Abstract
Laparoscopy is the gold standard for diagnosing endometriosis; however, it is an invasive and costly method. Recent studies offer a non-invasive approach based on extracellular vesicle miRNA. Despite this, no consensus diagnostic biomarker has been identified to date. For addressing this gap, we decided to investigate plasma derived extracellular vesicle associated candidate miRNAs. In order to identify candidate miRNAs, a comprehensive search was performed in PubMed database using the search terms “micro-RNA” and “endometriosis”. Then, bioinformatics analysis was performed utilizing the miRTarBase database, Enrichr, and relevant software. During the experimental phase, the presence of candidate miRNAs was assessed in blood samples of 13 women with severe endometriosis, confirmed through laparoscopy or doppler sonography, as well as in 11 endometriosis-free women, as control group. After literature review of 405 articles published between 2007 and 2023, followed by bioinformatics analysis, were identified five miRNAs (miR-451a, 148a, 23b, 100, and 154) as candidate miRNAs. Subsequently, the expression levels of miR-451a, 148a, 23b, and 100 found to exhibit differences between the case and control groups. Our study suggests to serve of these miRNAs as a potentially diagnostic biomarker panel for endometriosis, however it needs to be confirmed by future studies with large diagnostic validation.
Similar content being viewed by others
Introduction
Endometriosis is an inflammatory disorder that affects 10–15% of women of reproductive age and is characterized by the presence of endometrial tissue outside the uterus1. The symptoms are heterogeneous, with the most common including pelvic pain, dysmenorrhea, and infertility2. Often, Nonspecific symptoms may lead to misinterpretation, resulting in delays in diagnosis3. Visualization of ovarian endometriotic cysts and deep endometriotic lesions can be performed using ultrasonography and MRI; however, these methods are unable to identify peritoneal endometriosis or endometriosis-associated adhesions4. The American Society for Reproductive Medicine (ASRM) classification system categorizes endometriosis into four stages based on the number of lesions and the degree of infiltration. More advanced stages of endometriosis (III-IV) can lead to complications such as pelvic pain, which may lead to a hysterectomy. Moreover, Pearce et al. demonstrated that malignant transformation of ovarian endometrioma into ovarian cancer, particularly the clear cell and endometrioid subtypes5.
The gold standard diagnosis of endometriosis is performed by laparoscopy with direct visualization. This procedure is invasive, costly, and in endometrioma cases through the excision of ovaries can lead to subfertility or infertility in women since is associated with diminished ovarian reserve6. Therefore, the need for a non-invasive, biomarker-based approach is evident7. To the best of our knowledge, reliable diagnostic biomarkers are still unavailable.
Extracellular vesicles (EVs) refer to a diverse population of vesicles that vary in size and biogenesis mechanism, which are released into the extracellular space by various cell types8. Exosomes and microvesicles are two main subtypes of EVs that can transfer signals to recipient cells, establishing a new communication mechanism between cells, which they are considered as a novel communication paradigm9. mRNA and miRNA derived from EVs are functional and can exchange genetic material among cells10. Recently, there has been increasing interest in utilizing the content of EVs—especially miRNA—as a diagnostic biomarker11.
miRNAs are small non-coding RNA molecules, typically ranging from 20 to 24 nucleotides in length. It is estimated that approximately 30% of transcriptomes are regulated by miRNAs12,13. A miRNA can be found bound to proteins, such as Argonaute proteins, sorted in EVs, or circulating freely in plasma/ serum. miRNAs play vital functional roles in cells, such as differentiation, neoplastic transformation, regeneration, and cell replication14. Several studies have demonstrated that the dysregulation of miRNAs is involved in endometriosis and may develop lesion implantation, angiogenesis, proliferation, adhesion, and increased levels of cytokines, such as migration inhibitory factor (MIF)15. In a study conducted by Caroline Frisendahl et al., it was shown that the downregulation of miR-193 and miR-374 promotes cell migration and may serve as therapeutic targets for this condition16. Research has also evaluated the expression of miRNAs in the serum, plasma17and ectopic / eutopic tissues of endometriosis patients18.
Based on current knowledge, a consensus biomarker for the diagnosis of endometriosis has not yet been identified. However, studies have demonstrated that the number of EVs elevates in the early stages of the disease19. The spread and stability of EVs, make them an ideal biomarker14.
Consequently, we have chosen to measure the expression levels of candidate miRNAs found in plasma-derived EVs. This decision is based on the belief that these miRNAs could serve as non-invasive diagnostic biomarkers for endometriosis, representing a promising avenue for exploration.
Results
Results of effect sized analysis
Cohen’s d values for the significantly dysregulated miRNAs ranged from 0.61 to 0.92 indicating moderate to large effect sizes. Despite the limited sample size, these effect sizes suggest meaningful biological differences between the endometriosis and control groups (Supplementary Table S1).
Selection of MiRNAs and prediction of targets
Using bioinformatics tools, we evaluated potential targets and signaling pathways of microRNAs in the 60 selected articles. The analysis of the results ultimately yielded a list of 15 miRNAs (Table 1).
MiRTarBase database, enrichment, and pathway analysis
miR-148a, miR-451a, miR-154, miR-23b, and miR-100 were selected as candidate microRNAs for further investigation due to their potential involvement in the development of endometriosis. The selection criteria were based on the target genes regulated by these microRNAs and their established significance in the pathogenesis of endometriosis.
miR-451a
No pathogenic single nucleotide polymorphisms (SNPs)of miR-451a were identified. The five signaling pathways in which miR-451a received the highest scores were analyzed using Enrichr software (Supplementary Table S2).
miR-148a
The miR-148a molecule is an intergenic microRNA. The signaling pathways associated with this miRNA suggest a potential link to cancer. To date, no pathogenic SNPs have been reported for this miRNA (Supplementary Table S3).
miR-23b
miR-23b is located in intron 3 of the AOPEP (aminopeptidase O) gene on chromosome 9q, and no pathogenic SNPs associated with this miRNA have been reported (Supplementary Table S4).
miR-100
miR-100 is a secretory microRNA that is studied in various fluids, tissues, and EVs27,33. It is an intergenic RNA located on chromosome 11q (Supplementary Table S5).
miR-154
miR-154 is an intergenic microRNA, and no SNPs have been reported for it. Bioinformatics analysis indicates that miR-154 is involved in the Hippo signaling pathway (Supplementary Table S6).
Final list of MiRNAs selection
The dysregulation of miR-451a in plasma, serum, and endometriotic tissue has been investigated. miR-451a plays a role in regulating the expression of the MIF cytokine. miR-23b and miR-23a are downregulated in both ectopic and eutopic endometrium when compared to normal endometrium18. Microarray analysis has demonstrated that miR-100-5p is expressed differentially in ectopic tissue compared to eutopic tissue27. Furthermore, bioinformatics analysis has identified COX-2 as the target of miR-100.
Bioinformatics analyses have demonstrated that miR-154 plays a role in the Hippo signaling pathway. In addition, miR-154, in conjunction with other microRNAs such as miR-196b-5p, may serve as a potential diagnostic biomarker for endometriosis30. Research indicates that miR-148a regulates the expression of DNMT1 and HLA-G mRNA34 (Table 2). In the first step, studies conducted without any time limitations up to April 2023 were reviewed. Subsequently, five miRNAs were selected for the laboratory phase using bioinformatics analysis.
STRING model
Protein-protein interaction networks were analyzed using the STRING database (Fig. 1).
The expression level of Circulating and EV- associated candidate MiRNAs
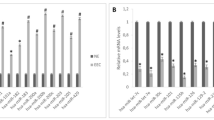
After successfully passing the normality test, the data were analyzed using a parametric test. The data were normalized with miR-16. Our findings indicated that miR-451, miR-23b, miR-148a, and miR-100 were significantly downregulated in the endometriosis group compared to the control group. Surprisingly, miR-154 was not detected in either group (Fig. 2).
qRT-PCR efficiency results
Amplification efficiency determined via Lin Reg PCR indicated a range between 1.91 and 1.92 for all candidate miRAN, except for miR-154, which indicate an efficiency of 1.0. This result supports the specificity and reliability of the qRT-PCR reactions, while indicating a lack of detectable expression for miR-154 in both case and control group (Supplementary Table S7).
Discussion
The aim of this study was to investigate whether the expression levels of plasma derived circulating and EV-associated candidate miRNAs changes and the end can serve as a potential biomarker for the disease. EV-miRNAs have higher stability, specificity and sensitivity compared other type biomarkers such as circulating cell- free DNA, making them potentially more effective for early disease detection and monitoring. The packaging of EVs is purposeful and they are tissue specific allowing for targeted diagnosis and monitoring. In additional they can more accurate and reliable than traditional biomarker like proteins35,36,37.
In the first step, we identified candidate miRNAs based on previous literature. Subsequently, through bioinformatics analysis, we performed a final selection of miRNAs. The expression levels of these miRNAs had been evaluated in serum, plasma, and tissue in previous studies17,18,30. The expression levels of individual microRNAs exhibit considerable variability across studies, attributable to multiple confounding factors, including: sample size, sample type (serum, plasma, tissue, or extracellular vesicles), menstrual cycle phase of subjects, experimental techniques used and sampling conditions. To minimize inter-individual variability, we collected all samples in the morning and restricted sampling to early proliferative phase (menstrual cycle days 3–5). Notably, the expression profile of EV-associated microRNAs diverges from cellular or tissue-derived patterns. This discrepancy arises from two biologically significant mechanisms: active sorting and cellular clearance.
miRNAs can regulate the expression levels of messenger RNAs (mRNAs) and play an essential role in both normal and abnormal biological processes. In addition to cellular miRNAs, they can secrete from different cell types via EVs38. EVs transport and also transduce miRNAs into target cells, even distant via the blood circulation. EVs- derived miRNA changes the expression levels and functions of recipient cells39. Therefore, investigating plasma-derived miRNAs will aid for endometriosis progression, therapy, and the identification of novel diagnostic biomarker. We chose candidate miRNAs based on their targets and signaling pathway, which play an important role in endometriosis.
The macrophage migration inhibitory factor (MIF) is one of the targets of miR-451a. Endometriotic cells secrete the MIF cytokine and has mitogenic properties that promote the growth of these cells. MIF can stimulate the secretion of factors associated with cell proliferation and angiogenesis. Elevated levels of MIF have been shown in endometriotic tissues and peritoneal fluid of patients with endometriosis40. The expression level of miR-451a is inversely related to the extent of lesions17. Also, the miR-451a serves as a reproducible diagnostic serum biomarker1. Our findings indicate that miR-451a is downregulated in EV-derived plasma, which may influence the progression of the disease.
The steroidogenic factor-1 (SF-1) regulates through miR-23a and miR-23b. SF-1 and other estrogenic enzymes regulate estrogen formation that has a significant role in endometriosis. An increase in the expression level of SF-1 is associated with a reduction in the expression levels of miR-23a and miR-23b in endometrial tissue. The upregulation of SF-1 leads to an increase in the expression of steroidogenic acute regulatory protein (stAR) and cytochrome P450 aromatase (CYP19A1), both of which are essential components of the estrogen synthesis pathway18. Also, our study indicates that the expression level of miR-23b reduce in EV-plasma derived can lead to the dysregulation of target genes and contribute to the progression of endometriosis.
Bioinformatics analysis has demonstrated that COX-2 is a validated target of miR-100. COX-2 is expressed during proliferation, differentiation, and inflammation, which are prominent indexes of endometriosis. COX-2 has a significant role in pain associated with endometriosis41. Despite the downregulation of miR-100 in ectopic versus eutopic endometrium38,22 our study indicates that the expression level of EV-derived plasma miR-100 decreases in the endometriosis group.
The expression level of miR-154 is not affected by the menstrual cycle. It can be considered, either alone or in combination with hsa-miR-196b-5p, hsa-miR-378a-3p, and hsa-miR-33a-5p, as a non-invasive potential diagnostic biomarker30. In addition, miR-154 is involved in the Hippo signaling pathway which has crucial roles in both normal and abnormal functions of endometrial cells in endometriosis. This pathway regulates cell proliferation, apoptosis and epithelial mesenchymal transition (EMT) process. Therefore, dysregulation of Hippo signaling pathway can develop endometriosis42. We could not detect miR-154 in the EVs derived from plasma of our participants, including both the case and control groups. We performed amplification efficiency analysis with LinReg PCR software. While all other miRNAs showed amplification above 1.91, miR-154 exhibited an efficiency of 1.0 consistence with the true absence of miR- 154. This finding indicates that the lack of detectable expression of miR-154 in circulating and EV- associated RNA derived from plasma of case and control groups in this study is probably due to inter- individual or population specific variability, differences in RNA isolation methods or sample processing and technical limitation.
Hypoxia is a hallmark of endometriosis that leads to altered methylation of the promoters of several genes involved in the development of endometriosis. The miR-148a/AUF complex suppresses the function of DNMT1 mRNA, resulting in hypomethylation34. HLA-G can protect the fetus from maternal immune surveillance and rejection. Finding has shown that serum levels of HLA-G are higher in women with endometriosis compared to a control group, that indicatea the runaway power of ectopic endometrial tissue of the immune system34. Additionally, miR-148a is an element of the G protein-coupled estrogen receptor (GPER)/miR-148a/HLA-G signaling pathway in ovarian endometriosis. The expression level of miR-148a is reduced in both the endometrium and endometriosis-associated ovarian cancer32. Furthermore, miR-148a can affect the expression of BCL-2 and the activity of caspase-3/9, making it a potential candidate for treatment of endometriosis32.
We showed that the expression levels of miR-451a, miR-23b, miR-148 and miR-100 significantly decrease in plasma derived EV of endometriosis patients. We propose that plasma-derived EV-miRNAs could serve as a potential diagnostic biomarker for endometriosis. However, further studies with a larger case and control groups are suggested to validate these findings.
Materials and methods
Eligibility criteria and search strategy
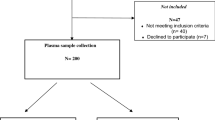
At the first, we reviewed all published studies in the PubMed database, without any time limitations, up to April 2023. The search terms applied were (“MicroRNAs“[Mesh]) OR (“microRNA“[Title/Abstract]) OR (“miR“[Title/Abstract]) AND (“endometriosis“[Title]) OR (“endometriosis“[Title/Abstract]) OR (“Endometriosis“[Mesh]). We assessed studies involving patients with endometriosis that examined miRNAs in relation to their role in disease development or their potential as diagnostic biomarkers. The study population consisted of women with endometriosis at any stage, diagnosed through laparoscopy and/or Doppler sonography (Fig. 3).
Study selection
All obtained references were independently screened on the keywords in title/abstract or Mesh terms. Five miRNAs were selected for our next in vitro phase using bioinformatics analysis. Finally, the expression level of candidate miRNAs were evaluated in the plasma derived EV in the case (n = 13) and control group (n = 11).
Bioinformatics analysis
By utilizing the miRTarBase database (https://mirtarbase.cuhk.edu.cn), the validated targets of miRNAs were acquired. Followed by the identification of associated miRNA pathway using the lists of uploaded genes in the Enrichr software (https://maayanlab.cloud/Enrichr/). Ultimately, five miRNAs were selected for the invitro phase.
Criteria for MiRNAs selection
Two criteria for selection of miRANs is considered. First, the role of miRNA in progrresion of endometriosis or intredused it as a potentially diagnostic biomarker based on previous litreture. Second, specific targets or pathway miRNAs in roled in endometriosis based on bioinformatics analysis.
STRING model
The validated targets of candidate miRNAs were acquired from the miRTarBase database based on reports with more consensus assay including q-PCR, micro array and NGS. After removing repetitive targets among five miRNAs the overlapping targets between miRTarBase and TargetScan (https://www.targetscan.org) were identified. Then protein- protein interaction achieved by the STRING (https://string-db.org/) database.
Patients and control groups
Twenty-four women aged 20–37 years old with normal body mass index (BMI) were enrolled in this study between June and august 2019 at the Royan Institute (Tehran, Iran). The study was approved by the Research Ethics Committee of the Royan Institute (Code No.: IR.ACECR.ROYAN.REC.1398.144), and written informed consent was obtained from all participants.
The presence of the disease was confirmed either by the laparoscopy and histopathological examinations (n = five) or doppler sonography( nn = eight). All cases who selected by laparoscopy suffered of an ovarian endometrioma at stages III–IV according to the Revised American Fertility Society Classification System (ARMS).
The control group was composed of 11 normal ovulatory women without endometriosis who were infertile due to male factors. The absence of the endometrioma cysts was confirmed with the doppler sonography. Women with a history of an irregular menstruation, cancers, inflammatory and autoimmune disease, fibroma, and adenoma were excluded from the study.
At the sampling time, all women were on days two-five of the menstrual cycle phase. No subjects had taken any hormonal treatments at least three months ago. All of the samples were collected in the morning.
Effect sized analysis
To show that sample size cannot affect the validity of the results, Cohen’s d was calculated to estimate effect size, using conventional thresholds (α = 0.05, two tail).
Plasma isolation
Peripheral blood samples (6 ml) were collected into a tube containing citrate as the anticoagulant agent. Whole blood was centrifuged at 2500 g at room temperature for 15 min. The plasma was transferred into a new microtube, and then centrifuge procedure was repeated. Platelet-free plasma (PFP) was isolated by a high-speed centrifugation at 13,000 g for 5 min. Finally, the PFPs were aliquoted and stored at −80 °C until use.
RNA extraction of plasma and EV isolation
Total RNA including circulating and EV- associated RNA was extracted from 2 ml of plasma. we used the Plasma/Serum Circulating and Exosomal RNA Purification Mini Kit (Cat. No. 51000, Norgen Biotek). Briefly 2 ml of plasma was mixed with lysis buffer A to disrupt EV membranes. And release their RNA contents. Ethanol was then added, and the lysate was passed through spin column to bind RNA. After several washing steps, RNA was eluted in 50 ml of elution solution A and sorted at −80 °C until further analysis. This kit designed to extract total RNA from plasma/ serum including both circulating and EV-associated RNA. The kit lyses extracellular vesicles during the extraction process, allowing access to vesicle-contained RNA without requiring physical EV isolation (e.g., ultracentrifugation or NTA).
Quantitative reverse transcription PCR assay
Immediately after extracting RNA, cDNA was synthesized using reverse transcription (RT) enzyme (Takara, USA) following the manufacturer’s protocol. Stem-loop primers were used for more specifics. Primers were designed based on miRNA sequences obtained from the miRbase (https://mirbase.org/) database as follows (Supplementary Table S8). The prepared master mix contained 10ng RNA, 4 ml enzyme buffer, 1 ml RT enzyme, and 1 ml stem loop master primer. The expression level of the candidate miRNAs was measured by the RT-PCR SYBR Green. Then, the expression levels of miRNAs were normalized with the expression level of hsa-miR-16.
Assessment of PCR efficiency
LinReg PCR software was used to calculate the efficiency of each qRT-PCR reaction. This software analysis row fluorescence data, adjust the base line automatically, and identifies the exponential phase (Window of linearity) for each sample. Then it uses linear regression to estimate the slope of that phase, which is used to determine the reaction efficiency for each sample.
Statistical analysis
The expression level of plasma derived EVs candidate miRNAs were assessed between the normal and endometriosis group. All data was analyzed for normal distribution. After successfully passing the normality parametric test, the SPSS software was used. A P value < 0.05 was considered as statistically significant.
Data availability
The archived datasets analyzed in support of the conclusions of this article will be made available upon request by the first or corresponding authors.
References
Moustafa, S., Burn, M., Mamillapalli, R., Nematian, S. & Flores, V. Accurate diagnosis of endometriosis using serum MicroRNAs. Am. J. Obs Gynecol. 223 (4), 557 (2020).
Parasar, P., Ozcan, P. & Endometriosis, T. K. Epidemiology, diagnosis and clinical management. Curr. Obs Gynecol. Rep. 6 (1), 34–41 (2017).
Ballweg, M. L. Impact of endometriosis on women’s health: Comparative historical data show that the earlier the onset, the more severe the disease. Best Pr Res. Clin. Obs Gynaecol. 18 (2), 201 (2004).
Dunselman, G. A. J. et al. ESHRE guideline: Management of women with endometriosis. Hum. Reprod. 29 (3), 400 (2014).
Pearce, C. L. et al. Association between endometriosis and risk of histological subtypes of ovarian cancer: A pooled analysis of case–control studies. Lancet Oncol. 3 (4), 385–389 (2012).
Coccia, M. E. & Nardone, L. Endometriosis and infertility: A long-life approach to preserve reproductive integrity. Int. J. Environ. Res. Public Health. 19 (10), 616 (2022).
Mehdizadeh Kashi, A., Chaichian, S., Ariana, S., Fazaeli, M. & Moradi, Y. The impact of laparoscopic cystectomy on ovarian reserve in patients with unilateral and bilateral endometrioma. Int. J. Gynaecol. Obs. 136 (2), 200 (2017).
Raposo, G. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell. Biol. 200 (4), 373 (2013).
Raposo, G. Extracellular vesicles: A new communication paradigm? Nat. Rev. Mol. Cell. Biol. 20 (9), 509 (2019).
Valadi, H., Ekström, K., Bossios, A., Sjöstrand, M. & Lee, J. J. Exosome-mediated transfer of mRNAs and MicroRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell. Biol. 9 (6), 654–659 (2007).
Liu, Y. Extracellular vesicle micrornas: Biomarker discovery in various diseases based on RT-qPCR. Biomark. Med. 9, 791–805 (2015).
Friedländer, M. R. et al. Kagerbauer B, G. J. Evidence for the biogenesis of more than 1000 novel human MicroRNAs. Genome Biol. 15, R57 (2014).
Bartel, D. P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116, 281–297 (2004).
Sanz-Rubio, D. et al. MiRNAs in H. Stability of circulating exosomal MiRNAs in healthy subjects. Sci. Rep. 8, 103–106 (2018).
Chekini, Z., Shahhoseini, M. & Aflatoonian, R. The relationship between functional promoter variants of macrophage migration inhibitory factor and endometriosis. Cell. J. 22, 450–456 (2020).
Frisendahl, C. et al. miR-193b-5p and miR-374b-5p are aberrantly expressed in endometriosis and suppress endometrial cell migration in vitro. Biomolecules 14, 1400 (2024).
Nothnick, W. B., Falcone, T., Joshi, N. & Fazleabas, A. T. Serum miR-451a levels are significantly elevated in women with endometriosis and recapitulated in baboons (Papio anubis) with experimentally-induced disease. Reprod. Sci. 24, 1195–1202 (2017).
Shen, L. et al. L. Y. MicroRNA23a and microRNA23b deregulation derepresses SF-1 and upregulates estrogen signaling in ovarian endometriosis. J. Clin. Endocrinol. Metab. 98, 1572–1582 (2013).
Munrós, J. et al. B. J. Total circulating microparticle levels are increased in patients with deep infiltrating endometriosis. Hum. Reprod. 32, 325–331 (2017).
Graham, A., Falcone, T. & Nothnick, W. B. The expression of microRNA-451 in human endometriotic lesions is inversely related to that of macrophage migration inhibitory factor (MIF) and regulates MIF expression and modulation of epithelial cell survival. Human Reprod. 30, 642–652 (2015).
Hirakawa, T. et al. miR-503, a MicroRNA epigenetically repressed in endometriosis, induces apoptosis and cell-cycle arrest and inhibits cell proliferation, angiogenesis, and contractility of human ovarian endometriotic stromal cells. Hum. Reprod. 31 (11), 2587–2597. https://doi.org/10.1093/humrep/dew217 (2016).
Hawkins, S. M. et al. M. M. Functional MicroRNA involved in endometriosis. Mol. Endocrinol. 25, 821–832 (2011).
Abe, W. et al. miR-196b targets c-myc and Bcl-2 expression, inhibits proliferation and induces apoptosis in endometriotic stromal cells. Human Reprod. 28, 750–761 (2013).
Rekker, K. et al. Circulating miR-200-family micro-RNAs have altered plasma levels in patients with endometriosis and vary with blood collection time. Fertil. Steril. 104, 938–946 (2015).
Wang, W., Zhao, Y., Han, B., Hong, S. & Chen, Y. Circulating MicroRNAs identified in a genome-wide serum MicroRNA expression analysis as noninvasive biomarkers for endometriosis. J. Clin. Endocrinol. Metab. 98, 1–9 (2013).
Liu, S., Gao, S., Wang, X. Y. & Wang, D. B. Expression of miR-126 and Crk in endometriosis: miR-126 May affect the progression of endometriosis by regulating Crk expression. Arch. Gynecol. Obs. 285, 1065–1072 (2012).
Filigheddu, N. et al. Differential expression of microRNAs between eutopic and ectopic endometrium in ovarian endometriosis. J. Biomed. Biotechnol. 2010, 369549 (2010).
Takebayashi, K. et al. Hsa-miR-100-5p, an overexpressed MiRNA in human ovarian endometriotic stromal cells, promotes invasion through attenuation of SMARCD1 expression. Reprod. Biol. Endocrinol. 18, 1–10 (2020).
Dai, L. & Gu, L. MiR-199a attenuates endometrial stromal cell invasiveness through suppression of the IKKβ/NF-κB pathway and reduced interleukin-8 expression. Mol. Hum. Reprod. 18, 136–145 (2012).
Pateisky, P. et al. hsa-miRNA-154-5p expression in plasma of endometriosis patients is a potential diagnostic marker for the disease. RBMO 38, 449–466 (2018).
Wu, D., Lu, P. & Mi, X. Exosomal miR-214 from endometrial stromal cells inhibits endometriosis fibrosis. Mol. Hum. Reprod. 27, 357–365 (2018).
He, S. Z. H. I. et al. G protein–coupled estrogen receptor/miR–148a/human leukocyte antigen–G signaling pathway mediates cell apoptosis of ovarian endometriosis. Mol. Med. Rep. 18, 1141–1148 (2018).
Tan, Q., Shi, S., Liang, J., Cao, D. & Wang, S. Endometrial cell-derived small extracellular vesicle miR-100-5p promotes functions of trophoblast during embryo implantation. Mol. Ther. Nucleic Acids. 5, 217–231 (2020).
Hsiao, K. Y. et al. Coordination of AUF1 and miR-148a destabilizes DNA methyltransferase 1 mRNA under hypoxia in endometriosis. Mol. Hum. Reprod. 21, 894–904 (2015).
Cheng, L. et al. Exosomes provide a protective and enriched source of MiRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 3078, 23743 (2014).
Condrat, C. E. et al. MiRNAs as biomarkers in disease: Latest findings regarding their role in diagnosis and prognosis. Cells 9, 1–32 (2020).
Felekkis, K. & Papaneophytou, C. The circulating biomarkers league: Combining miRNAs with cell-free DNAs and proteins. Int. J. Mol. Sci. 25, 3403. (2024).
Takahashi, R. U., Prieto-Vila, M., Hironaka, A. & Ochiya, T. The role of extracellular vesicle MicroRNAs in cancer biology. Clin. Chem. Lab. Med. 55, 648–656 (2017).
Ueta, E., Tsutsumi, K., Kato, H., Matsushita, H. & Shiraha, H. Extracellular vesicle–shuttled MiRNAs as a diagnostic and prognostic biomarker and their potential roles in gallbladder cancer patients. Sci. Rep. 11, 1–13. https://doi.org/10.1038/s41598-021-91804-0 (2021).
Graham, A. & Falcone, T. The expression of microRNA-451 in human endometriotic lesions is inversely related to that of macrophage migration inhibitory factor (MIF) and regulates MIF expression and modulation of epithelial cell survival. Hum. Reprod. 30, 642–652 (2015).
Lai, Z. Z. et al. Cyclooxygenase-2 in endometriosis. Int. J. Biol. Sci. 15, 2783–2797 (2019).
Jin, T. et al. The inactivation of Hippo signaling pathway promotes the development of adenomyosis by regulating EMT, proliferation, and apoptosis of cells. Reprod. Sci. 30, 2715–2727 (2023).
Acknowledgements
This study was financially sponsored by the genetic department of Royan Institute Tehran, Iran. The authors dedicate this article to the memory of Dr. Saeid Kazemi Ashtiani, the late founder of Royan Institute.
Funding
No funds, grants, or other support was received except full financial support from the genetic department of Royan Institute Tehran, Iran. Institutional Review Board Statement: The study was conducted in accordance with the Declaration of Helsinki, and approved by the Research Ethics Committee of the Royan Institute (Code No.: IR.ACECR.ROYAN.REC.1398.144).
Author information
Authors and Affiliations
Contributions
SAS, PA, FSh, AM, MT, MSh, Contributed to conception and design; SAS, Contributed to all experimental work, data and statistical analysis, and interpretation of data; PA, MT, Were responsible for overall supervision; MH, AM Cooperated in the sampling section; MH, ZCh, Was responsible for patients assessment and diagnosis, also for patient recruitment to study; SAS Collected the samples; AGh, Calculated the number of samples; ZCh Cooperated with some experimental tests; FSh helped in design of E.V, approval tests and supervision of manuscript in EV isolation part; AGh analyzed the row data; MSh, AGh, RA helped with interpretation of results; SAS, ZCh Drafted the initial manuscript and wrote the manuscript; PA, MT, AM Contributed to revise and edit the manuscript; All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Informed consent
Informed consent was obtained from all subjects involved in the study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sadati, S.A., Chekini, Z., Shekari, F. et al. Expression analysis of plasma extracellular vesicle associated candidate MiRNAs in endometriosis using integrative bioinformatics and experiential data. Sci Rep 15, 24970 (2025). https://doi.org/10.1038/s41598-025-09660-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-09660-1