Abstract
The skeletal muscle tissue has a remarkable capacity of growth and regeneration. Fusion of myoblasts and myotubes elongation are fundamental processes in muscle development. Previous studies have depicted impaired myogenic processes in animal models and myoblast from human patients with muscle diseases. Here, we evaluated the myogenesis in patients with Limb-girdle Muscle Dystrophy 2B (LGMD2B). Aiming to explain why dysferlin-deficient muscle cells lose its myogenic potential, we used immortalized myoblasts from LGMD2B patients and a cellular DYSF knocking-out model. Myotubes from patients were smaller and containing less myonuclei than control myotubes. Main muscle regulatory factors expression were not altered in these cells. The analysis of the expression of newly described genes associated with muscle fusion and growth, such as MYMK, MYMX, PALMD, SHISA2, COL25A, didn´t show any difference with controls, which is a novel finding. It was also observed that dysferlin deficiency doesn’t alter the expression of FAM65B and HDAC6 genes, components of a proposed protein complex that needs to be formed to allow muscle differentiation. Interestingly, morphometric analysis of DYSF knock-out myotubes induced by CRISPR/Cas9 also revealed reduced myogenic capacity with formation of smaller myotubes. These findings suggest that the absence of DYSF itself is sufficient to impair muscle formation in vitro, and that downstream gene and protein expression related to muscle development might depend on the presence and proper function of dysferlin.
Similar content being viewed by others
Introduction
The skeletal muscle is a highly specialized tissue with a remarkable ability for development and growth. Due to its mechanical properties, the skeletal muscle is prone to lesion, and its striking regenerative capacity allows it to recover after lesion and restore its anatomic and physiological functions. Processes involved in muscle formation, such as stem cell activation, myoblast proliferation, and fusion into new muscle fibers, are reactivated during muscle repair1. However, some conditions can impair proper muscle regeneration, leading to muscle degeneration, malfunction, and disease. That is the case of muscle dystrophies, a group of genetically diverse disorders characterized by intense and progressive degenerative process and inefficient regeneration.
Limb-girdle muscular dystrophy 2B (LGMD2B or LGMDR2) is an example of these disorders. Patients experience a progressive loss of muscle strength, particularly in the proximal limb muscles, due to intense muscle degeneration, inflammation, and replacement of muscle tissue with fat and fibrotic tissues2. LGMD2B is caused by mutations in the DYSF gene resulting in reduced or absent expression of the dysferlin (DYSF) protein.
Dysferlin is a member of the ferlin family of proteins, which includes proteins with a single short extracellular C-terminal domain and five to seven C2 intracellular domain, plus some Fer and DysF domains3. Another member of this family, myoferlin, shares great similarity with dysferlin and acts on myoblast fusion and muscle regeneration during skeletal muscle development4,5, suggesting a correlate function with DYSF.
The impact of the deficiency of dysferlin has been demonstrated previously. Further than the well-known histological features observed in muscle biopsies from LGMD2B patients, including degeneration and presence of small immature fibers, damage to the muscle fiber membrane and accumulation of subsarcolemmal small vesicles were observed on fibers from patients and dysferlin-deficient mice as well6,7,8,9. Additionally, it has been shown that lack of DYSF impairs myogenesis in vitro in both human and animal primary myoblast10,11,12.
Many functions are designated to dysferlin, such as vesicle traffic, calcium homeostasis, T-tubule biogenesis, and membrane repair after lesion. It was also reported that dysferlin participates in a transient protein complex that binds FAM65B and HDAC6 proteins to promote myoblast fusion and differentiation13. In this model, the formation of the DYSF-HDAC6-FAM65B protein complex would inhibit the deacetylation of the α-tubulin monomers by HDAC6, allowing the polymerization and elongation of the tubulin microtubules, which is necessary for myotubes fusion and growth. Nevertheless, it has not been demonstrated so far what are the consequences to the protein complex with the dysferlin protein withdrawal, and how it could impact on myogenesis.
Hence although many of the dysferlin attributes are well known, the molecular mechanisms by which dysferlin acts need to be further studied, especially those mechanisms that are associated with muscle development and regeneration.
In the past two decades, a significant number of genes and proteins have been associated with skeletal muscle development and membrane fusion. Myomaker (MYMK) and Myomixer (MYMX) are two muscle specific proteins, expressed and needed during membrane fusion and that act jointly to perform muscle differentiation during development and regeneration14,15,16,17,18,19. Another protein, palmdelphin (PALMD) was shown to be necessary for muscle formation and regeneration, as its expression increases during these processes, and its inhibition impairs myogenesis20. SHISA2 is a protein found in the endoplasmic reticulum that regulates satellite cells derived myoblast fusion in the first stages of cell differentiation21. The last one is the protein COL25A1, that was shown to be expressed during muscle embryogenesis and its expression increases with myogenin (MYOG) at the same time22.
It is so far unclear whether these newly identified proteins participate in muscle development and regeneration with dysferlin altogether. There are no studies that evaluate if dysferlin interacts physically with these proteins, mainly MYMK and MYMX that reside in the same subcellular compartment and perform similar functions, nor if dysferlin modulates the expression of its coding genes somehow. Hence, in this present study, we propose to evaluate if these other proteins participate in the process and are altered by dysferlin deficiency, or, if dysferlin deficiency by itself is sufficient to cause the observed myogenic impairment.
Results
LGMD2B muscle cells have impaired differentiation
Differentiated myotubes from the control and the three LGMD2B patients cell lines (here identified as P1, P2 and P3) were stained against alpha-actinin protein and measured. The myonuclei were also counted to perform the fusion index and proportion analysis.
The area of the myotubes (µm2) from the dysferlin-deficient lines was much smaller than that of the myotubes from the control line after three days of differentiation (Fig. 1A). Mean ± standard deviation of control was 72.356 μm² ± 57.879; and from LGMD2B = 28.027 μm² ± 28.091. This difference was statistically significant (Mann-Whitney test statistic, p < 0.0001; control = 35 myotubes, LGMD2B = 77 myotubes) (Fig. 1B). Individually, with exception of P2, the area of the myotubes from P1 and P3 also showed a statistically significant reduced measure (P1 x Control: Mann-Whitney test statistic, p = 0.0008; control = 35 myotubes, P1 = 31 myotubes; P3 x Control: Mann-Whitney test statistic, p < 0.0001; control = 35 myotubes, P3 = 25 myotubes). Such difference was not observed between P2 and Control (Mann-Whitney test statistic, p = 0.1285; control = 35 myotubes, P2 = 21 myotubes), although the mean size of the P2 myotubes were approximately 60% of the mean size of the control myotubes (P2: mean ± standard deviation = 44.382 μm² ± 37.395). 90% (P1), 71% (P2) and 100% (P3) of myotubes were smaller than 50,000µm2 compared with 46% in control cells, being 32% of the myotubes > 100.000 μm² or larger (Fig. 1C). A Pearson’s chi-square test was performed to verify whether the distribution of myotube area differed among control and LGMD2B patients’ cells. The overall analysis revealed a statistically significant difference in the frequency distribution across groups (χ² = 31.467; df = 12; p = 0.0017). Pairwise comparisons showed a significant difference between Control and P1 (χ² = 15.13; df = 4; p = 0.0044), which was confirmed by Fisher’s exact test due to expected counts less than 5 (p = 0.0014). A highly significant difference was found between Control and P3 (χ² = 19.861; df = 4; p = 0.00053; Fisher p = 3.08 × 10⁻⁵). However, no significant difference was observed between Control and P2 (χ² = 4.95; df = 4; p = 0.292; Fisher p = 0.252) Multiple comparisons using the Bonferroni method were performed, and the differences between Control x P1 (adjusted p = 0.0133) and Control x P3 (adjusted p = 0.0016) remained statistically significant, whereas Control x P2 was not significant (adjusted p = 0.876).There was no correlation in myotube size and DYSF levels in myotubes from LGMD2B cell lines, as cells from P1, with total absence or P2 and P3 with reduction of dysferlin protein in the muscle (analysis done in routine diagnostic analysis, Table 1), presented similar altered sizes.
Myotubes from the LGMD2B lines are smaller than control. (A) Myotubes from control and the LGMD2B cell lines differentiated for three days stained with anti-alpha-actinin antibody (green labelling). (B) Mean and SD values of myotube area showing that in the mean, the size of the three LGMD2B cell lines is significantly smaller than those from the control cell line. (C) Proportion of myotube’s size classes, indicating that most measured myotubes from the LGMD2B cell lines are smaller than 50,000 μm², while more than half of the myotubes from the control cell line are larger than 50,000 μm2.
Despite the variation on the myotubes size, the fusion index, calculated by the ratio between myonuclei within myotubes and total myonuclei number in the field of view, was similar comparing control and LGMD2B with no statistical significance (two-sample Student’s t-test, p = 0.8380) (Fig. 2A). However, there was a difference concerning the proportion of the number of myonuclei per myotube: the highest percentage of myotubes from LGMD2B cell lines were from myotubes with fewer myonuclei (< 20 or between 21 and 40 myonuclei/myotube), while in the control, the major proportion of myotubes were formed containing more than 40 myonuclei. (Fig. 2B).
Fusion parameters from myotube differentiation are not altered in the LGMD2B, but there is a predominance of myotubes with fewer myonuclei. (A) Fusion index, a percentage calculated by the ratio between the number of myonuclei inside myotubes and the total number of myonuclei in the field, indicates no differences between control and LGMD2B myotubes. (B) Distribution of the proportion of myotubes containing different number of myonuclei, showing that most myotubes from the control cell line have more nuclei than the myotubes from the LGMD2B cell lines.
Expression of muscle regulatory factors is normal despite the impaired myogenic process in patient cells
RT-qPCR analysis was performed to quantify and compare the expression of the genes MYF5, MYOD and MYOG between control and LGMD2D cells.
The genes MYF5, MYOD and MYOG were similarly expressed in both control and LGMD2B cells in all cell stages. No statistical significances were observed in any time point (Fig. 3), as well as for the comparison between the control cell line and each of the patients’ cell lines.
Relative expression of the muscle regulatory factors MYF5, MYOD and MYOG shows that the expression profile of these genes in the LGMD2B cell lines follow the normal pattern of the expression seen in the control cell line, without differences between them in any of the stages of cell fusion. Bars represent standard deviation.
Dysferlin deficiency does not change the expression of the genes associated to muscle fusion
RT-qPCR analysis was performed to quantify and compare the expression of the genes MYMK, MYMX, PALMD, SHISA2, COL25A1 between control and LGMD2D cells. Immunofluorescence staining was also performed to detect and localize their expressed proteins. The relative expression of the genes MYMK, MYMX, PALMD, SHISA2 and COL25A1 during muscle differentiation has a similar pattern in both control and LGMD2B cells (Fig. 4A), with no statistically significant difference among the levels of expression in each of the cell stages. An exception was the difference of MYMX expression levels, which was lower in LGMD2B myoblasts (two-sample Student’s t-test, p = 0.041). The expression of COL25A1 was higher in all cell stages in LGMD2B, although the difference was not statistically significant. MYMX, PALMD, SHISA2 and COL25A1 proteins were detected by immunofluorescence in both undifferentiated (myoblast) and differentiated (myotube) stages of control and LGMD patients (Fig. 4B), with cytoplasmatic localization in fused myotubes in PALMD, and COL25A1. A nuclear labelling was observed for SHISA2 in fused myotubes, both in controls and LGMD lines. For MYMX, however, the nuclear labelling was observed only in the affected LGMD2B lines.
MYMK, MYMX, PALMD, SHISA2 and COL25A1 genes and proteins are normally expressed and translated in the LGMD2B cell lines. (A) The relative mRNA expression profile of these genes in the LGMD2B cell lines follows the same pattern as in the control cell line, without differences between them in any of the stages of cell fusion. Exception was observed for MYMX in myoblasts from LGMD2B presenting lower expression than controls. Bars represent standard deviation. (B) Immunofluorescence staining of the proteins coded from these genes, showing that they are expressed in both undifferentiated and differentiated stages, and without qualitative differences between control and LGMD2B cell lines.
Dysferlin deficiency does not change the expression of the components of the DYSF-HDAC6-FAM65B protein complex
RT-qPCR analysis was performed to quantify and compare the expression of the genes HDAC6 and FAM65B between control and LGMD2D cells. Immunofluorescence staining was performed to qualitatively determine the expression of acetylated α-tubulin and myogenin, as an indirect marker of the formation of the protein complex.
The mean expression of HDAC6 and FAM65B was similar in LGMD2B during the differentiation process in all time points compared to the control, with no statistical difference (Fig. 5A). It was also qualitatively observed that cytoskeleton microtubules of acetylated α-tubulin were present in the formed myotubes from both controls and LGMD2B lines, mainly in P1 and P2, showing that the polymerization of the microtubules keeps happening regardless of dysferlin being presence or not. Furthermore, the myotubes from LGMD2B cell lines also expressed nuclear myogenin, and it has been showed elsewhere that myogenin expression depends on the acetylation of α-tubulin (see Discussion session; Fig. 5B).
The expression of the genes and the make-up of the protein complex FAM65B-HDAC6-DYSF formed by them seems to be unaltered in LGMD2B cell lines. (A) The relative expression profiles of HDAC6 and FAM65B genes in the LGMD2B cell lines follow the pattern of expression seen in the control cell line, without differences between them in any of the stages of cell fusion. (B) Immunofluorescence staining of acetylated α-tubulin and myogenin, showing that they are expressed in LGMD2B cells. Although these cells form smaller myotubes, they retain the expression of acetylated α-tubulin and MYOG, indicating that the mechanisms of formation of the protein complex are ongoing.
DYSF knocking-out reproduces LGMD2B cells phenotype
CRISPR/Cas9 approach was performed to knock-out the DYSF gene in the control cell line in order to reproduce the LGMD2B cells phenotype. Differentiated myotubes from the control (DYSF+/+) and the edited (DYSF−/−) cell lines were stained against myosin protein and measured. The dysferlin protein levels were detected using Western Blotting. The myonuclei were also counted to perform the fusion index and proportion analysis.
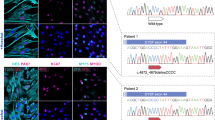
We could successfully knock-out the DYSF gene (DYSF−/−) using the CRISPR/Cas9 approach on cells from the control (DYSF+/+) cell line. Several different clones were obtained, and after subsequent steps of purification by clonal selection, one clone was selected in which the dysferlin gene was successfully silenced. Molecular analysis showed that this clone harbored two pathogenic mutations in the exon 4 of the DYSF gene in compound heterozygosis, identified from the analysis of the whole exome sequencing of the edited cell line: NM_003494:DYSF: c.564_565insT; p.Leu97Serfs*51 with an in silico prediction of mRNA degradation by nonsense-mediated decay (NMD) mechanism, and an inversion that breaks the exon 4 coding sequence. Immunofluorescence staining revealed absence of the DYSF protein in myotubes from the DYSF−/− cell line (Fig. 6). The transformed myoblasts were able to differentiate in myotubes, expressing myosin (MY32), indicating that these cells did differentiate and that DYSF absence was not due to lack of differentiation. Protein detection by western blot also showed DYSF reduction in DYSF−/−. This reduction is like what was found in the cell lines from LGMD2B patients. The DYSF−/− cell line expressed the dystrophin (DMD) protein, another finding that indicated that the cells were differentiated by the time that the proteins were extracted. It was also found a secondary reduction in the expression of the calpain 3 (CAPN3) protein in DYSF−/−, a finding that has been reported in other LGMD2B patients (Fig. 6B).
Silencing of the DYSF gene by CRISPR/Cas9 successfully reduces the synthesis of the dysferlin protein in myotubes from the control cell line. (A) Immunofluorescence staining indicating the absence of DYSF in the edited cell line, and the presence of myosin, showing that the cells maintain their capacity to differentiate. (B) Protein detection by western blotting highlights the reduction of the dysferlin protein to the levels of the LGMD2B myotubes and shows a secondary reduction of the calpain-3 protein, a finding already observed in other LGMD2B patients. Internal WB control for protein content in each sample can be observed with labelling to dystrophin (DMD), dysferlin (DYSF) and calpain3 (CAPN3).
The area of the myotubes (µm²) from the DYSF−/− line was largely reduced compared to that of the myotubes from the DYSF+/+ line after three days differentiation (mean ± standard deviation: DYSF+/+ = 87.035 μm² ± 86.962; DYSF−/− = 31.132 μm² ± 22.945; Fig. 7A). A comparison between these two groups showed that this difference is statistically significant (Mann-Whitney test statistic, p = 0.005; DYSF+/+ = 50 myotubes, DYSF−/− = 50 myotubes; Fig. 7B). Almost every myotube from the DYSF−/− cell line has an area smaller than or equal to 50,000 μm² (82%), whilst only half myotubes from the DYSF+/+ have this range of size (Fig. 7C). A Pearson’s chi-square test was performed to verify whether the distribution of myotube area differed among DYSF+/+ and DYSF−/− cells, revealing a statistically significant difference in the frequency distribution (χ² = 19.227; df = 4; p = 0.0007092), which was confirmed by Fisher’s exact test due to expected counts less than 5 (p = 0.0001616).
Dysferlin knocking-out reproduces the impaired myogenic phenotype observed in the LGMD2B myotubes. (A) Immunofluorescence staining against anti-alpha-actinin antibody shows smaller myotubes in the DYSF−/− cell line. (B) Mean and SD values of myotube area showing that the size of the DYSF−/− myotubes are significantly smaller than those from the control cell line. (C) Proportion of myotube’s size classes, indicating that over 80% measured myotubes from the DYSF−/− cell lines are smaller than 50,000 μm², while about half of the myotubes from the control cell line are larger than 50,000 μm².
Discussion
The observation of the formation of smaller myotubes from the LGMD2B patients’ cell lines during in vitro differentiation compared to a control cell line was what first motivated the execution of this work. Such finding was related to the lack of dysferlin, as all the dysferlin-deficient cell lines from this work, which come from different and unrelated affected donors, and with different mutations in the DYSF gene, showed a reduced capacity to form myotubes with a normal size. It is noteworthy point out that different levels of the dysferlin protein (absent or reduce) in LGMD2B cell lines did not correlate with the myotube phenotype, as regardless of the resulting amount of dysferlin, similar morphometrical changes were observed among the three LGMD2B cell lines.
Previous studies with two dystrophic and dysferlin-reduced mice models (SJL/J and A/J) of LGMD2B also reported that their myotubes presented smaller diameter, an altered pattern of expression of MRFs including MYOG, and increased expression of pro-inflammatory pathways including the NFκB pathway, which has been shown to be a negative regulator of myogenesis11,23 and keep together these findings, providing evidence to a cross-species fundamental role of dysferlin in muscle development.
Similar results were found by de Luna et al., 200610 from fusion analyses of human primary myoblasts derived from three DYSF−/− patients, in which a delayed fusion was noted in the patients’ cells from the fifth day of differentiation and onwards and a reduction of the muscle regulatory factor MYOG. The cells used in our work, both control and patient, were genetically modified with the constitutional expression of the CDK4 gene that induces a more efficient proliferative rate24,25, making cells able to proliferate and differentiate earlier compared to primary non-modified myoblast. However, differently from what was reported by de Luna, cells from this study showed the same proliferative and fusion capacity indicated by the fusion index. These data can suggest that, even though one of the main DYSF roles is in cell membrane dynamics, the lack of dysferlin might be more significant to the processes of myotube growth and maturation than to fusion mechanisms itself.
Followed by the impaired myotube formation, we expected that the expression of the main muscle regulatory factors could be also altered, as they are involved in the main processes of muscle differentiation. Nevertheless, the expression of MYOD and MYF5 is quite similar to the control in the LGMD2B cell lines. MYF5 and MYOD are normally expressed in the muscle progenitor cells driven themselves to become myoblasts and proliferate, and their expression reduces greatly as soon as differentiation processes start26. Hence, our analysis showed that the expression of these two MRFs in our cells follow their conventional pattern of expression, indicating that the myoblasts behavior might not be altered in the LGMD2B cell lines, adding another piece of evidence that the myogenic impairment in dysfelin deficient cells is a later stage problem.
The third analyzed MRF is MYOG that is associated with myotube differentiation, so its expression increases and keeps higher during most part of the differentiation stage26. Here, we also observed that the expression in the control and LGMD2B increases after cells receive the stimuli to differentiate, with no statistical difference between them, although the mean fold change was lower in LGMD2B cells. As afore mentioned, it was shown a reduced expression of MYOG in primary myoblasts from dysferlin deficient patients10 and in muscle cells from Dysf deficient mice strains11. A plausible explanation could be the fact that while primary myoblasts take about two weeks to differentiate, the immortalized myoblasts have a more efficient cell cycle and faster differentiation process (3 to 6 days). This faster process could be more stable and resistant to fluctuations in the expression of the MRFs.
The modulation of the expression of MYOG seems to be disturbed by many factors in muscle development. It was shown previously that the protein FAM65B acts during muscle differentiation and that its silencing causes reduction on the expression of MYOG and impairs myotube formation27. We evaluated if there were any changes in FAM65B in the LGMD2B cells, and we found that its expression was similar to the control, increasing during the process of differentiation. This was, therefore, compatible with the levels of expression of MYOG in those dysferlin deficient cell lines.
A more recent study proposed a mechanism by which FAM65B would be related to muscle differentiation and it involves the formation of a transient protein complex composed of FAM65B, HDAC6 and DYSF13. In this interesting model, FAM65B would be necessary to bind HDAC6 to DYSF, blocking its role of deacetylate α-tubulin monomers, allowing them to polymerizate into cytoskeleton microtubules that help with cell elongation and fusion. The expression analysis of FAM65B and HDAC6 showed no significant difference between the control and LGMD2B. Using immunofluorescence staining, we could identify the presence of acetylated α-tubulin inside the myotubes from the LGMD2B patients, showing that α-tubulin acetylation keeps undergoing even in the deficiency of dysferlin. Therefore, the role of the formation of the tricomplex (or bi-complex) allowing the acetylation of α-tubulin is apparently maintained. One explanation could be that even a small amount of this complex would be sufficient to sequester some HDAC6 and therefore allow the polymerization of a-tubulin microtubules. In this regard, less formation of the complex could explain the smaller size of myotubes observed in LGMD2B patients. A second evidence of the formation of acetylated α-tubulin in the LGMD2B myotubes is the clear presence of nuclear MYOG in those myotubes, since it has also be reported that the expression of MYOG depends on the acetylation of α-tubulin28.
Hence, regarding the formation of the FAM65B-HDAC6-DYSF protein complex, we could elaborate two possible scenarios: first, in which residual dysferlin levels would still allow the complex formation and promote muscle differentiation by the formation of smaller myotubes; second, if we consider that an alternate protein could occupy the role of dysferlin, such as myoferlin, which seems to play a role in the first steps of myoblast fusion5. We could hypothesize that myoferlin could play the role of dysferlin in this context, allowing the formation of an alternative tri-complex, sufficient to allow the formation of myotubes. Both hypotheses can fit as explanations to the impaired myogenesis observed in LGMD2B: the formation of a residual protein complex, maintaining some unbound HDAC6; or a complex that is less stable in the presence of a dysferlin substitute.
At the conception of this work, we were more interested in the possible role that genes recently associated with muscle development could play when dysferlin is reduced. As far as we know, these new genes haven’t been studied in muscle dystrophies. The MYMK and MYMX genes are the newest muscle specific genes identified with an important role in the fusion of myoblasts into myotubes29. For efficient cell fusion, bilateral MYMK expression has been shown to be essential, whereas MYMX expression is required in only one of two adjacent muscle cells. In the proposed mechanism, MYMK would act first, promoting membrane competence for fusion, and MYMX would act afterwards destabilizing and creating pores in the cell membrane so that cells can fuse their cytoplasm14,30,31,32. Therefore, both are extra nuclear proteins. The expression of both genes increases at the beginning of the muscle cell differentiation under the control and regulation of MYOD and MYOG29,33,34. In our findings, the expression of MYMK increases during differentiation in the control and LGMD2B cells, following the same pattern of MYOG expression, in agreement with the proposed regulation of MYMK by MYOG. The mean fold change of MYMK expression in LGMD2B is about half of the control expression during the proliferation and differentiation, and although this difference is not statistically significant, it might have a biological consequence to the muscle cell differentiation. As for MYMX, myoblasts from the LGMD2B cell lines have a lower, statistically different, MYMX expression during the proliferation in the undifferentiated stage, but the mean expression values become similar to the control as cells differentiate. Additionally, immunofluorescence analysis identified a higher presence of this protein in the nucleus of affected myotubes. So far, there are no evidence of an alternative role of MYMX in other cell compartments rather than the cellular membrane, hence this is a new observation that could point towards novel MYMX functions. However, the implications of this cellular relocation are not clear, so we suggest that this new finding should be further addressed.
The remaining genes evaluated are PALMD, SHISA2 and COL25A1. The PALMD gene, responsible for encoding the cytosolic protein palmdelphin, was also associated with the process of muscle differentiation, with gradually increased expression during development and in the process of muscle regeneration after injury20. In line with what has been reported in the literature, we observed an increase in PALMD expression during development in all lineages, with higher expression in myotubes than in myoblasts. This pattern was similar in the LGMD2B cell lines compared to the control. Therefore, the expression of this gene does not seem to vary as a consequence of the DYSF reduction. SHISA2 was identified encoding an endoplasmic reticulum protein that regulates the fusion of satellite cell-derived myoblasts, whose expression is also increased in myogenic differentiation21. In fact, in our experiments we also observed an increase in the expression of this gene in myotubes in relation to myoblasts, without differences between control and LGMD2B. Therefore, the expression of the SHISA2 gene also does not appear to vary as a consequence of the DYSF deficiency. The COL25A1 gene is responsible for synthesizing collagen XXV, a transmembrane protein. This gene is highly transcribed during the beginning of myogenesis, acting in the initial formation of myofibers. Its expression is reduced when multinucleated myotubes are already formed by a muscle-specific microRNA22. In general, there was a slightly higher expression of COL25A1 in LGMD2B lines compared to control in myoblasts, with a reduction of expression with the formation of myotubes. Therefore, the pattern of expression was similar in the patients and control.
Finally, we sought to investigate if we could reproduce the impaired myogenesis observed in the patients’ muscle cells by knocking-out solely the DYSF gene in the control cell line. By doing this, we could evaluate the effects of the reduction of DYSF without the interference of the genetic background variability that we were dealing with as we were analyzing cell lines from unrelated patients. We could successfully knock-out the DYSF gene, and by measuring the developed myotubes after three days under differentiation stimuli, it was clear that myotubes from the edited cell line also presented a reduction in its myogenic capacity.
Intriguingly, we also observed a secondary reduction of the calpain-3 protein. The CAPN3 gene, which codes for the calpain-3 protein, is associated with LGMD2A35. Previous studies have reported this secondary CAPN3 reduction in LGMD2B patients36,37, although this reduction couldn’t be related to phenotype modulation. Calpain-3 has been proposed to modulate the dysferlin protein complex, which is established during the dysferlin-dependent membrane repair mechanism38. However, it remains necessary to better understand this modulation, and it is still unclear how and why dysferlin reduction can also provoke calpain-3 reduction.
In conclusion, our findings suggest that the absence of DYSF itself is sufficient to impair muscle formation in vitro, and that downstream gene and protein expression related to muscle development might depend on the presence and proper function of dysferlin somehow. We also highlight that many more complex factors, further than the modulation of muscle regulatory factors, can be involved in the pathophysiology of the LGMD2B phenotype regarding muscle development, so additional studies addressing this matter should be performed. As far as we know, this work is the first to evaluate the consequences of dysferlin reduction in the expression of the genes recently associated with muscle differentiation; hence, we contribute to the characterization of their functions.
Methods
We confirm that all the used methods were carried out in accordance with the scientific guidelines in the area, and the used protocols are approved by the scientific committee of the Department of Genetic of the University of Sao Paulo.
Cell culture
Immortalized cell lines with mutation in the dysferlin gene and normal controls were kindly obtained from the Institut de Myologie cell bank- AFM-MyoLine. In this bank, all human samples were obtained in accordance with European recommendations and written informed consent were obtained from all subjects.
The muscle sample used for generating the control cell line was also supplied by MyoBank. MyoBank is the tissue bank associated with the Institut de Myologie in Paris and affiliated with EuroBioBank. MyoBank is authorized by the French Ministry of Higher Education, Research, and Innovation to distribute human samples for research purposes (Authorization code AC-2019-3502).
In total, four different strains were used, one being a normal control cell line that was used in all experiments, and three DYSF deficient cell lines (Table 1). P1 cell line comes from the vastus lateralis muscle of a 36 years old female carrier of the mutations c.107T > A/c.1448 C > A in compound heterozygosis in DYSF, which causes total absence of the DYSF protein; P2 cell line comes from the vastus lateralis muscle of a 57 years old male homozygous for the mutation c.4022T > C, which causes reduction of the DYSF protein; and P3 cell line comes from the quadriceps of a 17 years old male homozygous for the mutation c.4882G > A, which causes reduction of the DYSF protein.
Human myoblasts were immortalized with the addition of the human hTERT and CDK4 genes and were gently provided by Professor Vincent Mouly of the Institut de Myologie.
The myoblasts were seeded and let to proliferate in a cell culture media containing DMEM, high glucose, GlutaMAX™ Supplement medium (Gibco, 10566016), 199 medium (Gibco, 11150-059), 20% fetal bovine serum (Gibco, 12657-029), 25 µg/mL fetuin (Life Technologies, 10344026), 5ng/mL hEGF (Life Technologies, PHG0311), 0,5ng/mL bFGF (Life Technologies, PHG0026), 5 µg/mL insulin (Sigma, 91077 C), 0,2 µg/mL dexametasone (Sigma, D4902) and 1% antibiotics penicillin e streptomycin (Gibco, 15140-122). After reaching 80-90% confluence, cells were differentiated in a 1% Matrigel coated surface for three and six days. Culture media was replaced by a differentiation media containing DMEM, high glucose, GlutaMAX™ Supplement medium (Gibco) with 2% horse serum (Gibco, 26050-070), 1% insulin (Sigma, 91077 C) and 0.1% antibiotic gentamycin (Gibco, 15710-072). The myoblasts and myotubes were (1) fixated with 4% paraformaldehyde for immunostaining and (2) harvested for protein and RNA extraction.
Immunofluorescence and morphometric analysis
Immunofluorescence reaction was performed on cells to (1) delimitate myotube’s size for measurement, and (2) detect proteins of interest. The delimitation of myotubes and nuclear labeling was done following a standard protocol, using the primary antibody Monoclonal Anti-alpha-actinin (Sigma A2543) 1/500 and diluted DAPI (2 µg/mL) in Vectashield® Mounting Medium (Vector Laboratories). The images were taken using a Zeiss LSM 880 confocal microscope at 200x magnification. At least ten fields from each strain/plate were photographed. The ImageJ software was used to measure the myotube area (in µm²) and myonuclei count. The parameter used to evaluate fusion efficiency was the calculation of the fusion index, estimated as the ratio between myonuclei within myotubes and total myonuclei number in the field of view. For protein detection, the cells were stained with the following antibodies: Anti-ESGP [MYMX] (ab214934, Abcam), 1/150; PALMD (PA5-53678, Invitrogen), 1/200; Anti-COL25A1 (SAB1409894, Sigma), 1/150; Anti-SHISA2 (HPA050172, Sigma), 1/100; Acetyl-α-Tubulin (Lys40) (D20G3) XP® (#5335, Cell Signaling), 1/500; Anti-Myogenin (556358, BD Biosciences), 1/100. Slides were mounted with DAPI (2 µg/mL) and Vectashield® Mounting Medium (Vector Laboratories). The images were taken using a Zeiss LSM 880 confocal microscope at 400x magnification using a Plan-APOCHROMAT 40x/1,4 Oil DIC (UV) VIS-IF (Zeiss) lens.
RT-qPCR analysis
For gene expression analyses, RNA was extracted from cell lines at the myoblast and myotubes (three and six days) stages. The TRIzol™ Reagent RNA extraction protocol (Invitrogen) was used following the manufacturer’s recommendation. The obtained RNA was quantified in a NanoDrop spectrophotometer and stored at -70ºC until later use. The cDNA synthesis was performed using the SuperScript™ VILO™ Master Mix kit (Invitrogen) and 1 µg of total RNA. The samples were incubated in a thermocycler for 10 min at 25ºC, 42ºC for 60 min and 85ºC for 5 min. The obtained cDNA was quantified in a NanoDrop spectrophotometer and diluted for downstream applications. Quantification and comparative analysis of the expression of the genes MYMK, MYMX, PALMD, SHISA2, COL25A1, MYOG, MYOD, MYF5, HDAC6 and FAM65B were performed using 10 µg/mL cDNAs. Three independent growth and differentiation experiments were performed to obtain experimental triplicates of each of the cell lines. TBP gene expression values were used as endogenous reference. The reaction was carried out using the PowerUp SYBR Green Master Mix (Applied Biosystems) on the QuantStudio™ 5 Real-Time PCR System equipment (Applied Biosystems), following the thermocycling program 95ºC for 10 min, 95ºC for 15 s, 60ºC for 1 min, repeating the last two for 40 cycles. Expression values were obtained according to the 2^-ΔΔCт method.
CRISPR/Cas9 dysferlin Silencing
We established a knock-out model of the DYSF protein by silencing the DYSF gene in the control myoblast line through gene editing based on the CRISPR/Cas9 system. We used the enzyme TrueCut™ Cas9 Protein v2 (Invitrogen) and the sgRNA (TrueGuide™ Synthetic sgRNA, ThermoFisher Scientific) with the sequence “TGAAGCTGGCGGACAGACTA”, juxtaposed to a PAM sequence “GGG” targeting the chr.2:71503259–71,503,281 region (GRCh38/hg38 version of the human genome), which corresponds to the exon 04 of the DYSF gene. Transfection of the sgRNA (0.75 µg) + Cas9 (1.25 µg) complex was carried out through electroporation using the Neon™ Transfection System equipment (ThermoFisher Scientific) applying the following electroporation parameters: Pulse voltage (v) = 1650v; Pulse width = 10ms; Number of pulses = 3; Number of cells = 5 × 104. Sanger sequencing was performed to identify possible mutations and estimate the efficiency of gene editing, and we analyzed the .abi resulting files in the online software ICE CRISPR Analysis Tool (Synthego, www.synthego.com/products/bioinformatics/crispr-analysis). Whole Exome by Next-Generation sequencing was also performed and the detection of variants and reads alignment to the reference genome was carried out by analyzing the .bam files in the Integrative Genomics Viewer (IGV) software. Validation of the success and efficiency of DYSF gene silencing was carried out in situ through immunofluorescence analysis in myotubes differentiated after three days of the edited line with labelling for the DYSF protein, using primary antibody Recombinant Anti-Dysferlin [JAI-1-49 -3] (Abcam ab124684) 1/200. Protein detection analysis was also performed using Western blot. Total protein was extracted from differentiated myotubes by resuspending cells in lysis and extraction buffer RIPA (R0278, Sigma-Aldrich) supplemented with 1% protease and phosphatase inhibitors (Sigma-Aldrich). Protein quantification was done using the Pierce™ BCA Protein Assay Kits (Thermo Scientific) and diluted to 30 µg in 2x Laemmli buffer in a 1:1 proportion. The samples were separated by unidirectional gel electrophoresis and transferred to a PVDF membrane using iBlot® Gel Transfer Device (Invitrogen). After washing and blocking with 5% low fat dried milk solution for 60 min at room temperature, the membrane was subjected to incubation with primary antibodies for the proteins DYSF Anti-Dysferlin clone HAM1/7B6 (Vector Laboratories VP-D503), dystrophin Dystrophin (Rod Domain) clone Dy4/6D3 (Vector Laboratories VP-D508) and calpain Calpain clone Calp3c/12A2 (Vector Laboratories VP-C304), all produced in mice and used at a concentration of 1/100. Secondary antibodies conjugated to horseradish peroxidase were used. The membranes were developed using the Novex™ ECL Chemiluminescent Substrate Reagent kit (Invitrogen) and images were obtained using a photodocumentator (ChemiDoc Imaging System, Bio-Rad) The edited lineage was subsequently differentiated into myotubes on a 22 mm Tissue Culture Coverslips plastic coverslips (Sarstedt No.83.1840.001) and we proceeded to morphometric analysis, following the protocol described previously.
Whole-exome sequencing
Complete exome sequencing was carried out using Next-generation Sequencing (NGS), using the library preparation and capture kit SureSelectQXT V6 Reagent Kit (Agilent), and the sequencing reaction was carried out in a HiSeq2500 sequencer (Illumina). The generated fastq files were aligned against the GRCh38/hg18 version of the human genome using the BWA-MEM tool. Picard Tools 2.18.7 was used in the conversion of sam files to bam files and to mark PCR duplicates. The genotypes were called following the GATK best practices and we used the Genome Analysis Toolkit (GATK) UnifiedGenotyper, version 3.7. The ANNOVAR tool was used for variant annotation.
Data availability
The datasets generated and/or analyzed during the current study are available at request, to Prof. Mariz Vainzof, mvainzof@usp.br.
References
Turner, N. J. & Badylak, S. F. Regeneration of skeletal muscle. Cell. Tissue Res. 347, 759–774. https://doi.org/10.1007/s00441-011-1185-7 (2012).
Bashlr, R. et al. D. A gene for autosomal recessive Limb-Girdle muscular dystrophy maps to chromosome 2p. Hum. Mol. Genet. 3, 455–457. https://doi.org/10.1093/hmg/3.3.455 (1994).
Cárdenas, A. M., González-Jamett, A. M., Cea, L. A., Bevilacqua, J. A. & Caviedes, P. Dysferlin function in skeletal muscle: possible pathological mechanisms and therapeutical targets in dysferlinopathies. Exp. Neurol. 283, 246–254. https://doi.org/10.1016/j.expneurol.2016.06.026 (2016).
Davis, D. B. Myoferlin, a candidate gene and potential modifier of muscular dystrophy. Hum. Mol. Genet. 9, 217–226. https://doi.org/10.1093/hmg/9.2.217 (2000).
Doherty, K. R. et al. Normal Myoblast Fusion Requires Myoferlin Development 132, 5565–5575, https://doi.org/10.1242/dev.02155. (2005).
Selcen, D., Stilling, G. & Engel, A. G. The earliest pathologic alterations in dysferlinopathy. Neurology 56, 1472–1481. https://doi.org/10.1212/WNL.56.11.1472 (2001).
Bansal, D. et al. Defective membrane repair in Dysferlin-Deficient muscular dystrophy. Nature 423, 168–172. https://doi.org/10.1038/nature01573 (2003).
Cenacchi, G., Fanin, M., De Giorgi, L. B. & Angelini, C. Ultrastructural changes in dysferlinopathy support defective membrane repair mechanism. J. Clin. Pathol. 58, 190–195. https://doi.org/10.1136/jcp.2004.018978 (2005).
Hornsey, M. A., Laval, S. H., Barresi, R., Lochmüller, H. & Bushby, K. Muscular dystrophy in Dysferlin-Deficient mouse models. Neuromuscul. Disord.. 23, 377–387. https://doi.org/10.1016/j.nmd.2013.02.004 (2013).
de Luna, N. et al. Absence of dysferlin alters Myogenin expression and delays human muscle differentiation in vitro. J. Biol. Chem. 281, 17092–17098. https://doi.org/10.1074/jbc.M601885200 (2006).
Cohen, T. V., Cohen, J. E. & Partridge, T. A. Myogenesis in Dysferlin-Deficient myoblasts is inhibited by an intrinsic inflammatory response. Neuromuscul. Disord. 22, 648–658. https://doi.org/10.1016/j.nmd.2012.03.002 (2012).
Ishiba, R. et al. Faster regeneration associated to high expression of Fam65b and Hdac6 in Dysferlin-Deficient mouse. J. Mol. Histol. 50, 375–387. https://doi.org/10.1007/s10735-019-09834-y (2019).
Balasubramanian, A. et al. Fam65b is important for formation of the HDAC6-Dysferlin protein complex during myogenic cell differentiation. FASEB J. 28, 2955–2969. https://doi.org/10.1096/fj.13-246470 (2014).
Millay, D. P. et al. Myomaker is a membrane activator of myoblast fusion and muscle formation. Nature 499, 301–305. https://doi.org/10.1038/nature12343 (2013).
Millay, D. P., Sutherland, L. B., Bassel-Duby, R. & Olson, E. N. Myomaker is essential for muscle regeneration. Genes Dev. 28, 1641–1646. https://doi.org/10.1101/gad.247205.114 (2014).
Millay, D. P. et al. Structure–Function Analysis of Myomaker Domains Required for Myoblast Fusion. Proc. Natl. Acad. Sci. 113, 2116–2121, (2016). https://doi.org/10.1073/pnas.1600101113
Gamage, D. G. et al. Insights into the localization and function of myomaker during myoblast fusion. J. Biol. Chem. 292, 17272–17289. https://doi.org/10.1074/jbc.M117.811372 (2017).
Ishiba, R. Muscle Degeneration and Regeneration in Dysferlin-Deficient Murine Models (University of Sao Paulo, 2017).
Bi, P. et al. Control of muscle formation by the fusogenic micropeptide myomixer. Science 356, 323–327. https://doi.org/10.1126/science.aam9361 (2017).
Nie, Y. et al. Palmdelphin promotes myoblast differentiation and muscle regeneration. Sci. Rep. 7, 1–9. https://doi.org/10.1038/srep41608 (2017).
Liu, Z., Wang, C., Liu, X. & Kuang, S. Shisa2 regulates the fusion of muscle progenitors. Stem Cell. Res. 31, 31–41. https://doi.org/10.1016/j.scr.2018.07.004 (2018).
Gonçalves, T. J. M. et al. Collagen XXV promotes myoblast fusion during myogenic differentiation and muscle formation. Sci. Rep. 9, 5878. https://doi.org/10.1038/s41598-019-42296-6 (2019).
Peterson, J. M. & Guttridge, D. C. Skeletal muscle diseases, inflammation, and NF-ΚB signaling: insights and opportunities for therapeutic intervention. Int. Rev. Immunol. 27, 375–387. https://doi.org/10.1080/08830180802302389 (2008).
Mamchaoui, K. et al. Immortalized pathological human myoblasts: towards a universal tool for the study of neuromuscular disorders. Skelet. Muscle. 1 https://doi.org/10.1186/2044-5040-1-34 (2011).
Philippi, S. et al. Dysferlin-Deficient immortalized human myoblasts and myotubes as a useful tool to study dysferlinopathy. PLoS Curr. 4, RRN1298. https://doi.org/10.1371/currents.RRN1298 (2012).
Perry, R. L. & Rudnicki, M. A. Molecular mechanisms regulating myogenic determination and differentiation. Front. Biosci. 5, 148. https://doi.org/10.2741/Perry (2000).
Yoon, S., Molloy, M. J., Wu, M. P., Cowan, D. B. & Gussoni, E. C6ORF32 is upregulated during muscle cell differentiation and induces the formation of cellular filopodia. Dev. Biol. 301, 70–81. https://doi.org/10.1016/j.ydbio.2006.11.002 (2007).
Conacci-Sorrell, M., Ngouenet, C., Eisenman, R. N. & Myc-Nick A cytoplasmic cleavage product of Myc that promotes α-Tubulin acetylation and cell differentiation. Cell 142, 480–493. https://doi.org/10.1016/j.cell.2010.06.037 (2010).
Zhang, H. et al. Human myotube formation is determined by MyoD–Myomixer/Myomaker Axis. Sci. Adv. 6 https://doi.org/10.1126/sciadv.abc4062 (2020).
Quinn, M. E. et al. Myomerger induces fusion of Non-Fusogenic cells and is required for skeletal muscle development. Nat. Commun. 8, 15665. https://doi.org/10.1038/ncomms15665 (2017).
Leikina, E. et al. Myomaker and myomerger work independently to control distinct steps of membrane remodeling during myoblast fusion. Dev. Cell. 46, 767–780. https://doi.org/10.1016/j.devcel.2018.08.006 (2018).
Petrany, M. J., Millay, D. P. & Cell Fusion Merging membranes and making muscle. Trends Cell. Biol. 29, 964–973. https://doi.org/10.1016/j.tcb.2019.09.002 (2019).
Chen, B., You, W., Wang, Y. & Shan, T. The regulatory role of myomaker and Myomixer–Myomerger–Minion in muscle development and regeneration. Cell. Mol. Life Sci. 77, 1551–1569. https://doi.org/10.1007/s00018-019-03341-9 (2020).
Adhikari, A., Kim, W. & Davie, J. Myogenin is required for assembly of the transcription machinery on muscle genes during skeletal muscle differentiation. PLoS One. 16, e0245618. https://doi.org/10.1371/journal.pone.0245618 (2021).
Richard, I. et al. Mutations in the proteolytic enzyme Calpain 3 cause Limb-Girdle muscular dystrophy type 2A. Cell 81, 27–40. https://doi.org/10.1016/0092-8674(95)90368-2 (1995).
Anderson, L. V. B. et al. Secondary reduction in Calpain 3 expression in patients with limb girdle muscular dystrophy type 2B and Miyoshi myopathy (Primary Dysferlinopathies). Neuromuscul. Disord.. 10, 553–559. https://doi.org/10.1016/S0960-8966(00)00143-7 (2000).
Vainzof, M. et al. Dysferlin protein analysis in Limb-Girdle muscular dystrophies. J. Mol. Neurosci. https://doi.org/10.1385/JMN:17:1:71 (2001).
Huang, Y. et al. Calpain 3 is a modulator of the dysferlin protein complex in skeletal muscle. Hum. Mol. Genet. 17, 1855–1866. https://doi.org/10.1093/hmg/ddn081 (2008).
Acknowledgements
The authors would like to thank the scientific and technical support of the team of the Human Genome Research Center and Myoline, the platform for immortalization of human cells from the Institute of Myology. We would like to thank Eduard Gallardo and Simone Spuler for authorizing the use of the LGMD2B cell lines. We would like to thank the support from the NGS Diagnostic Laboratory and the help from the professionals of the Human Genome and Stem Cells Research Center. This work was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo - Centro de Pesquisa, Inovação e Difusão (FAPESP-CEPID, Grant 2013/8028-1), Conselho Nacional de Desenvolvimento Cientıfico e Tecnológico (CNPq-INCT, Grant 465355/2014-5), Financiadora de Estudos e Projetos (FINEP). CAPES – Coordenação de aperfeiçoamento de Pessoal de Nivel Superior.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: L.S.S., A.F.R.J. and M.V. Implementation of the methodologies: L.S.S. and A.F.R.J. Performed experiments: L.S.S. and I.A.Z. Contributed reagents/materials/analysis tools: L.S.S., R.I., A.L.F.S., D.B. and V.M. Prepared and/or revised the manuscript. L.S.S., A.B., and M.V.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Souza, L.S., Ishiba, R., Ribeiro-Junior, A.F. et al. Impaired myogenesis in limb girdle muscular dystrophy type 2B. Sci Rep 15, 33948 (2025). https://doi.org/10.1038/s41598-025-10205-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-10205-9