Abstract
Tumor-associated macrophages (TAMs) are pivotal immunosuppressive components of the tumor microenvironment (TME) in gastric cancer (GC), yet their heterogeneity and metabolic crosstalk with tumor cells remain poorly understood. Here, we performed single-cell RNA sequencing (scRNA-seq) on 75,743 cells from 11 GC tissues and identified four distinct TAM subsets (TAM-APOE, TAM-IDO1, TAM-SKAP1, TAM-POLB), each exhibiting unique functional signatures. Among these, the TAM-APOE subset, enriched in lipid metabolism and complement pathways, showed the strongest interaction with tumor cells via the MIF-CD74 axis. Functional studies revealed that GC-derived MIF promotes TAM-APOE polarization, upregulating immunosuppressive genes and enhancing tumor progression. In vivo, MIF overexpression in GC cells increases subcutaneous tumor volume, while MIF knockdown decreases tumor weight. Crucially, Milatuzumab, a CD74-targeting antibody, reversed MIF-induced TAM immunosuppression in vitro. Our research provides a comprehensive map of GC-TAM heterogeneity; reveals that the MIF-CD74 axis is a key driver of TAM-mediated immune suppression; and proposes that CD74 blockade is a new therapeutic strategy for GC.
Similar content being viewed by others
Introduction
Gastric cancer (GC) ranks as the sixth most prevalent malignancy and the third leading cause of cancer-related deaths globally, with a 5-year survival rate below 30% despite therapeutic advancements1,2,3,4,5,6. This dismal prognosis is attributed not only to tumor aggressiveness but also the immunosuppressive tumor microenvironment (TME), where tumor-associated macrophages (TAMs) emerge as pivotal regulators of tumor progression7,8,9,10,11,12,13,14,15.
TAMs, abundant in the TME, exhibit remarkable plasticity and can adopt distinct functional states that are determined by macrophage polarization16,17. Classically activated M1 macrophages exert anti-tumor effects via pro-inflammatory cytokines (e.g., TNF-α, IL-12), while alternatively activated M2 macrophages promote immunosuppression, angiogenesis, and metastasis through mediators like IL-10 and TGF-β8,12,16,18,19,20,21,22. However, this M1/M2 dichotomy oversimplifies macrophage heterogeneity, as single-cell RNA sequencing (scRNA-seq) has uncovered context-dependent TAM subsets with unique transcriptomic profiles in cancers such as NSCLC and colorectal cancer23,24. In GC, TAMs are implicated in chemotherapy resistance and metastasis, yet their heterogeneity and the mechanisms underlying interaction with tumor cells remain poorly defined8.
Emerging evidence suggests that metabolic crosstalk between TAMs and tumor cells, such as lipid metabolism and complement signaling, fuels TAM-mediated immunosuppression24,25. However, the molecular drivers of these interactions in GC are unknown. Moreover, while scRNA-seq has revolutionized TME analysis in other malignancies26its application to dissect GC TAM heterogeneity is limited.
Here, we combined scRNA-seq of clinical GC samples with functional validation to delineate TAM subsets and their tumor-promoting mechanisms. We identified a novel MIF-CD74 axis through which GC cells reprogram TAMs into immunosuppressive APOE + subsets, driving tumor progression. Our findings provide a high-resolution map of GC TME and highlight CD74 as a therapeutic target to disrupt TAM-mediated immunosuppression.
Materials and methods
Patient and sample collection
Gastric cancer tissues form patient donors (supplement Table S1) were used for single-cell analysis. This study was authorized by the Ethics Committee of Guangdong Provincial People’s Hospital (KY2024-123-01). In addition, the study complied with all applicable guidelines and regulations. All methods were performed in accordance with the relevant guidelines and regulations. Each participant provided a written informed consent for participation.
Single-cell RNA-seq data processing
According to the manufacturer’s protocol, the preparation of single cells was conducted in Chromium single-cell gene expression solution utilizing Chromium single cell 5 ‘gel beads, chips, and Library Kits v2 (10X Genomics). Each channel was loaded with 8000-10,000 cells, with an average cell recovery rate. The cells were divided into Gel Beads in the Chromium apparatus for barcode reverse transcription of the mRNA and cell lysis, subsequently amplification, shearing, 5’splice, as well as sample index attachment. Due to the cryopreservation of samples, when screening cells, the number of transcripts detected in each cell is controlled between 100 and 100,000, and cells with less than 100 detected genes are excluded. In data processing, the proportion of mitochondrial genes that cells are allowed to detect is less than 25%.
Cell-cell communication analysis
Intercellular networks were inferred using CellChat (v1.1.3). CellChat objects were created from normalized expression matrices, followed by standard preprocessing. Ligand-receptor interactions were quantified via law of mass action using cluster-averaged expression (computeCommunProb()), filtered for clusters with ≥ 10 cells (min.cells = 10), and aggregated into global networks (aggregateNet()).
Gene ontology and pathway enrichment analysis
The DEGs were used to identify over-represented gene ontology categories and KEGG pathways using DAVID 6.7 (https://david-d.ncifcrf.gov/). KEGG network was visualized by Cytoscape software v. 3.7.2 54.
Animal research
The study is reported in accordance with ARRIVE guidelines. Animal experiments were approved by the Guangdong Medical Laboratory Animal Center. All methods were performed in accordance with the relevant guidelines and regulations. All BALB/c thymus nude mice were purchased from the Guangdong Medical Laboratory Animal Center and placed in a specific pathogen-free (SPF) environment. A subcutaneous xenograft gastric tumor model was created in BALB/c nude mice that were 3-4 weeks old. HGC27 or AGS GC cells were injected subcutaneously to create the subcutaneous tumor model. At the end of the experiment, mice were injected intraperitoneally with 100 to 200ul of 1.25% avertin.
Cell line and cell culture
The human gastric cancer cell lines including MGC803, N87, HGC27, AGS and human gastric epithelial cell line GES-1, were supplied by the American Type Culture Bank (ATCC) and kept in the Department of Pathology, Guangdong Provincial People’s Hospital. All cell lines were characterized and detected for mycoplasma contamination in DMEM medium with 10% fetal bovine serum (FBS) as well as RPMI 1640 medium with 10% FBS.
THP-1 cell culture
Human monocytes THP-1 cells were cultured in RPMI 1640 medium (BI, USA) with 10% bovine fetal bovine serum (Gibco, USA). After being treated with PMA (100 ng/mL) for 48 h, THP-1 monocytes are differentiated into macrophages.
Cell proliferation experiment
In the CCK8 experiment, tumor cells were inoculated into 96 Wells and CCK8 solution (100 µl) was added at 0, 24, 48, 72, and 96 h, respectively (Biosharp, China). After 2 h of incubation, the absorbance was determined by Epoch Biotek (USA) at 450 nm.
Real-time quantitative PCR
Total RNA was extracted using TRIzol reagent, and quantification was performed using NanoDrop to assess RNA concentration and purity. The purified RNA was reverse transcribed into cDNA using a reverse transcription kit. This cDNA was then amplified with SYBR Green PCR Master Mix (Takara) on the LightCycler 480 Fast Real-Time PCR system (Roche).The amplification protocol consisted of 40 cycles, comprising 10 s at 95 °C and 30 s at 60 °C. GAPDH were served as an internal control, and the resulting data were analyzed employing the 2-ΔΔCt method. The sequences of the primers are detailed in supplement Table S2.
Western blotting
Cells were lysed using RIPA buffer, which was supplemented with protease and phosphatase inhibitors, and this procedure was carried out on ice for thirty minutes. The resultant cell lysates were clarified by centrifugation at 12,000 g for twenty minutes at 4 °C. Equal amounts of protein (20 µg) were subjected to SDS-PAGE and subsequently transferred onto PVDF membranes (Millipore, USA) using a wet transfer system (Bio-Rad, USA). The membranes were initially blocked with 5% BSA in TBST before incubation with specific primary antibodies. After three washes with TBST, the membranes were incubated with HRP-conjugated secondary antibodies at room temperature for one hour. Protein visualization was performed using the GelView 6000 Plus (BLT Photon Technology, China), following the manufacturer’s instructions. Each experiment was replicated three times. The primary antibodies we use are as follows: anti-MIF (1:1000; Proteintech, 20415-1-AP) and anti-GAPDH (1:1000; Proteintech, 10494-1-ap).
Immunohistochemistry
Tissue samples were fixed, embedded in paraffin, and cut into 3 μm thick sections. After de-paraffinization, an EDTA-based solution was used for antigen retrieval. Hydrogen peroxide was then applied to suppress endogenous peroxidase activity. The sections were washed three times with PBS, and then incubated with 5% goat serum for one hour at room temperature to block non-specific binding sites. The sections were then incubated overnight at 4 °C with primary antibodies. After three subsequent PBS washes, HRP-conjugated secondary antibodies were introduced at room temperature for one hour. After another three washes with PBS, the sections were developed with a DAB kit, and the chromogenic reaction was monitored for 5–10 min until the desired color intensity was reached. The nuclei were counterstained with hematoxylin, and the sections were then dehydrated, cleared, and mounted. Finally, microscopy was employed to capture images, which were subsequently analyzed using ImageJ software(ImageJ 1.54, https://imagej.nih.gov/ij/). We used the following antibodies: CD74 (Santa Cruz, ln2), MIF (Proteintech, 20415-1-AP), Goat anti-rabbit antibody (Servicebio, G1213) and goat anti-mouse antibody (Servicebio, G1214).
Immunofluorescence staining
Immunofluorescence staining was operated and analyzed as described27. The antibodies used were anti-CD68 (rabbit, 1:2000, GB113150) and anti-CD74 (mouse, 1:50, Proteintech, cat. no. GB121179).
Statistical analysis
All experiments, except for those involving mice, were performed in at least three independent biological replicates, with technical replicates for each experiment. Data are expressed as the mean ± SD. For data with a normal distribution, unpaired or paired two-tailed Student’s t-tests were used to compare the significance of differences between two groups of independent samples. For the in vivo experiments related to animals, the number of biological replicates is 4. Animals were allocated to control experimental groups using a blinding and randomization method. Statistics were performed by GraphPad Prism 8.0. A P-value less than 0.05 was considered statistically significant. All P-values are indicated in the figures (#P > 0.05; *P < 0.05; **P < 0.01;***P < 0.001).
Result
Single-cell atlas of non-malignant and malignant gastric tissues
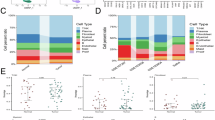
To fully characterize the single-cell profile of gastric malignancies, a total of 14 samples were collected, including 11 patients with primary tumor (GC) gastric cancer and 3 patients with non-atrophic gastritis (NAG) from previous studies [PMID: 31067475]. The clinical and histopathological features are presented in supplementary information (Table S1). After quality control and removal of batch effect, we further performed downstream analysis on 75,743 cells. To recognize distinct cell populations according to the patterns of gene expression, we identified the cell populations of these cells by descending and unsupervised clustering. Batch effects between multiple samples were successfully eliminated (Fig. 1A). Based on the UMAP map, nine cell types were identified and isolated from selected cells for downstream analysis (Fig. 1B), including B cells (labeled by MS4A1, CD79A, and CD19), T cells (labeled by CD2, CD3E, and CD3D), Plasma B cells (labeled by MZB1, IGHG1, and SDC1), epithelial cells (labeled by EPCAM, KRT19, and KRT7), Fibroblasts (labeled by DCN, COL1A1, and COL1A2), endothelial cells (labeled by VWF, ENG, and FLT1), macrophages (labeled by CD68, CD163, and CD14), Monocytes (labeled by IFITM2, NAMPT, and TREM1), Mast cells (labeled CPA3, TPSB2, and TPSAB1) (Fig. 1C, D). The proportion of each cell type in the samples indicates that macrophages, epithelial cells, T cells and B cells, in the PT samples have a high degree of patient specificity compared with the NAG (Fig. 1E). Gastric cancer tissues were then analyzed separately (Fig, S1).
Single-cell RNA-seq of gastric tumor and non-malignant samples. (A) UMAP maps of non-atrophic gastritis (NAG, n = 3) and primary tumor (PT, n = 11). (B) UMAP mapped the single-cell identification of different cell clusters in samples. (C) Dot plots of marker gene expression levels in each cell type. (D) UMAP plot showing the expression levels of canonical marker genes for nine cell types. (E) The proportion of each cell type in 14 samples.
Identification and molecular characterization of four macrophage subtypes in gastric cancer
Macrophage analysis revealed higher levels of invasion-related genes (CTSB, SPP1, VEGFA, and VEGFB), immunosuppressive factors (LGALS3, STAT4, STAT3, APOC1 and APOE), and M2-related genes (CD163, ARG1 and MSR1) in PT. However, M1 genes (CD86, CD80, CD83 and CD14) were significantly expressed in NAG (Fig. 2A). This trend suggests that with the progression of NAG to PT, macrophage clusters evolved from having an antitumor and inflammatory phenotype to a pro-tumorigenic phenotype. Through unsupervised cluster analysis (with an accuracy of 0.04), we found that macrophages in tumor tissues could be divided into four different subgroups, and each subgroup showed different transcriptome characteristics (Fig. 2B, C). Subgroup 0, designated as TAM-APOE, constituted the predominant TAM population and exhibited elevated expression of complement components (C1QB, C1QA) and lipid metabolism markers (APOE, APOC1). APOE, a key regulator of lipid homeostasis, has been implicated in promoting immunosuppression by modulating cholesterol efflux and enhancing TAM survival within the TME. Additionally, the upregulation of MMP9 and CTSD suggests a role in extracellular matrix remodeling, further facilitating tumor invasion and metastasis(Fig. 2B)24,25,28. Furthermore, GO and KEGG analyses of this subgroup correlated with complement and lipid metabolic pathways (Fig. 2D, E). Therefore, we name them TAM-APOE clusters. Subgroup 1 expresses high levels of ENOX1, IDO1 and BIRC3. GO and KEGG analyses of this subgroup revealed a notable enrichment of pathways linked to protein processing (Fig. 2D, E). Therefore, we designated them as TAM-IDO1. Subgroup 2 expresses SKAP1, CD247, IL32, and CCL5, which are typically linked to T-cell differentiation, modulation of T-cell activation, and characterization of T-cell receptor signaling pathways. Therefore, we named them TAM-SKAP1. GO and KEGG analyses also showed significant enrichment of T cell differentiation and T cell receptor signaling pathways (Fig. 2D, E). High levels of genes related to RNA processing, including SOX4, IRF4, and POLB, were expressed by subgroup 3. GO and KEGG of this subgroup also suggest that this subgroup may be associated with RNA splicing and nucleocytoplasmic transport (Fig. 2D, E). As a result, the subgroup was named TAM-POLB. To determine the potential interaction between GC cells and TME cells, we conducted CellChat analysis to map the intercellular signals. The results showed that GC cells had the strongest interaction with T cells, Fibroblasts, and TAMs (Fig. S2A). The interaction between tumor cells and TAMs can alter the quantity, phenotype, functional localization, and metabolic state of macrophages. This ultimately promotes immune escape and malignant progression of tumors24. Subsequent CellChat analysis revealed that, compared with other TAM clusters, the interaction between GC cells and TAM-APOE was stronger (Fig. S2B). The above results indicate that GC cells may promote tumor progression primarily through interaction with TAM-APOE. Next, we examined whether and how tumor cells control macrophage activity. To do this, we examined ligand-receptor pairs that are expressed on tumor cells and four distinct macrophage subtypes; these may indicate potential pathways for recruitment or interaction between particular macrophage subtypes and tumors. Analysis shows that among the interactions between TAM-APOE and tumor cells, MIF -(CD74 + CD44) is the strongest, followed by APP-CD74 (Fig. 2F). However, Kaplan-Meier survival analysis showed that higher APP level in gastric cancer patients associated with higher survival rate (Fig. S2C, D). Additionally, the higher the expression of MIF is, the lower the survival rate of patients is (Fig. 3E, F).Therefore, we speculated that macrophages may be affected by CD74 ligand MIF and we then investigated whether the expression of MIF in tumor cells changes the immune composition of the microenvironment to affect tumor progression. An additional CellChat plot of a separate MIF-CD74 axis highlights various specific interactions involving the CD74 gene (Fig. S2E).
Different macrophage subpopulations in gastric cancer. (A) Differential expression analysis of genes associated with M2 polarization, immunosuppression, invasion, recruitment, and M1 polarization in macrophage cluster. (B) Heatmap showing the top DEGs in each macrophage subtype. (C) UMAP plots show marker genes in different macrophage clusters. (D) KEGG analysis in different macrophage subtypes. (E) GO analysis in different macrophage subtypes. (F) Ligand receptor pairs of the signaling pathway network from tumor to macrophage were analyzed by CellChat. Dot color reflects communication probabilities, and dot size represents computed p-values. Empty space means the communication probability is zero.
Expression of MIF and CD74 in gastric cancer. (A) Expression of MIF mRNA in GEPIA and TCGA-GC databases. (B–D) mRNA levels of MIF in the GSE54129、GSE65801and GSE29272 dataset from the GEO database. (E, F) Kaplan-Meier survival analysis of GC patients based on MIF expression levels. (G) Representative IHC images of MIF protein expression in 71 paired GC tumor tissues and adjacent nontumor tissues. Statistical analysis of immunohistochemical score of MIF expression in GC tumor tissues and paired adjacent tissues is shown on the right. (H) Expression of CD74 in gastric cancer. (I) Representative immunofluorescence staining of CD74 (red), CD68 (green), and nuclei (blue) in paired GC samples.*P < 0.05, **P < 0.01, ***P < 0.001.
Expression and localization of MIF and CD74 in gastric cancer
We examined MIF expression in a public database to investigate the role of MIF-CD74 in GC. These include Gene Expression Atlas (GEO), Cancer Genome Atlas (TCGA), and GEPIA. The GEPIA, TCGA, and GEO databases (GSE54129, GSE65801, and GSE29272) all detected abnormally raised MIF expression in tumor tissues of GC patients (Fig. 3A-D). Besides, Kaplan-Meier survival analysis indicated that higher MIF expression was associated with lower patient survival rate (Fig. 3E, F). In addition, immunohistochemical results further showed that in comparison to adjacent non-tumor tissues, MIF was also expressed at a higher level in tumor tissues (Fig. 3G). We then determined the CD74 expression in GC and found that it was mainly expressed in TME (Fig. 3H). To evaluate the expression of CD74 in tissues, we performed combined staining of CD74 and CD68 in gastric cancer tissue sections. It was found that the number of CD74 + CD68 + cells in tumor tissues increased significantly compared to neighboring non-tumor tissues. The CD74 expression in tumor-infiltrating macrophages was verified by these findings (Fig. 3I).
Deciphering tumor-macrophage interaction supports the MIF-CD74 axis as a new therapeutic target for GC
We analyzed the MIF expression in four distinct GC cell lines and normal gastric epithelial cells in order to determine the biological significance of the MIF-CD74 axis in GC. We observed that MIF had high expression in GC cell lines in contrast to normal gastric epithelial cell GES-1 (Fig. 4A). On the basis of these results, we selected HGC27 and AGS to construct overexpression and knockdown cell lines respectively (Fig. 4B). We then examined whether MIF acted independently of the CD74 receptor in gastric cancer cells. Experiments involving CCK-8 and colony formation demonstrated that the growth and proliferation potential of GC cells was unaffected by MIF knockdown or overexpression. Colony formation and CCK-8 experiments showed that overexpression or knockdown of MIF did not affect the growth and proliferation capacity of gastric cancer cells (Fig. 4C, D). Therefore, MIF is unlikely to contribute to gastric cancer progression in a cellular manner. After excluding the fact that MIF can promote tumor growth through the autonomic role of cells, we investigated whether MIF expressed by tumor cells can change the composition of TME and promote the progression of gastric cancer through a subcutaneous tumorigenesis model. Compared to the control group, the subcutaneous tumors generated by MIF overexpressing cells were bigger and heavier (Fig. 4E). At the same time, GC cells with MIF knockdown grew markedly smaller tumors than the MIF wild-type (Fig. 4F). These findings indicate that MIF may promote GC progression by altering the composition of TME. To confirm this hypothesis, we co-stained subcutaneous tumor sections with CD74 and CD68. CD74 + CD68 + cells were markedly increased in tumors overexpressing MIF and markedly decreased in tumors with MIF knockdown compared to the control group (Fig. 4G). Our findings suggest that MIF may promote the accumulation of TAM-APOE clusters in gastric tumors, potentially through the MIF-CD74 axis. However, further studies are needed to rule out the contribution of other factors. We then investigated the effect of MIF on macrophages in vitro and found that adding rMIF to macrophages upregulated macrophage immunosuppressive genes. This change was abolished with the addition of Milatuzumab, a naked, fully-humanized monoclonal antibody targeting CD74 (Fig. 4H). Furthermore, the transwell experiment demonstrated that rMIF treatment inhibited the migration of macrophages, while Milatuzumab could eliminate the migration of macrophages (Fig. 4I). These results suggest that gastric cancer cells can promote the accumulation of TAM-APOE cluster in tumors through the MIF-CD74 axis, enhance the immunosuppressive function of TAM and promote tumor growth.
The MIF-CD74 axis remodeled the TME. (A) Expression of MIF in normal gastric epithelial cells and 4 different GC cell lines by Western blot. (B) Western blot assays were performed to verify the successful construction of MIF overexpression and knockdown GC cells. (C) Colony formation assays were performed to determine the effects of MIF on the growth of GC cells. The number of colonies (> 50 cells) was scored (mean ± SD, n = 3). (D) CCK-8 assays were performed to determine the effects of MIF on the proliferation of GC cells (mean ± SD, n = 3). (E) Control and MIF overexpressed HGC27 cells formed subcutaneous tumors in nude mice. The image on the right shows the weight of the subcutaneous tumors. (F) Control and MIF silenced AGS cells formed tumors subcutaneously in nude mice. The image on the right shows the weight of the subcutaneous tumors. (G) Representative immunofluorescence staining of CD74 (red), CD68 (green) and nucleus (blue) in subcutaneous tumor tissue sections. The numbers of CD74 + CD68 + cells per FOV are represented. (H) The effect of MIF on macrophages was analyzed by qRT-PCR. Differences were analyzed using the unpaired test two-sided t test (n = 3). (I) Transwell was used to analyze MIF and its influence on macrophage migration. Differences were analyzed using the unpaired test two-sided t test (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ns indicates P > 0.05.
Conclusions
In this study, we constructed a comprehensive single-cell atlas of gastric cancer (GC) and adjacent non-tumor tissues, revealing significant intertumor heterogeneity in the tumor microenvironment (TME). Through scRNA-seq analysis of 75,743 cells, we identified four distinct TAM subpopulations (TAM-APOE, TAM-IDO1, TAM-SKAP1, TAM-POLB), each exhibiting unique functional signatures. Among these, the TAM-APOE subset, which is enriched in lipid metabolism and complement pathways, emerged as the dominant immunosuppressive population interacting with tumor cells via the MIF-CD74 axis. Mechanistically, GC-derived MIF binds to CD74 on TAMs, triggering upregulation of immunosuppressive genes (e.g., IL-10, ARG1) and promoting tumor progression. In vivo, MIF overexpression amplified tumor growth by 2.5-fold, while MIF knockdown suppressed tumorigenesis by 60%. Crucially, the CD74-blocking antibody Milatuzumab reversed MIF-induced TAM immunosuppressive function in vitro, highlighting its therapeutic potential.
This study provides a high-resolution map of TAM heterogeneity in GC and determines that the MIF-CD74 axis is a key regulator of TME immunosuppression. However, our findings are limited by the sample size and reliance on xenograft models. Future work should validate these mechanisms in larger clinical cohorts and humanized models, while exploring spatial transcriptomics to resolve TAM dynamics. Targeting CD74 with Milatuzumab, alone or in combination with immune checkpoint inhibitors, represents a promising strategy to reprogram the immunosuppressive TME and improve GC treatment outcomes.
Data availability
All raw data are in the Genome Sequence Archive for Human (GSA-Human) at the National Genomics Data Center under the accession number HRA009590 (http://bigd.big.ac.cn/gsa-human).
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209 (2021).
Siegel, R. L., Miller, K. D., Fuchs, H. E. & Jemal, A. Cancer statistics, 2021. CA Cancer J. Clin. 71, 7 (2021).
Newton, A. D., Datta, J., Loaiza-Bonilla, A., Karakousis, G. C. & Roses, R. E. Neoadjuvant therapy for gastric cancer: current evidence and future directions. J. Gastrointest. Oncol. 6, 534 (2015).
Wang, X. et al. Postoperative chemoradiotherapy in gastric cancer: a phase I study of radiotherapy with dose escalation of oxaliplatin, 5-fluorouracil, and leucovorin (FOLFOX regimen). Med. Oncol. 28 (Suppl 1), 274 (2011).
Yang, L. et al. Incidence and mortality of stomach cancer in china, 2014. Chin. J. Cancer Res. 30, 291 (2018).
Galletti, G. et al. Microtubule engagement with taxane is altered in taxane-resistant gastric cancer. Clin. Cancer Res. 26, 3771 (2020).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423 (2013).
Gambardella, V. et al. The role of tumor-associated macrophages in gastric cancer development and their potential as a therapeutic target. Cancer Treat. Rev. 86, 102015 (2020).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12, 453 (2015).
Bai, R. et al. The hypoxia-driven crosstalk between tumor and tumor-associated macrophages: mechanisms and clinical treatment strategies. Mol. Cancer. 21, 177 (2022).
Erbani, J., Boon, M. & Akkari, L. Therapy-induced shaping of the glioblastoma microenvironment: macrophages at play. Semin Cancer Biol. 86, 41 (2022).
Mantovani, A., Marchesi, F., Malesci, A., Laghi, L. & Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 14, 399 (2017).
Kielbassa, K., Vegna, S., Ramirez, C. & Akkari, L. Understanding the origin and diversity of macrophages to tailor their targeting in solid cancers. Front. Immunol. 10, 2215 (2019).
Biswas, S. K. Metabolic reprogramming of immune cells in cancer progression. Immunity 43, 435 (2015).
Pittet, M. J., Michielin, O. & Migliorini, D. Clinical relevance of tumour-associated macrophages. Nat. Rev. Clin. Oncol. 19, 402 (2022).
Murray, P. J. Macrophage polarization. Annu. Rev. Physiol. 79, 541 (2017).
Okabe, Y. & Medzhitov, R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell 157, 832 (2014).
Chen, Y. et al. Tumor-associated macrophages: an accomplice in solid tumor progression. J. Biomed. Sci. 26, 78 (2019).
Condeelis, J. & Pollard, J. W. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell 124, 263 (2006).
Cassetta, L. & Pollard, J. W. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov. 17, 887 (2018).
DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19, 369 (2019).
Hallowell, R. W. et al. mTORC2 signalling regulates M2 macrophage differentiation in response to helminth infection and adaptive thermogenesis. Nat. Commun. 8, 14208 (2017).
Casanova-Acebes, M. et al. Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature 595, 578 (2021).
Cheng, S. et al. A pan-cancer single-cell transcriptional atlas of tumor infiltrating myeloid cells. Cell 184, 792 (2021).
Afshar-Kharghan, V. The role of the complement system in cancer. J. Clin. Invest. 127, 780 (2017).
Zhang, M. et al. Single-cell transcriptomic architecture and intercellular crosstalk of human intrahepatic cholangiocarcinoma. J. Hepatol. 73, 1118 (2020).
Li, J. et al. Mir-20a-5p induced WTX deficiency promotes gastric cancer progressions through regulating PI3K/AKT signaling pathway. J. Exp. Clin. Cancer Res. 39, 212 (2020).
Podgornik, H., Sok, M., Kern, I., Marc, J. & Cerne, D. Lipoprotein lipase in non-small cell lung cancer tissue is highly expressed in a subpopulation of tumor-associated macrophages. Pathol. Res. Pract. 209, 516 (2013).
Acknowledgements
This project was supported by grants of the National Natural Science Foundation of China (Ql. Z, 8197227 and 82173033; YW. X, 82102712 ); China Postdoctoral Science Foundation (YW. X, 2021M690751);Key-Area Research and Development Program of Guangdong Province (2021B0101420005). High-level Hospital Construction Project (Ql. Z, DFJHBF202108 and YKY-KF202204). Guangdong Provincial Key Laboratory of Artificial Intelligence in Medical Image Analysis and Application(No. 2022B1212010011),The figures were drawn by Figdraw.
Author information
Authors and Affiliations
Contributions
Q. Z. designed the study and prepared the manuscript. W.L., and C.W. performed experiments. Y.L.,J.L and C.Z. performed the statistical analyses. Q.H.,Z.L.,R.Z. and J.X. assisted with tissue sample collection. L.L performed the data analysis and interpretation.L.Z performed the data analysis and interpretation. All authors approved the final version of the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, W., Wang, C., Liang, L. et al. Single-cell RNA sequencing analysis revealed the immunosuppressive remodeling of tumor-associated macrophages mediated by the MIF-CD74 axis in gastric cancer. Sci Rep 15, 26883 (2025). https://doi.org/10.1038/s41598-025-10301-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-10301-w