to evaluate and improve the model based on multicenter and large sample prospective clinical data in the future. We will also focus on increasing data diversity, selecting appropriate predictors (e.g., social support, socioeconomic status) optimizing model training methods, and exploring integrated learning models. In addition, we will actively seek external validation opportunities to further expand the extrapolation of the results of this study and promote model updating.
Conclusion
In summary, this study developed six ML models based on the analysis of clinical indicators for patients with PSD, and the model developed using the XGBoost algorithm showed the best performance, with an AUC of 0.941 and an accuracy of 0.876. This model has robust predictive performance and broad applicability, enabling clinicians to accurately measure the risk of depression in stroke patients based on their unique clinical diagnosis and treatment characteristics, enabling them to make prompt and effective clinical diagnosis and treatment decisions.
Data availability
Data sets cannot be made publicly available due to the privacy of research participants being compromised. However, minimal data can be shared with interested individuals upon reasonable request from the corresponding author.
References
Luo, S. et al. Establishment and verification of a nomogram model for predicting the risk of post-stroke depression. PeerJ 11, e14822 (2023).
WHO. Global Health estimates 2016: Deaths by cause, age, sex, by country and by region, 2000–2016 (World Health Organization, 2018).
Ayerbe, L., Ayis, S., Wolfe, C. D. & Rudd, A. G. Natural history, predictors and outcomes of depression after stroke: Systematic review and meta-analysis. Br. J. Psychiatry 202, 14–21 (2013).
Frank, D. et al. Pathophysiology and current drug treatments for post-stroke depression: A review. Int. J. Mol. Sci. 23(23), 15114 (2022).
Williams, L. S., Ghose, S. S. & Swindle, R. W. Depression and other mental health diagnoses increase mortality risk after ischemic stroke. Am. J. Psychiatry 161(6), 1090–1095 (2004).
Pan, A. et al. Depression and risk of stroke morbidity and mortality: A meta-analysis and systematic review. JAMA 306, 1241–1249 (2011).
Castilla-Guerra, L., Fernandez Moreno, M. D. C., Esparrago-Llorca, G. & Colmenero-Camacho, M. A. Pharmacological management of post-stroke depression. Expert Rev. Neurother. 20(2), 157–166 (2020).
Das, J. & Rajanikant, G. K. Post stroke depression: The sequelae of cerebral stroke. Neurosci. Biobehav. Rev. 90, 104–114 (2018).
Guo, J., Wang, J., Sun, W. & Liu, X. The advances of post-stroke depression: 2021 update. J Neurol. 269(3), 1236–1249 (2022).
Wolff, R. F. et al. PROBAST: A tool to assess the risk of bias and applicability of prediction model studies. Ann. Intern. Med. 170(1), 51–58 (2019).
Ladwig, S., Ziegler, M., Südmeyer, M. & Werheid, K. The post-stroke depression risk scale (PoStDeRiS): Development of an acute-phase prediction model for depression 6 months after stroke. J. Acad. Consult. Liaison Psychiatry 63(2), 144–152 (2022).
Gong, J. et al. Liver function test indices-based prediction model for post-stroke depression: A multicenter, retrospective study. BMC Med. Inform. Decis. Mak. 23(1), 127 (2023).
Chen, Y. M. et al. Predicting new-onset post-stroke depression from real-world data using machine learning algorithm. Front. Psychiatry 14, 1195586 (2023).
Wang, M. et al. Establishment and evaluation of early prediction model for severe disease risk of COVID-19 patients based on XGBoost. J. Army Med. Univ. 44(03), 195–202 (2022).
Yang, C. et al. Clinical research and application of a machine learning-based prognostic model for predicting aneurysmal subarachnoid haemorrhage. Chin. J. Hosp. Pharm. 44(03), 257–262 (2024).
Luo, S. et al. Nomogram model for predicting the risk of post-stroke depression based on clinical characteristics and DNA methylation. PeerJ 11, e16240 (2023).
Huang, W. L., Liao, X. Q. & Fang, X. Quality evaluation of clinical practice guidelines related to post-stroke depression and content analysis of nursing practice. Chin. J. Nurs. Manag. 20(10), 1570–1576 (2020).
Ma, C. C. et al. Associations of objectively measured sleep characteristics and incident hypertension among police officers: The role of obesity. J. Sleep Res. 29, e12988 (2020).
Li, Z. T. & Sillanpää, M. J. Overview of LASSO-related penalized regression methods for quantitative trait mapping and genomic selection. Theor. Appl. Genet. 125(3), 419–435 (2012).
Misumi, K. et al. Derivation and validation of a machine learning-based risk prediction model in patients with acute heart failure. J. Cardiol. 81(6), 531–536 (2023).
Chen, G. et al. A machine learning model based on MRI for the preoperative prediction of bladder cancer invasion depth. Eur. Radiol. 33(12), 8821–8832 (2023).
Dai, A. et al. Incorporating intraoperative blood pressure time-series variables to assist in prediction of acute kidney injury after type a acute aortic dissection repair: An interpretable machine learning model. Ann. Med. 55(2), 2266458 (2023).
He, W. & Ruan, Y. Poor sleep quality, vitamin D deficiency and depression in the stroke population: A cohort study. J. Affect. Disord. 308, 199–204 (2022).
Nguyen, T. T. P. et al. Post-stroke depression in Vietnamese patients is associated with decreased sleep quality and increased fatigue: A one-institution cross-sectional analysis. Sleep Breath 27(4), 1629–1637 (2023).
Dong, L. et al. Pre-stroke sleep duration and post-stroke depression. Sleep Med. 77, 325–329 (2021).
Tsuno, N., Besset, A. & Ritchie, K. Sleep and depression. J. Clin. Psychiatry 66(10), 1254–1269 (2005).
Robinson, R. G. & Jorge, R. E. Post-stroke depression: A review. Am. J. Psychiatry 173, 221–231 (2015).
Ojagbemi, A., Akinyemi, R. & Baiyewu, O. Cognitive dysfunction and functional limitations are associated with major depression in stroke survivors attending rehabilitation in Nigeria. NeuroRehabilitation 34(3), 455–461 (2014).
Li, S. W. et al. Study on the relationship between mild cognitive impairment and post-stroke depression. Chin. J. Stroke 13(06), 631–635 (2018).
Zhang, Z. H., Wu, L. N., Song, J. G. & Li, W. Q. Correlations between cognitive impairment and brain-derived neurotrophic factor expression in the hippocampus of post-stroke depression rats. Mol. Med. Rep. 6(4), 889–893 (2012).
Pascoe, M. C., Skoog, I., Blomstrand, C. & Linden, T. Albumin and depression in elderly stroke survivors: An observational cohort study. Psychiatry Res. 230(2), 658–663 (2015).
Hua, J., Lu, J., Tang, X. & Fang, Q. Association between geriatric nutritional risk index and depression after ischemic stroke. Nutrients 14(13), 2698 (2022).
Chen, H. et al. The role of indoleamine 2,3-dioxygenase 1 in early-onset post-stroke depression. Front. Immunol. 14, 1125634 (2023).
Maes, M. et al. Anthropometric and biochemical assessment of the nutritional state in depression: Evidence for lower visceral protein plasma levels in depression. J. Affect. Disord. 23(1), 25–33 (1991).
Maes, M. et al. Lower serum zinc in major depression is a sensitive marker of treatment resistance and of the immune/inflammatory response in that illness. Biol. Psychiatry 42(5), 349–358 (1997).
Maes, M. et al. Serotonin-immune interactions in major depression: Lower serum tryptophan as a marker of an immune-inflammatory response. Eur. Arch. Psychiatry Clin. Neurosci. 247(3), 154–161 (1997).
Elias, S. et al. Factors associated with post-stroke depression in the acute phase of ischemic stroke: A cross-sectional study. Clin. Neurol. Neurosurg. 223, 107505 (2022).
VanDerwerker, C. J. et al. Combining therapeutic approaches: rTMS and aerobic exercise in post-stroke depression: A case series. Top Stroke Rehabil. 25(1), 61–67 (2018).
Koen, J. D. & Yonelinas, A. P. The effects of healthy aging, amnestic mild cognitive impairment, and Alzheimer’s disease on recollection and familiarity: A meta-analytic review. Neuropsychol. Rev. 24(3), 332–354 (2014).
Joko, T. et al. Patterns of hippocampal atrophy differ among Alzheimer’s disease, amnestic mild cognitive impairment, and late-life depression. Psychogeriatrics 16(6), 355–361 (2016).
Metoki, N. et al. Relationship between the lesion location of acute ischemic stroke and early depressive symptoms in Japanese patients. Ann. Gen. Psychiatry 15, 12 (2016).
Tang, W. K. et al. Association of frontal subcortical circuits infarcts in poststroke depression: a magnetic resonance imaging study of 591Chinese patients with ischemic stroke. J. Geriatr. Psychiatry Neurol. 24, 44–49 (2011).
Shi, Y. Z. et al. The relationship between frontal lobe lesions, course of post-stroke depression, and 1-year prognosis in patients with first-ever ischemic stroke. PLoS ONE 9, e100456 (2014).
Hama, S. et al. Post-stroke affective or apathetic depression and lesion location: Left frontal lobe and bilateral basal ganglia. Eur. Arch. Psychiatry Clin. Neurosci. 257, 149–152 (2007).
He, L. & Shao, H. Y. Research progress of correlation between post-stroke depression and lesion location. Chin. J. Geriatr. Cardio Cerebrovasc. Dis. 18(10), 1115–1117 (2016).
Kim, S. W. & Chang, M. C. The usefulness of machine learning analysis for predicting the presence of depression with the results of the Korea National Health and Nutrition Examination Survey. Ann. Palliat. Med. 12(4), 748–756 (2023).
Xia, F., Li, Q., Luo, X. & Wu, J. Machine learning model for depression based on heavy metals among aging people: A study with National Health and Nutrition Examination Survey 2017–2018. Front. Public Health 10, 939758 (2022).
Abdulla, H., Maalouf, M., & Jelinek, H. F. Machine Learning for the Prediction of Depression Progression from Inflammation Markers. In Annual International Conference of the IEEE Engineering in Medicine & Biology Society Vol. 2023, 1–4 (2023).
Liu, H. et al. An optimization for postpartum depression risk assessment and preventive intervention strategy based machine learning approaches. J. Affect. Disord. 328, 163–174 (2023).
Zhang, X. et al. Machine learning algorithms assisted identification of post-stroke depression associated biological features. Front. Neurosci. 17, 1146620 (2023).
Acknowledgements
It is with great gratitude that I express my appreciation to my research partners and our funding agency for their unwavering encouragement and support throughout this study.
Funding
This work was supported by the National Natural Science Foundation of China (General Program; Grant 82474422), China Postdoctoral Science Foundation (Grant GZB20240036), and Peking University Medical Youth Science and Technology Innovation Yangfan Programme (Grant BMU2025YFJHPY006). The funders had no involvement in study conception, data acquisition, analysis, interpretation, or manuscript preparation and submission decisions.
Author information
Authors and Affiliations
Contributions
H.J. and J.L. were the main coordinators of the project and were responsible for the design of the study. T.Z. and X.Z. drafted the manuscript of this paper. S.L. and G.Z. were involved in the supervision of data collection and stratification. D.L. and X.Z. contributed to data compilation and analysis. All authors contributed intellectually to this manuscript and have approved this final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
This study was conducted in accordance with the ethical guidelines and approval of the Ethics Committee of the Affiliated Hospital of Shandong University of Chinese Medicine. All participants provided written informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhong, X., Zhao, T., Lv, S. et al. Development and interpretation of a machine learning risk prediction model for post-stroke depression in a Chinese population. Sci Rep 15, 28602 (2025). https://doi.org/10.1038/s41598-025-09322-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-09322-2