Abstract
Aging is a complex biological process, and some individuals are aging faster or slower than expected. This phenomenon of aging acceleration occurs when biological age exceeds chronological age and can be assessed by epigenetic clock estimation. As aging acceleration is known to occur in response to some environmental exposures as well as trauma, we hypothesized that World Trade Center (WTC) exposures may have led to epigenetic aging acceleration. WTC-exposed women were selected from the World Trade Center Environmental Health Center (WTC EHC) clinic, with peripheral blood collected during routine clinical monitoring visits. The reference group was selected from the NYU Women’s Health Study (NYUWHS), a prospective cohort study that collected blood samples before 9/11/2001. Epigenomes of WTC-exposed vs. unexposed women were profiled using the Infinium MethylationEPIC array. DNA-based epigenetic aging was estimated using Hannum, Horvath, PhenoAge and GrimAge epigenetic clocks. Age acceleration was defined as the residual from regressing estimated epigenetic age on chronological age. Ordinary least squares regression was used to investigate the relationship between WTC exposure and accelerated aging. After adjustment for race/ethnicity, smoking status, Body Mass Index (BMI), batch and cell type composition, WTC exposure was associated with epigenetic aging acceleration using the Hannum epigenetic clock (βWTC Exposed vs. Unexposed: 3.789; p-value: <0.001). WTC exposure was also associated with epigenetic aging acceleration when using other epigenetic clock types (Horvath and PhenoAge, but not GrimAge), and when stratifying by breast cancer case (βWTC Exposed vs. Unexposed: 3.473; p-value: <0.001) or cancer-free participant (βWTC Exposed vs. Unexposed: 4.369; p-value: 0.001) status. Among all participants, having a breast cancer diagnosis was statistically significantly associated with accelerated aging (βCancer vs. cancer-free: 1.658; p-value: 0.021). WTC exposure is statistically significantly associated with epigenetic aging acceleration. This was true even after stratifying on cancer status. WTC exposure was positively associated with epigenetic aging acceleration in the overall cohort, among only those women who were cancer-free, and among breast cancer cases.
Similar content being viewed by others
Introduction
Aging is characterized by the progressive decline in genomic and system integrity and is itself a major driver of many chronic diseases1. There are multiple mechanisms of aging, and hallmarks of aging can be characterized based on damage to the genome, the physiological response to this damage, and the results from accumulation of damage beyond a point that the body can compensate for; this complexity and nuance in the aging process is indicated by the heterogeneity in health among persons of the same chronological age2. Some individuals may actually be aging faster than expected. This phenomenon of aging acceleration occurs when biological age exceeds chronological age. Persons exposed to the World Trade Center (WTC) disaster, including rescue and recovery workers (responders) and community members (survivors) may be experiencing aging acceleration given that WTC exposure is associated with greater age-related vulnerability. This is evidenced by the excess burden of aging-related conditions in this group, including frailty. Almost one-third of the WTC responders meets the criteria for frailty under a cumulative deficit model, a syndrome of increased age-related vulnerability that manifests as mortality, morbidity, disability, hospitalizations, and unfavorable outcomes after surgical procedures3. Frailty can also be conceptualized as the diminished ability to maintain normal function/homeostasis4. Age-related morbidity is also increased among WTC responders. Specifically, cancer in general, and rates of certain cancers, including prostate, thyroid, melanoma and tonsil cancer, are elevated among responders5,6,7,8,9, and cancer, including breast cancer, is commonly observed among WTC survivors10,11. WTC-associated cancers may also be, on average, more aggressive than cancers of WTC-unexposed persons12,13. Responders and survivors also suffer an excess burden of other aging-related syndromes and morbidities such as compromised lung function and increased asthma incidence, decreased sleep quality, mental health, and cognitive functioning impairment, and increased risk of autoimmune diseases, among other adverse health impacts14,15. Increased WTC exposure is additionally associated with increased risk of mortality16. Moreover, WTC responders and survivors experienced intense psychological trauma. Risk of post-traumatic stress disorder (PTSD) is elevated among WTC responders17, and PTSD and trauma are associated with immune system dysregulation18 and premature aging19.
Despite mounting epidemiological evidence of the WTC-associated premature aging, studies are needed that explore this on the biological level. Epigenetic clocks are commonly used to estimate biological aging through DNA methylation-derived changes. DNA methylation occurs when there is an addition of a methyl group at a cytosine-phosphate-guanine (CpG) site20,21. When DNA methylation occurs at gene promotors, this will often result in silencing of gene expression21. Dynamic and actively maintained throughout the genome, DNA methylation is sensitive to environmental stimuli, acting as an interface between the environment and the genome21. Epigenetic clocks use DNA methylation levels at CpG sites throughout the genome to estimate biological age, and are generally useful predictors of biological aging across the lifespan22, with some epigenetic clocks, like GrimAge, specifically trained on mortality data23. Epigenetic aging acceleration can occur in response to environmental exposures1. Exposure to endocrine-disrupting chemicals1, air pollution24,25,26, metals26, and dioxins27, found in the WTC dust, have all been linked to aging acceleration. WTC dust comprised damaging toxicants including metals (Arsenic [As], Beryllium [Be], Cadmium [Cd], Chromium [Cr] and Nickel [Ni]), asbestos, polycyclic aromatic hydrocarbons (PAHs), persistent organic pollutants (POPs, including polychlorinated biphenyls [PCBs] and dioxins), volatile organic compounds (VOCs, including benzene), and endocrine disrupters like per- and polyfluoroalkyl substances (PFAS)5,9,28. These environmental exposures are thought toimpact aging by modifying molecular mechanisms, such as mitochondrial metabolism and inflammation1.
Aging acceleration has important health implications. It is inversely correlated with physical and cognitive fitness29,30,31,32, and accelerated aging increases risk of mortality, independent of chronological age33. For each 1-year increase in aging acceleration, there is a 6% risk of developing cancer within 3 years and a 17% increase in the risk of dying from cancer within 5 years34.
To summarize, we hypothesize that WTC exposure may be associated with premature aging given: (1) aging-related vulnerability and mortality is increased among WTC-exposed persons; (2) trauma and PTSD, which are associated with premature aging, are also common among WTC-exposed individuals; and (3) many of the toxicants found in the WTC dust are known to be associated with epigenetic dysregulation. The main objective of the current study was to investigate epigenetic aging acceleration of WTC-exposed persons using previously validated epigenetic clocks.
Materials and methods
Study participants and samples
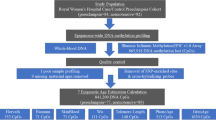
WTC-exposed women were selected from the World Trade Center Environmental Health Center (WTC EHC). The WTC EHC acts as the “Center of Excellence” for the treatment and surveillance of WTC-affected local community members, also known as survivors10,35. WTC survivors include “persons who were present in the dust or dust cloud on 9/11 or who worked, lived, or attended school, childcare centers, or adult day care centers in the NYC disaster area”35. Patients self-enroll into the WTC EHC program and are required by law to have a “certifiable condition”, such as a respiratory condition or cancer, or some other condition, in addition to WTC exposure15,35,36. Certifiable conditions are those determined by the WTC Health Program as WTC-related; more information can be found at https://www.cdc.gov/wtc/conditions.html. Only women who were cancer-free or who had breast cancer were included in this study. More than 6000 cancer patients, including 1300 breast cancer patients, have been diagnosed as of November 1, 202310,11. Physical examinations, mental health screens, blood tests, and chest X-rays are routinely performed at the WTC EHC Clinic. Participants were consented and blood was collected during routine clinical visits. WTC EHC subjects’ blood samples were collected in 2022–202337,38. Relevant clinical and sociodemographic data were collected from both initial and monitoring visits using interviewer-administered questionnaires10,11.
WTC-unexposed controls were selected from the NYU Women’s Health Study (NYUWHS). The NYUWHS is a prospective cohort in which women between the ages of 35 and 65 years old were enrolled between March 1985 and June 1991 at the Guttman Breast Diagnostic Institute in New York City39. Subjects completed a self-administered baseline questionnaire on demographic, medical, reproductive, regular physical activity, and recent medication use. All cohort participants donated peripheral blood, which was subsequently placed in long-term storage at − 80 °C. Because the sample collection predated 9/11/2001, all preserved NYUWHS blood samples are guaranteed to be WTC-unexposed. Participants have been followed up regularly since enrollment, to identify incident cancer cases. To match NYUWHS inclusion criteria, only those WTC EHC participants who were women between the ages of 35–65 years old, who were not pregnant or breastfeeding, and who had not been pregnant or breastfeeding in the 6 months preceding study enrollment, were included in this analysis. WTC-exposed and unexposed women were frequency matched on age. This study relied on convenience sampling of WTC-exposed women with breast cancer. Women being seen at the WTC EHC clinic for their annual monitoring visit were approached for study participation by study personnel (ST) provided they met these inclusion criteria. Study eligibility was determined by the clinical team (lead by JR). Women were consented and enrolled in the clinic until the target number of 96 was reached. Women who agreed to participate had an additional vial of blood collected during their routine bloodwork. This study had a 192-target final sample size (96 WTC-exposed and 96 unexposed participants).
Epigenome-wide profiling and epigenetic clock estimation
We used established methods for DNA processing and DNA methylation profiling as described more fully elsewhere37,38. Briefly, blood samples were sent to the NYU Langone Health Center for Biospecimen Research and Development (CBRD) laboratory for DNA extraction from white blood cells and treated with bisulfite to convert unmethylated cytosines to uracil, allowing for the identification of DNA methylation patterns at single-base resolution. Following the manufacturer’s protocol, the Illumina Infinium MethylationEPIC BeadChip version 2.0 (EPICv2) (Illumina®) was used to determine the DNA methylation status of 866,562 CpG sites. Participants samples were run using two EPIC BeadChips kits, each balanced for WTC-exposed and unexposed samples, randomized across array chips and scanned within the same time frame. We used the “ComBat” function from the “sva” R package to perform batch correction using the Sentrix slide ID as the batch variable. Each slide corresponds to a unique Illumina BeadChip processing 8 samples, and this correction effectively addresses both between-kit and within-kit (slide-level) technical variation. The R package “minfi” was used to process and analyze methylation data. Probes were quantile normalized and the background adjusted, using functions “preprocessQuantile” and “preprocessNoob40,41. Normalized DNA methylation data at various CpG sites was used to calculate epigenetic clocks, estimated using the DNA Methylation Age calculator (available at: https://dnamage.clockfoundation.org/). This included the Horvath42, Hannum43, PhenoAge32 and GrimAge23,44 epigenetic clocks. GrimAge version 2 was used. These epigenetic clock models are capturing different DNA methylation age-predictive factors. For instance, the Horvath and Hannum clocks have only 6 CpG sites in common22, and so utilizing various methodologies ensures a more robust investigation of the study hypothesis. Differences between these epigenetic clocks are described in Supplementary Table S1. In terms of presented results, the Hannum clock epigenetic age was prioritized as it is the original blood-based method of epigenetic age estimation and has been the widely validated and used type45, and because it is correlated with chronological age. In contrast, while Horvath is likewise correlated with chronological age, it is tissue-based, and PhenoAge and GrimAge correlate with aging-related morbidity and mortality, respectively (Supplementary Table S1). Later generation epigenetic clocks were not considered here but should be utilized in future.
Notably, the ability of epigenetic clocks to reflect biological aging is chronological age dependent. Specifically, epigenetic clocks are less effective at predicting biological age among older individuals, and may actually underestimate biological age due to the phenomenon of saturation, i.e., when CpG sites reach either full methylation or complete demethylation46,47. Defining age acceleration as the residual from regressing estimated biological age on chronological age (residuals (lm (epigenetic age - chronological age))) is one method to correct for this issue47. The full model is illustrated as follows: Y = β0 + Σj = 1.p βjXj + ε, where Y is the residual of epigenetic age on chronological age, β0 is the value of Y when X (WTC exposure) is equal to zero (unexposed), and βj is the slope, or change in Y when X (WTC exposure) is equal to 1 (exposed), adjusted for confounding factors. Epigenetic accelerated aging when calculated as the residual variation in epigenetic age independent of chronological age is “a measure of how much an individual is aging faster or slower than their chronological age”46.
Statistical analysis
Chronological and DNA methylation-derived age were summarized as continuous variables. T-tests were used to compare means for chronological and epigenetic ages between WTC-exposed and unexposed groups. Factors associated with WTC exposure status, including race, smoking status, BMI, were assessed using the Pearson test for normal and categorical variables and the Spearman test for continuous non-normal variables. These variables were chosen as each is known to influence DNA methylation48,49,50. Methylation-derived age acceleration is defined as positive residual variation in biological age independent of chronological age. Unadjusted and adjusted Ordinary Least Squares (OLS) regression was used to assess differences in accelerated aging according to the WTC exposure status (WTC-exposed vs. unexposed). In the adjusted model the following covariates were included: breast cancer status (post-diagnostic breast cancer case [WTC EHC] or pre-diagnosis case [NYUWHS] vs. control), race/ethnicity (Asian, Hispanic, Non-Hispanic Black, Non-Hispanic White, Other, Unknown), BMI (healthy [defined as BMI 18.5 to < 25], underweight [defined as BMI < 18.5], overweight [defined as BMI 25.0 to < 30], obese [defined as BMI ≥ 30, unknown), smoking status (ever, never), and education status (≤ high school/vocational school, attended college or graduate school, unknown). We used the estimateCellCounts() function in the minfi package to estimate the proportions of major immune cell types (CD8 + T, CD4 + T, NK, B cells, monocytes, and granulocytes). These estimates were then included as covariates in our regression models51. This approach—incorporating cell-type proportions as model covariates rather than adjusting methylation values directly—is a widely used and recommended strategy. Given the proportional nature of the cell type counts, granulocytes were not incorporated into the models. Data was also batch-adjusted (nbatches = 2) based on which EPIC BeadChip kit was used. Participants with missing data for covariates were not included in the adjusted models. All statistical analyses were done using R statistical software52.
Results
Initially samples were acquired for 192 participants, enough for two Illumnia BeadChips. DNA was successfully extracted, and epigenetic clocks were estimated for 189 participants, 4 of which had to be excluded as chronological age was unknown; of those remaining, 105 were WTC-exposed and 80 WTC-unexposed37,38,53. Characteristics of these participants have been described in detail elsewhere37,38,53. Notably, WTC-exposed vs. unexposed women differed meaningfully in race/ethnicity, smoking status, and BMI. Compared to the unexposed women, WTC-exposed participants were more racially and ethnically diverse (44.8% vs. 70.0% non-Hispanic white), were more likely to be never smokers (66.7% vs. 48.8%) and have higher BMIs (obese: 29.5% vs. 22.5%) (Table 1). These factors were included in the adjusted models, in addition to cell type composition, cancer status, and batch.
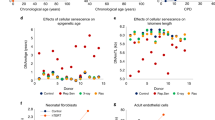
The mean chronological age at blood donation for the entire cohort was 56.4 years old. Although WTC-exposed and unexposed women were frequency-matched on age, women with WTC exposure were, on average, slightly older (mean chronological age: 58.4, standard deviation [SD]: 7.6 years) than; unexposed women (mean chronological age: 53.3, SD: 7.2 years). For each of the epigenetic clock methods, the mean estimated epigenetic age was higher among WTC-exposed vs. unexposed participants: Hannum: 68.9 vs. 63.0; Horvath: 65.3 vs. 61.1; PhenoAge: 52.0 vs. 48.0; GrimAge: 74.9 vs. 74.0. Univariate analysis showed that between WTC-exposed and unexposed women, mean chronological age, Hannum (p < 0.0001), Horvath (p < 0.0001) and PhenoAge (p = 0.0015) ages were statistically significantly different, but not for GrimAge (Table 2; Fig. 1). The average residual for aging acceleration was Hannum: 1.10 for WTC-exposed and − 1.10 for WTC-unexposed; Horvath: 0.367 for WTC-exposed and − 0.389 for WTC-unexposed; PhenoAge: 0.419 for WTC-exposed and − 0.025 for WTC-unexposed; and GrimAge: − 0.376 for WTC-exposed and 0.517 for WTC-unexposed.
Compared with WTC-unexposed women, women with WTC exposure had accelerated aging using Hannum clock both before (βWTC Exposed vs. Unexposed: 2.198; p-value: 0.001) and after (βWTC Exposed vs. Unexposed: 3.789; p-value: <0.001) adjustment. In adjusted models, WTC exposure was also associated with accelerated aging using Horvath (βWTC Exposed vs. Unexposed: 2.201; p-value: 0.009) and PhenoAge (βWTC Exposed vs. Unexposed: 2.185; p-value: 0.045) methods, but not GrimAge epigenetic clocks (βWTC Exposed vs. Unexposed: 0.449; p-value: 0.418) (Fig. 2). Among all participants, having a breast cancer diagnosis was statistically significantly associated with accelerated aging (βCancer vs. cancer-free: 1.658; p-value: 0.021; Hannum epigenetic clock). When stratified, WTC exposure was statistically significantly associated with accelerated aging in both breast cancer cases (adjusted βWTC Exposed vs. Unexposed: 3.473; p-value: <0.001) and cancer-free participants (adjusted βWTC Exposed vs. Unexposed: 4.369; p-value: 0.001). Full model results for the Hannum epigenetic clock are reported in Supplementary Table S2.
Discussion
Accelerated aging due to complex environmental exposures is an understudied phenomenon. We demonstrated that measuring biological aging using DNA methylation and epigenetic clocks can improve our understanding of age-related vulnerability among WTC-exposed individuals. We also present preliminary evidence that WTC exposure may be associated with accelerated aging. Women with WTC exposure, on average, appear to be aging more rapidly compared to unexposed persons according to several epigenetic clocks.
It is currently uncertain what aspects of aging epigenetic clocks represent. One possibility is that epigenetic clocks reflect an evolutionarily conserved and highly regulated process whereby DNA methylation rates change throughout the life course31,42. It’s also possible that epigenetic clocks are capturing cumulative lifetime damage to DNA54. These theories are not mutually exclusive, and accelerated aging could represent a deviation from normal processes and/or an accumulation of DNA damage. A better understanding of what biological aspects are being captured by different epigenetic clocks, and how they can be dysregulated, may help clarify how environmental exposures may accelerate aging. Notably, the different methods of epigenetic clock estimation showed considerable variability, especially the PhenoAge and GrimAge clocks. Future work is needed to better understand which type of clocks are best for studying the impact of environmental exposures on biological aging.
This work, while preliminary, is consistent with past findings. A large body of work from our and other groups has demonstrated that global and site-specific DNA methylation dysregulation is common among WTC-exposed individuals37,38,53,55. While this is the first study to use epigenetic clocks to compare rates of accelerated aging among WTC-exposed vs. unexposed participants, in a recent analysis of WTC responders with and without current PTSD, GrimAge methylation age acceleration was observed to be associated with PTSD diagnosis, although PhenoAge, Hannum, and Horvath methylation age acceleration were not robustly related to PTSD56. RNA-seq results corroborate this finding; responders with current PTSD, compared to those without, showed accelerated transcriptional aging, even after adjustment for chronological age56. An analysis of post-9/11 United States military veterans found that veterans with current PTSD were aging faster than those without current PTSD, assessed using DNA methylation measures of epigenetic aging19. Multiple other studies confirm the link between trauma, PTSD, and epigenetic aging acceleration57,58,59, possibly due to an increase in inflammation and oxidative stress, that induce cell division and lead to telomere shortening60. Biological aging acceleration has therefore previously been documented among WTC responders and similar groups; however, this is the first study to attempt to quantify the degree to which WTC-exposed vs. unexposed groups differ in their aging processes. Future work is needed to expand on these results to validate the findings reported here, and to elucidate the ways in which accelerated aging may be adversely affecting the health of WTC-exposed responders and survivors.
Strengths of our study included the use of a WTC-unexposed control group, the use of multiple epigenetic clock methods, a statistical design that incorporated baseline chronological age into the estimation of biological aging, and adjustment for important confounding factors, such as smoking status and BMI, as well as cell type composition. This is also the first study focused on WTC-exposed women. This is important as it’s well established that sex is an effect measure modifier of both DNA methylation profiles and the normal aging process61.
Several study limitations should be considered. This work was based on relatively small project epigenetically profiling WTC-exposed vs. unexposed women who were either cancer-free or had breast cancer53. Women with other cancer types were excluded, and thus we cannot address the impact of WTC exposure on epigenetic aging among other cancer phenotypes. While women with breast cancer had blood donated close to their diagnoses, and so should have been treatment naive, we cannot discount an impact of cancer treatment on epigenetics and therapy-induced aging62. Also notable, this study relied on convenience sampling, which has inherent limitations which could impact validity and generalizability. The possibility of selection bias cannot be excluded, and WTC survivors who were more health conscious, and more likely to visit the WTC EHC clinic, may be overrepresented in our study sample. A larger study based on a random sample of WTC survivors is warranted. A statistically significant relationship between WTC exposure and accelerated aging as measured by GrimAge was not observed. While the fact that the Hannum, Horvath and PhenoAge epigenetic clocks all yielded statistically significant results increases confidence in our study findings, it should be noted that these clocks were designed to capture different aspects of the biological aging process, and which clock, if any, is best for capturing the effect of environmental exposures on aging remains a matter of ongoing research. GrimAge, correlated to mortality data, may be capturing different aspects of the biological aging process that are less impacted by WTC exposure. A larger sample size of WTC-exposed individuals may also allow for an investigation into a dose-response relationship between WTC exposure and accelerated aging, which would further support the study’s hypothesis. Additionally, while our results highlight the need to adjust for confounding factors when investigating accelerated aging, unmeasured confounding may persist. Socioeconomic status, specifically, is likely an important consideration. Education status was available for a subset of subjects, which can be used as a proxy for income. In a sensitivity analysis of subjects with known education status, WTC exposure was still associated with accelerated aging, even after adjustment for education (Hannum, adjusted βWTC Exposed vs. Unexposed: 6.445; p-value: <0.001; data not shown). Future studies with more detailed clinical and sociodemographic data are nevertheless warranted. WTC-exposed and unexposed samples differed by sample storage time, with WTC-unexposed NYUWHS samples having been collected decades before the analysis. Stability of white blood cell DNA methylation profiles, however, is robust. It has been reported previously that blood samples stored for long periods at − 80 °C have stable DNA methylation profiles63. DNA extracted from the blood thawed 20 years after it was frozen at − 80 °C has been shown to have stable DNA methylation levels at different genomic regions (TSS, promoter, gene body, CGI, and CGI shore regions)64. It is a notable limitation that we cannot discern the effects of specific components of the WTC toxic dust, and trauma, on epigenetic aging, which should be the focus of future research. Future studies should explore this relationship in WTC-exposed males, including WTC rescue and recovery workers, who may have had higher levels of acute WTC dust exposure. Lastly, germline genetics likely also plays a role in accelerated aging, however, participants genotypes were unknown and sample size was too small to investigate gene-gene and gene-environment effects.
Conclusions
WTC-exposed persons appear to be experiencing accelerated epigenetic aging compared to unexposed individuals. Most WTC responders are now at a greater chronological age and at an increased risk of aging-related chronic diseases. To date, the biological mechanisms underpinning WTC exposure-disease relationships remain understudied, but premature aging may play a role. Improved understanding of how WTC exposure modifies the biological aging processes could improve both preventive and therapeutic approaches for WTC-exposed persons. Biomarkers of accelerated aging can also be used to screen for age-related vulnerability and syndromes. The results presented here, while preliminary, could have important implications not just for WTC-exposed persons but also for individuals exposed to other complex environmental exposures.
Data availability
The data that support the findings of this study are not openly available due to reasons of sensitivity and are available from the corresponding author upon reasonable request.
References
Prada, D., Belsky, D. & Baccarelli, A. A. Is this environment making you older? Molecular biomarkers and new approaches to investigate the influences of environmental chemicals through aging. Med. Lav. 112 (1), 8–14. https://doi.org/10.23749/mdl.v112i1.10826 (2021).
López-Otín, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. Hallmarks of aging: an expanding universe. Cell 186 (2), 243–278. https://doi.org/10.1016/j.cell.2022.11.001 (2023).
Bello, G. A. et al. Development and validation of a clinical frailty index for the world trade center general responder cohort. J. Aging Health. 33 (7–8), 531–544. https://doi.org/10.1177/0898264321997675 (2021).
Ab, M., Je, G. & Aj, M. Frailty, fitness and late-life mortality in relation to chronological and biological age. BMC Geriatr. 2 https://doi.org/10.1186/1471-2318-2-1 (2002).
Lioy, P. J. et al. Characterization of the dust/smoke aerosol that settled East of the world trade center (WTC) in lower Manhattan after the collapse of the WTC 11 September 2001. Environ. Health Perspect. 110 (7), 703–714 (2002).
Zeig-Owens, R. et al. Early assessment of cancer outcomes in new York City firefighters after the 9/11 attacks: an observational cohort study. Lancet Lond. Engl. 378 (9794), 898–905. https://doi.org/10.1016/S0140-6736(11)60989-6 (2011).
Solan, S. et al. Cancer incidence in world trade center rescue and recovery workers, 2001–2008. Environ. Health Perspect. 121 (6), 699–704. https://doi.org/10.1289/ehp.1205894 (2013).
Li, J. et al. Association between world trade center exposure and excess cancer risk. JAMA. 308 (23), 2479–2488. https://doi.org/10.1001/jama.2012.110980 (2012).
Yiin, L. M. et al. Comparisons of the dust/smoke particulate that settled inside the surrounding buildings and outside on the streets of Southern new York City after the collapse of the world trade center, September 11, 2001. J. Air Waste Manag. Assoc. 1995. 54 (5), 515–528. https://doi.org/10.1080/10473289.2004.10470935 (2004).
Shao, Y. et al. The development of a WTC environmental health center pan-cancer database. Int. J. Environ. Res. Public. Health. 18 (4), 1646. https://doi.org/10.3390/ijerph18041646 (2021).
Durmus, N. et al. Characteristics of cancer patients in the world trade center environmental health center. Int. J. Environ. Res. Public. Health. 17 (19), E7190. https://doi.org/10.3390/ijerph17197190 (2020).
Hashim, D. et al. Prostate cancer characteristics in the world trade center cohort, 2002–2013. Eur. J. Cancer Prev. Off J. Eur. Cancer Prev. Organ. ECP. 27 (4), 347–354. https://doi.org/10.1097/CEJ.0000000000000315 (2018).
Arslan, A. A. et al. Breast Cancer characteristics in the population of survivors participating in the world trade center environmental health center program 2002–2019. Int. J. Environ. Res. Public. Health. 18 (14), 7555. https://doi.org/10.3390/ijerph18147555 (2021).
Brackbill, R. M., Graber, J. M. & Robison, W. A. (eds) (Allen). Editorial for long-term health effects of the 9/11 disaster. Int. J. Environ. Res. Public Health. 16(18), 3289. https://doi.org/10.3390/ijerph16183289 (2019).
Reibman, J. et al. Destruction of the world trade center towers. Lessons learned from an environmental health disaster. Ann. Am. Thorac. Soc. 13 (5), 577–583. https://doi.org/10.1513/AnnalsATS.201509-572PS (2016).
Jordan, H. T. et al. Mortality among survivors of the Sept 11, 2001, world trade center disaster: results from the world trade center health registry cohort. Lancet Lond. Engl. 378 (9794), 879–887. https://doi.org/10.1016/S0140-6736(11)60966-5 (2011).
Berninger, A. et al. Trends of elevated PTSD risk in firefighters exposed to the world trade center disaster: 2001–2005. Public. Health Rep. 125 (4), 556–566 (2010).
Smith, A. K. et al. Cell-type-specific and inflammatory DNA methylation patterns associated with PTSD. Brain Behav. Immun. 128, 540–548. https://doi.org/10.1016/j.bbi.2025.04.031 (2025).
Bourassa, K. J. et al. Posttraumatic stress disorder, trauma, and accelerated biological aging among post-9/11 veterans. Transl. Psychiatry. 14 (1), 1–8. https://doi.org/10.1038/s41398-023-02704-y (2024).
Martin, E. M. & Fry, R. C. Environmental influences on the epigenome: Exposure- associated DNA methylation in human populations. Annu. Rev. Public. Health. 39 (1), 309–333. https://doi.org/10.1146/annurev-publhealth-040617-014629 (2018).
Moore, L. D., Le, T. & Fan, G. DNA methylation and its basic function. Neuropsychopharmacology. 38 (1), 23–38. https://doi.org/10.1038/npp.2012.112 (2013).
Jylhävä, J., Pedersen, N. L. & Hägg, S. Biological age predictors. EBioMedicine. 21, 29–36. https://doi.org/10.1016/j.ebiom.2017.03.046 (2017).
Lu, A. T. et al. DNA methylation grimage strongly predicts lifespan and healthspan. Aging. 11 (2), 303–327. https://doi.org/10.18632/aging.101684 (2019).
Gao, X. et al. Accelerated DNA methylation age and the use of antihypertensive medication among older adults. Aging. 10 (11), 3210–3228. https://doi.org/10.18632/aging.101626 (2018).
Ward-Caviness, C. K. et al. Long-term exposure to air pollution is associated with biological aging. Oncotarget. 7 (46), 74510–74525. https://doi.org/10.18632/oncotarget.12903 (2016).
Malecki, K. M. C. et al. Integrating environment and aging research: opportunities for synergy and acceleration. Front. Aging Neurosci. https://doi.org/10.3389/fnagi.2022.824921 (2022).
Nwanaji-Enwerem, J. C. et al. Serum dioxin levels and sperm DNA methylation age: findings in Vietnam war veterans exposed to agent orange. Reprod. Toxicol. 96, 27–35. https://doi.org/10.1016/j.reprotox.2020.06.004 (2020).
Lioy, P. J. & Georgopoulos, P. The anatomy of the exposures that occurred around the world trade center site: 9/11 and beyond. Ann. N Y Acad. Sci. 1076, 54–79. https://doi.org/10.1196/annals.1371.002 (2006).
Horvath, S. & Raj, K. DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat. Rev. Genet. 19 (6), 371–384. https://doi.org/10.1038/s41576-018-0004-3 (2018).
Marioni, R. E. et al. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian birth cohort 1936. Int. J. Epidemiol. 44 (4), 1388–1396. https://doi.org/10.1093/ije/dyu277 (2015).
Horvath, S. et al. Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging. 7 (12), 1159–1170. https://doi.org/10.18632/aging.100861 (2015).
Levine, M. E. et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging. 10 (4), 573–591. https://doi.org/10.18632/aging.101414 (2018).
Chen, B. H. et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging. 8 (9), 1844–1865. https://doi.org/10.18632/aging.101020 (2016).
Zheng, Y. et al. Blood epigenetic age may predict cancer incidence and mortality. EBioMedicine. 5, 68–73. https://doi.org/10.1016/j.ebiom.2016.02.008 (2016).
Azofeifa, A. World trade center health Program—United states, 2012–2020. MMWR Surveill. Summ. 70 https://doi.org/10.15585/mmwr.ss7004a1 (2021).
Reibman, J. et al. Characteristics of a residential and working community with diverse exposure to world trade center dust, gas, and fumes. J. Occup. Environ. Med. 51 (5), 534–541. https://doi.org/10.1097/JOM.0b013e3181a0365b (2009).
Arslan, A. A. et al. Genome-wide DNA methylation profiles in community members exposed to the world trade center disaster. Int. J. Environ. Res. Public. Health. 17 (15), 5493. https://doi.org/10.3390/ijerph17155493 (2020).
Tuminello, S. et al. Global DNA methylation profiles in peripheral blood of WTC-Exposed community members with breast Cancer. Int. J. Environ. Res. Public. Health. 19 (9), 5104. https://doi.org/10.3390/ijerph19095104 (2022).
Toniolo, P. G. et al. A prospective study of endogenous estrogens and breast cancer in postmenopausal women. J. Natl. Cancer Inst. 87 (3), 190–197. https://doi.org/10.1093/jnci/87.3.190 (1995).
Aryee, M. J. et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of infinium DNA methylation microarrays. Bioinformatics. 30 (10), 1363–1369. https://doi.org/10.1093/bioinformatics/btu049 (2014).
Moran, S., Arribas, C. & Esteller, M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 8 (3), 389–399. https://doi.org/10.2217/epi.15.114 (2016).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14 (10), R115. https://doi.org/10.1186/gb-2013-14-10-r115 (2013).
Hannum, G. et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol. Cell. 49 (2), 359–367. https://doi.org/10.1016/j.molcel.2012.10.016 (2013).
Lu, A. T. et al. DNA methylation grimage version 2. Aging. 14 (23), 9484–9549. https://doi.org/10.18632/aging.204434 (2022).
Bell, C. G. et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 20 (1), 249. https://doi.org/10.1186/s13059-019-1824-y (2019).
Jain, P. et al. Analysis of epigenetic age acceleration and healthy longevity among older US women. JAMA Netw. Open. 5 (7), e2223285. https://doi.org/10.1001/jamanetworkopen.2022.23285 (2022).
El Khoury, L. Y. et al. Systematic underestimation of the epigenetic clock and age acceleration in older subjects. Genome Biol. 20, 283. https://doi.org/10.1186/s13059-019-1810-4 (2019).
Adkins, R. M., Krushkal, J., Tylavsky, F. A. & Thomas, F. Racial differences in gene-specific DNA methylation levels are present at birth. Birt Defects Res. Clin. Mol. Teratol. 91 (8), 728–736. https://doi.org/10.1002/bdra.20770 (2011).
Maas, S. C. E. et al. Smoking-related changes in DNA methylation and gene expression are associated with cardio-metabolic traits. Clin. Epigenetics. 12 (1), 157. https://doi.org/10.1186/s13148-020-00951-0 (2020).
Samblas, M., Milagro, F. I. & Martínez, A. DNA methylation markers in obesity, metabolic syndrome, and weight loss. Epigenetics. 14 (5), 421–444. https://doi.org/10.1080/15592294.2019.1595297 (2019).
Houseman, E. A., Kim, S., Kelsey, K. T. & Wiencke, J. K. DNA methylation in whole blood: uses and challenges. Curr. Environ. Health Rep. 2 (2), 145–154. https://doi.org/10.1007/s40572-015-0050-3 (2015).
R: The R Project for Statistical Computing. https://www.r-project.org/ (accessed 13 Nov 2022).
Tuminello, S. et al. Genome-wide DNA methylation profiles and breast cancer among world trade center survivors. Environ. Epidemiol. 8 (3), e313. https://doi.org/10.1097/EE9.0000000000000313 (2024).
Sinclair, D. A. & Oberdoerffer, P. The ageing epigenome: damaged beyond repair? Ageing Res. Rev. 8 (3), 189–198. https://doi.org/10.1016/j.arr.2009.04.004 (2009).
Yu, H. et al. Global DNA methylation of WTC prostate cancer tissues show signature differences compared to non-exposed cases. Carcinogenesis. 43 (6), 528–537. https://doi.org/10.1093/carcin/bgac025 (2022).
Kuan, P. F. et al. PTSD is associated with accelerated transcriptional aging in world trade center responders. Transl. Psychiatry. 11, 311. https://doi.org/10.1038/s41398-021-01437-0 (2021).
Bourassa, K. J. & Sbarra, D. A. Trauma, adversity, and biological aging: behavioral mechanisms relevant to treatment and theory. Transl. Psychiatry. 14 (1), 1–13. https://doi.org/10.1038/s41398-024-03004-9 (2024).
Wang, Z. et al. Association between posttraumatic stress disorder and epigenetic age acceleration in a sample of twins. Psychosom. Med. 84 (2), 151–158. https://doi.org/10.1097/PSY.0000000000001028 (2022).
Smith, A. K. et al. Epigenetic age acceleration and disparities in posttraumatic stress in women in Southeast louisiana: NIMHD social epigenomics program. JAMA Netw. Open. 7 (7), e2421884. https://doi.org/10.1001/jamanetworkopen.2024.21884 (2024).
Wolf, E. J. & Morrison, F. G. Traumatic stress and accelerated cellular aging: from epigenetics to cardiometabolic disease. Curr. Psychiatry Rep. 19 (10), 75. https://doi.org/10.1007/s11920-017-0823-5 (2017).
Yusipov, I. et al. Age-related DNA methylation changes are sex-specific: a comprehensive assessment. Aging. 12 (23), 24057–24080. https://doi.org/10.18632/aging.202251 (2020).
Nikita, N. et al. Epigenetic landscapes of aging in breast cancer survivors: unraveling the impact of therapeutic Interventions—A scoping review. Cancers. 17 (5), 866. https://doi.org/10.3390/cancers17050866 (2025).
Gosselt, H. R. et al. Global DNA (hydroxy)methylation is stable over time under several storage conditions and temperatures. Epigenetics. 16 (1), 45–53. https://doi.org/10.1080/15592294.2020.1786318 (2021).
Li, Y. et al. Stability of global methylation profiles of whole blood and extracted DNA under different storage durations and conditions. Epigenomics. 10 (6), 797–811. https://doi.org/10.2217/epi-2018-0025 (2018).
Acknowledgements
First, we would like to thank all the enrollees of the WTC EHC and NYUWHS cohorts and participants of this study. We would like to acknowledge healthcare providers at the WTC EHC Clinic and other colleagues at the WTC EHC Data Center for assistance with data and sample collection. We also extend our thanks to all our colleagues at the NYU Center for Biospecimen Research and Development as well as at the Genome Technology Core.
Funding
This work was partially funded through NYU Laura & Isaac Perlmutter Comprehensive Cancer Center Support Inter-Disciplinary Population Research Pilot Grant Program (P30CA016087) and CDC/NIOSH grant 5R21OH012238. The WTC EHC clinic and the WTC EHC Data Center are funded by CDC/NIOSH contracts 200–2017 – 93327 and 200–2017 – 93427. ST was supported in part by NYU’s T32 Training Program in Healthcare Delivery Science and Population Health Research, funded by the Agency for Healthcare Research and Quality (AHRQ) grant HS026120. YAA and YS were also partially funded by CDC/NIOSH grant U01OH012486. YS and AA were partially supported by CDC/NIOSH Grant: U01OH012778.
Author information
Authors and Affiliations
Contributions
Conceptualization: A.A.A., M.S. and Y.S.; Data curation: S.T.; Formal analysis: S.T., Y.A., C.S.; Funding acquisition: A.A.A., J.R.; Investigation: S.T., Y.A., C.S., S.R; Methodology: S.T., A.A.A., Y.A., Y.S., R.S., C.S., and M.S.; Project administration: S.T., A.A.A.; Resources: A.A.A., M.S., and J.R.; Supervision: A.A.A., M.S. and Y.S.; Visualization: S.T. and Y.A. All authors have made substantial contributions to Writing - original draft and Writing - review & editing. All have final approval of the version to be submitted.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Boards of NYU Grossman School of Medicine and Bellevue Hospital (IRB numbers: s17-01207 and i21-00717). Informed consent was obtained from all study participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tuminello, S., Ashebir, Y.A., Schroff, C. et al. Epigenetic aging acceleration among World Trade Center-exposed community members. Sci Rep 15, 33942 (2025). https://doi.org/10.1038/s41598-025-10141-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10141-8