Abstract
The recruitment of participants for clinical trials has traditionally been a passive and challenging process, leading to difficulties in acquiring a sufficient number of qualified participants in a timely manner. This issue has impeded advancements in medical research. However, recent years have seen the evolution of knowledge graphs and the introduction of large language models (LLMs), providing innovative approaches for the pre-screening and recruitment phases of clinical trials. These developments promise enhanced recruitment efficiency and increased participant involvement. To ensure the safety and efficacy of clinical trials, it is crucial to establish precise inclusion and exclusion criteria for participant selection. This paper introduces a method to optimize the pre-recruitment stage by utilizing these criteria in conjunction with the cutting-edge capabilities of knowledge graphs and LLMs. The enhanced strategy includes the automated generation of questionnaires, algorithmic evaluation of eligibility, supplemental query-response functions, and a broader participant screening reach. The application of this framework yielded a detailed clinical trial recruitment questionnaire that accurately encompasses all necessary criteria. Its JSON output is noteworthy for its precision and reliability, achieving an impressive 90% accuracy rate in summarizing patient responses. Additionally, the questionnaire’s ancillary question-and-answer feature complies with stringent legal and ethical standards, meeting the requirements for practical deployment. This study validates the practicality and technological soundness of the presented approach. Utilizing this framework is expected to enhance the efficiency of trial recruitment and the level of patient participation.
Similar content being viewed by others
Introduction
Inclusion and exclusion criteria are essential to ensure the validity of clinical trials. These criteria specify the eligibility of potential participants, distinguishing between appropriate and inappropriate candidates. Often, the complexity of the eligibility criteria communicated in public recruitment materials can discourage patient participation due to a lack of understanding of the details of the trial. In addition, patient recruitment is based on referrals from healthcare professionals and direct recruitment efforts. Busy healthcare providers may find it challenging to devote extra time to recruitment activities1,2. Patient recruitment presents a significant obstacle to clinical trial progress, with roughly 80% of trials facing enrollment deficits that undermine statistical power and increase financial costs3,4,5. Recent strategies have incorporated social media platforms to enhance traditional recruitment methods, using their expansive reach and interactive nature to enlarge the pool of participants and improve sample diversity6,7. Nonetheless, current recruitment approaches for clinical trials remain rather passive, signaling a pressing need for innovative developments in the pre-recruitment phase to facilitate the seamless progression of clinical trials.8,9,10,11.
The rapid advancement of large language model (LLM) technology has led researchers and healthcare professionals to explore its vast potential applications in clinical practice12. Jimyung Park13and his team introduced the C2Q 3.0 system, which employs three GPT-4 prompts to extract concepts, generate SQL queries, and facilitate reasoning. The system’s reasoning prompts underwent extensive evaluations by three experts, who assessed them based on readability, accuracy, coherence, and usability. Hamer14 et al. implemented a strategy that combined one-shot prompts, selection reasoning, and thought-chaining techniques to evaluate LLM performance across ten patient records, focusing on identifying eligibility criteria, judging patient compliance with specific criteria, classifying overall patient suitability for clinical trials, and determining the necessity of further physician screening. Mauro Nievas15 and his team used GPT-4 to create a dedicated synthetic dataset, enabling effective fine-tuning under constrained data settings. They released an annotated evaluation dataset and the fine-tuned clinical trial LLM-Trial-LLAMA for public utilization. Qiao Jin16 et al. proposed TrialGPT, an innovative framework that utilizes LLMs for predicting patient eligibility levels and providing rationales, ranking, and filtering clinical trials based on patient free-text notes. This framework was validated using three public cohorts, which included 184 patients and 18,238 annotated clinical trials.
In terms of the effectiveness of prerecruitment questionnaires, several studies have demonstrated their value17,18. Weiss19 developed the COPD Screening Questionnaire (SCSQ), which identifies high-risk patients through a self-assessment tool and effectively pre-screens individuals who need pulmonary function tests, thus improving the early detection rate of COPD. Melo20 evaluated the usability and patient feedback of the STO tool in public dental university clinics and confirmed that the questionnaire is an important tool in dental pre-screening. It helps optimize the dental pre-screening process, reducing travel and saving costs. Mark21 validated the STOP-Bang questionnaire as a preoperative screening tool for obstructive sleep apnea (OSA), confirming its high sensitivity and discriminatory ability in effectively ruling out severe OSA. This paper primarily demonstrates the feasibility of the framework from a technical perspective.
In the initial planning phase, we conducted field surveys and discovered that patient data in electronic health records (EHR) or hospital systems may have timeliness issues. Additionally, the level of detail in the records varies depending on the attending physician. This heterogeneity complicates the direct matching of data with specific participation criteria. Furthermore, large language models are constrained by input length limits and may only process a finite subset of data, which introduces potential inaccuracies in the analysis, posing a challenge for practical application22,23.
To address these issues, this paper proposes a solution to establish a one-to-one correspondence among “participation criteria-questionnaire-answers”. Knowledge graphs are used to aid large language models in constraining the scope of knowledge, ensuring the accuracy of generated content24,25,26. With the aid of a knowledge graph, an LLM constructs a questionnaire tailored to each recruitment criterion and subsequently evaluates patient eligibility based on the submitted responses. This integrated solution combines the capabilities of large language models with knowledge graph technology to streamline processes, enabling automated questionnaire generation, automatic analysis and summarization of responses. This framework aims to implement automated and semi-automated methods to assist recruitment personnel in screening patients more quickly and effectively. By accelerating the initiation of clinical trials and shortening recruitment cycles, the framework seeks to save time and costs for research projects. In addition, medical professionals can also make appropriate adjustments to their recruitment strategies based on the collected questionnaire results.
Methods
This section provides a detailed overview of the proposed solution’s architecture and workflow, including the construction of a clinical trial knowledge graph, the formulation of prompts, the automated generation of questionnaires, the assessment of participation eligibility, and the development of an automated QA system empowered by the knowledge graph.
Scheme design
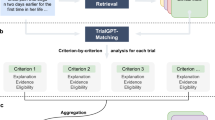
As depicted in Fig. 1, During the recruitment process, patients are initially screened based on structured data from the hospital’s patient management system or EHR (such as age, gender, and symptoms) to determine a certain number of candidates. Recruiters can then expand the recruitment scope based on the relationships between symptoms in the knowledge graph to ensure sufficient patient participation in the clinical trial. Subsequently, a large language model (LLM) transforms the inclusion and exclusion criteria for clinical trials into questionnaires, which are then disseminated to potential participants through phone calls, text messages, or emails. In order to ensure informed consent and privacy protection for the patients, we make sure to obtain clear consent from the patients before starting the questionnaire, and their privacy and data security will be fully protected. This helps to build trust with the patients and lessen their concerns. Additionally, we will clearly inform the patients about the voluntary and anonymous nature of the questionnaire, ensuring they know they have the option not to participate and that their responses will be treated confidentially. At the same time, we plan to provide links to resources for psychological support and counseling within the questionnaire, so that patients can obtain additional help if needed.
The rationale for employing a knowledge graph in this paper is twofold: first, it limits the scope of information to minimize inaccuracies generated by large language models; secondly, the versatility of the knowledge graph extends to its application in intelligent Q&A and recommendation systems27,28.
Figure 2 illustrates the primary functions of the graph service, notably symptom correlation, knowledge acquisition, and intelligent question-answering. The large language model service features capabilities such as questionnaire creation and evaluation of eligibility based on entry criteria. For data storage, two databases are utilized: Neo4j, which stores clinical trial information, and MySQL, which holds patients’ personal data. Furthermore, the data layer is positioned to incorporate future assets, such as training sets of question-answer pairs for private models, personal patient details, and other clinical trial data. On the operational front, the services offered include management of the questionnaire database, clinical trial data administration, and the handling of Electronic Health Records (EHR).
Knowledge graph construction
This study obtained clinical trial data related to Fudan University by sending HTTP requests to the API of clinicaltrials.gov, totaling 579 recruitment records. These records are stored in JSON file format and serve as the data foundation for constructing a knowledge graph.
Based on the data collected, we designed nine entity types: Recruitment_Project, Condition, Sponsor, Collaborator, Intervention_Product, Combination_Product, Intervention_Devices, Intervention_Drugs, and Other_Interventions. Correspondingly, nine relationship types were established as indicated in Table 1. These types of entities are the core features of clinical trial registration data. The entity type Recruitment_Project is characterized by attributes such as Inclusion Criteria, Exclusion Criteria, Funding Type, Study Type, Brief Summary, Study Title, Primary Outcome Measures, and Phases. The resulting knowledge graph is illustrated in Fig. 3.
The knowledge graph operates as the foundational infrastructure for the proposed solution. It specializes in retrieving relevant information from the ‘Inclusion Criteria’ and ‘Exclusion Criteria’ attributes of the ‘Recruitment_Project’ entity, facilitating data provision for the construction of recruitment questionnaires by the Large Language Modelling (LLM) mechanism.
Neo4j is a graph database that uses a graphical data structure to express knowledge as a set of nodes and edges. In this structure, nodes represent entities, while edges represent relationships between entities. The construction and storage process of knowledge graph are as follows29:
-
1.
Collect relevant professional data from ClinicalTrials.gov, and parse it according to the types of the obtained data, then store it as structured data.
-
2.
The Neo4j database is accessed via the Py2neo interface.
-
3.
Key entities and relationships are identified and extracted from the organized data. Nodes and relationships are then created utilizing Py2neo’s ‘graph.merge()‘ function.
-
4.
Corresponding entities and relationships are generated in the database.
-
5.
The established entities and relationships are then stored within the Neo4j database.
Large language model
The proposed solution utilizes prompt learning techniques in conjunction with large language models (LLMs), which have shown remarkable capabilities in reasoning and comprehension tasks. By employing prompt engineering, LLMs efficiently automate a variety of tasks. Specifically, this solution integrates the capabilities of both GLM-3-Turbo, GLM-4, qwen-turbo and llama3-70b-instruct models to leverage their respective strengths. It experiments with different prompts and adjusting parameters like top_p and temperature to improve output accuracy and stability.30,31.
Furthermore, this study employs Prompt Engineering to design and adjust input prompts, guiding the LLM to generate more accurate and targeted output. The Participation Criteria section can be dynamically input, allowing the generation of different questionnaires based on various Participation Criteria. Taking the recruitment of liver disease with the number NCT05442632 as an example. The participation criteria can be dynamically replaced with other participation criteria. As shown in Table 2 The design of the prompts in this study involves four key components:
-
1.
Task Description.
-
2.
Participation criteria.
-
3.
Output format example.
-
4.
Examples of output results.
Application process
The Questionnaire Generation and Application Process, depicted in Fig. 4, entails identifying patients with corresponding diseases and symptoms, followed by disseminating LLM-generated clinical trial recruitment questionnaires to these patients via email, SMS, and other communication methods32. Upon completion of the questionnaire, an LLM evaluates the patients’ responses to ascertain their compliance with the recruitment criteria. Patients who satisfy the criteria are advanced to preliminary recruitment, where they undergo additional examinations to verify their eligibility based on more comprehensive criteria33,34,35,36.
Preliminary screening of patients based on symptoms
Medical researchers or team institutions can utilize the structured information already present in electronic health record systems or hospital information systems, such as age and gender, to preliminarily screen patients and identify a suitable group of patients.
After the knowledge graph is constructed, staff members can visualize the relationships between entities through Neo4j’s intuitive graphical interface. Based on their expertise, they can appropriately expand the recruitment scope or adjust the recruitment strategies in accordance with relevant information.
Generation of recruitment questionnaires
The development of recruitment questionnaires harnesses the synergistic application of knowledge graphs combined with advanced large language model (LLM) technologies. This integrated system is adept at autonomously producing customized questionnaires for diverse clinical trial endeavors. It benefits from the dynamic enhancements and extensions of the knowledge graph database. The completion of the recruitment questionnaire serves not only to ascertain patients’ willingness to participate but also allows the feedback gathered to assist clinical trial personnel in promptly adjusting their recruitment strategies. Figure 4 elucidates the process of creating recruitment questions for clinical trials through steps 1 to 4, which are described subsequently:
Step 1: Clinical trial inclusion and exclusion criteria for specific diseases and medications are rapidly retrieved via a search within the knowledge graph.
Step 2 to 3: Input the participation criteria into the ‘Participation Criteria’ field as indicated in Table 2, then formulate a prompt that directs the LLM to accurately generate a questionnaire in the JSON format. The resulting questionnaires from various models are exhibited in Table 3.
Step 4: The “question” field contains the generated question, and “type” indicates the question category, where 0 represents a fill-in-the-blank question and 1 represents a true-or-false (judgment) question. For true-or-false questions, provide corresponding “options”. The finalized questionnaire is displayed via web or mobile application interfaces in a format that is accessible and convenient for users.
Admission and qualification criteria assessment
To achieve a precise match with the admission criteria, the system produces inquiries that align seamlessly with each specific criterion. This method enhances the large language models’ capacity to understand and assess information, improving the accuracy in evaluating a patient’s eligibility. The evaluation procedure, depicted in steps 5 to 7 in Fig. 4, unfolds as follows:
Step 5: After a patient completes and submits the initial screening questionnaire prepared for the clinical study, the page data is parsed via JSON and concatenated into the “Patient’s responses” section within the prompt shown in Table 4. This, together with the criteria, serves as the context for a large language model to determine whether the patient is eligible for participation. Table 4 presents an example of using Qwen-Turbo and GLM-3-Turbo to assess whether the patient fulfills the participation criteria based on the large language model.
Step 6: The patient’s responses are combined with the corresponding inclusion and exclusion criteria for each query and fed into the large language model for comprehensive analysis. This prompt configuration is detailed in Table 4.
Step 7: After processing the data, the large language model yields a JSON-formatted response that determines the patient’s adherence to the trial’s selection norms. Should the patient qualify, the recruitment team will prompt further assessments to confirm their suitability for the trial. Table 4 illustrates the analytical outcomes and data outputs generated by different models.
Results
This paper employs four models: GLM-3-turbo, GLM-437, llama3-70b-instruct38 and qwen_turbo39, with parameter adjustments and prompt modifications specifically for the GLM-3-turbo model. Evaluation metrics include accuracy, BLEU, ROUGE-1, ROUGE-2, and ROUGE-L. The evaluation involves simulating patient responses using a large model and assessing patient eligibility with input from 2 medical professionals and 3 medical students.
JSON format accuracy represents the correctness of the questionnaire’s JSON format and indicates whether the questionnaire can be successfully visualized in practical applications. BLEU, ROUGE-1, ROUGE-2, and ROUGE-L are widely employed evaluation metrics in natural language processing40,41, primarily for assessing the quality of machine-generated text. Higher values for these metrics generally indicate better text quality and less need for manual revision.
Among the 579 clinical trial registration entries retrieved by searching for Fudan University as the sponsor on ClinicalTrials.gov, 17 entries were found to have incomplete data. Consequently, during the experimental validation phase of this study, 562 corresponding pre-recruitment questionnaires were automatically generated based on the remaining 562 clinical trial registration entries. Each questionnaire contains approximately 10 questions on average, facilitating the staff’s swift design of a patient-friendly final version.
In terms of accuracy in JSON format, the performance of these four models was 98.57%, 89.33%, 100% and 99.28%, respectively. The llama3-70b-instruct model performs better in terms of questionnaire generation quality and accuracy in determining participation eligibility.
In Table 6, a comparison is presented of the GLM-3-turbo model’s accuracy in Json format, total number of questions, BLEU score, ROUGE-1, ROUGE-2, and ROUGE-L performance under various temperature values and top values. According to Table 6, reducing the temperature parameter judiciously can lead to a lower number of generated questions. Additionally, integrating sample questions into the prompts markedly improves the accuracy of the JSON file and the total quantity of generated questions.
The summary accuracy reflects the precision of the system’s automatic evaluation of patients’ eligibility to participate after they complete the questionnaire. To more comprehensively assess the accuracy of test eligibility, this method employed Large Language Model (LLM) technology to simulate 85 patients independently completing questionnaires. These 85 questionnaires, each reflecting the unique conditions of the patients, were used to evaluate the accuracy of LLMs in eligibility assessment. As shown in Table 5, the GLM-3-turbo, GLM-4, llama3-70b-instruct and qwen_turbo models achieved accuracy rates of 89.28%, 91.39%, 92.85% and 91.66%, respectively, in summarizing the responses to the questionnaires.
When assessing patient eligibility for participation, the llama3-70b-instruct model exhibited superior performance, with its accuracy generally meeting the requirements of practical applications.
Discussion
The method introduced in this article can achieve automatic generation of pre recruitment questionnaires for clinical trials. After patients complete the questionnaire, the Large Language Model (LLM) evaluates whether they meet the recruitment criteria and quantifies the number of patients who meet the criteria. Subsequently, eligible patients will be invited by recruiters to undergo professional examinations to confirm their suitability. This method combines LLM with knowledge graph technology to improve the efficiency of clinical trial recruitment and simplify workflow.
The robust generation of questionnaires and the effectiveness of their response judgments have been satisfactorily validated in practical applications. In addition, this method enables precise customization of questionnaires for various diseases and clinical trial environments. By adjusting the prompt appropriately, such as adding examples, the output quality of the questionnaire can be significantly improved. Adjusting the parameters of the questionnaire appropriately will result in a more detailed breakdown based on the inclusion and exclusion criteria.
Regarding the accuracy of summarization, there are issues with unclear descriptions and ambiguous content in the inclusion and exclusion criteria of the questionnaire. Although this does not impact the generation of the questionnaire, it will directly reduce the accuracy of patient participation in eligibility assessment.
Overall, the JSON format questionnaire generated by large language models has high stability and can comprehensively cover all inclusion and exclusion criteria. In addition, the accuracy of patient participation qualification assessment based on the content of the questionnaire is also high. Therefore, the methods introduced in this article are valuable for practical scenarios.
Conclusions
This solution significantly improves recruitment efficiency, primarily by expediting the development of recruitment questionnaires. By incorporating prompts to guide Large Language Models (LLMs), it enables automatic eligibility assessment based on preset criteria and patient responses, ensuring a high degree of accuracy.
To further implement the solution proposed in this article, we plan to introduce a human feedback mechanism. Through this mechanism, patients can offer valuable insights and suggestions regarding the questionnaire survey process, thereby facilitating continuous improvement and optimization of recruitment methods. Additionally, to safeguard patient privacy, our questionnaire employs an anonymous and multiple-choice design, ensuring that patients’ answers are independent of their personal identity. This approach not only protects patient privacy but also provides valuable information for healthcare professionals, assisting them in more effectively recruiting patients.
Looking ahead, our goal is to refine the use of EHRs and strengthen privacy protections via enhanced LLMs. By standardizing the input of inclusion and exclusion criteria, we will improve the precision of questionnaire analysis, thereby facilitating the conduct of clinical trials and advancing medical research.
Data availability
The datasets used and / or analyzed during the current study are available from the corresponding author on reasonable request. The partialtial code can be obtained by contacting author.thor.
References
Ismail, A., Al-Zoubi, T., El Naqa, I. & Saeed, H. The role of artificial intelligence in hastening time to recruitment in clinical trials. Bone Joint Res. 5(1), 20220023 (2023) https://doi.org/10.1259/bjro.20220023
Chaudhari, N., Ravi, R., Gogtay, N. J. & Thatte, U. M. Recruitment and retention of the participants in clinical trials: challenges and solutions. Perspect. Clin. Res. 11, 64–69. https://doi.org/10.4103/picr.PICR_206_19 (2020).
Su, Q., Cheng, G. & Huang, J. A review of research on eligibility criteria for clinical trials. Clin. Exp. Med. 23(6), 1867–1879. https://doi.org/10.1007/s10238-022-00975-1 (2023).
Darmawan, I., Bakker, C., Brockman, T. A., Patten, C. A. & Eder, M. The role of social media in enhancing clinical trial recruitment: Scoping review. J. Med. Internet Res. 22(10), 22810. https://doi.org/10.2196/22810 (2020).
Barten, T. R. M., Staring, C. B., Hogan, M. C., Gevers, T. J. G. & Drenth, J. P. H. Expanding the clinical application of the polycystic liver disease questionnaire: determination of a clinical threshold to select patients for therapy. HPB 25(8), 890–897. https://doi.org/10.1016/j.hpb.2023.04.004 (2023).
Tsai, M.-L., Ong, C. W. & Chen, C.-L. Exploring the use of large language models (llms) in chemical engineering education: Building core course problem models with chat-gpt. Education for Chemical Engineers 44, 71–95. https://doi.org/10.1016/j.ece.2023.05.001 (2023).
Brøgger-Mikkelsen, M., Ali, Z., Zibert, J. R., Andersen, A. D. & Thomsen, S. F. Online patient recruitment in clinical trials: Systematic review and meta-analysis. J. Med. Internet Res. 22(11), 22179. https://doi.org/10.2196/22179 (2020).
Weng, C. & Embi, P.J. In: Richesson, R.L., Andrews, J.E., Fultz Hollis, K. (eds.) Informatics Approaches to Participant Recruitment, pp. 219–229. Springer, Cham (2023). https://doi.org/10.1007/978-3-031-27173-1_12
Applequist, J. et al. A novel approach to conducting clinical trials in the community setting: utilizing patient-driven platforms and social media to drive web-based patient recruitment. BMC Med. Res. Methodol. 20(1), 58. https://doi.org/10.1186/s12874-020-00926-y (2020).
Wang, D., Wang, L., Zhang, Z., Wang, D., Zhu, H., Gao, Y., Fan, X. & Tian, F. “brilliant ai doctor” in rural clinics: Challenges in ai-powered clinical decision support system deployment. In: Proceedings of the 2021 CHI Conference on Human Factors in Computing Systems. CHI ’21. Association for Computing Machinery, New York, NY, USA (2021). https://doi.org/10.1145/3411764.3445432
Cai, L. Z. et al. Performance of generative large language models on ophthalmology board-style questions. Am. J. Ophthalmol. 254, 141–149. https://doi.org/10.1016/j.ajo.2023.05.024 (2023).
Peng, Y., Rousseau, J. F., Shortliffe, E. H. & Weng, C. Ai-generated text may have a role in evidence-based medicine. Nat. Med. 29(7), 1593–1594. https://doi.org/10.1038/s41591-023-02366-9 (2023).
Park, J. et al. Criteria2query 3.0: Leveraging generative large language models for clinical trial eligibility query generation. Available at SSRN 4637800
Hamer, D.M., Schoor, P., Polak, T.B. & Kapitan, D. Improving patient pre-screening for clinical trials: Assisting physicians with large language models. arXiv preprint arXiv:2304.07396 (2023)
Nievas, M., Basu, A., Wang, Y. & Singh, H. Distilling Large Language Models for Matching Patients to Clinical Trials. J. Am. Med. Inform. Assoc. 31 (9), 1953–1963(2023)
Jin, Q., Wang, Z., Floudas, C.S., Sun, J. & Lu, Z. Matching patients to clinical trials with large language models. arXiv:2307.15051ArXiv, 2307–150512 https://doi.org/10.1038/d41586-019-02871-3 (2023).
Trombini-Souza, F. et al. Knee osteoarthritis pre-screening questionnaire (kops): cross-cultural adaptation and measurement properties of the brazilian version—kops brazilian version. Advances in Rheumatology 62, 40 (2022).
Yingchankul, N. et al. Functional-belief-based alcohol use questionnaire (fbaq) as a pre-screening tool for high-risk drinking behaviors among young adults: A northern thai cross-sectional survey analysis. Int. J. Environ. Res. Public Health. 18(4), 1536 (2021).
Weiss, G. et al. Development and validation of the salzburg copd-screening questionnaire (scsq): a questionnaire development and validation study. NPJ Prim. Care Respir. Med. 27(1), 4 (2017).
De Melo, T. S., De Albuquerque Pacheco, G., Souza, De Castro De. & M.I., Figueiredo, K.,. A dental pre-screening system: usability and user perception. The International Society for Telemedicine and eHealth 11(1), 1–7 (2023).
Hwang, M. et al. Validation of the stop-bang questionnaire as a preoperative screening tool for obstructive sleep apnea: a systematic review and meta-analysis. BMC Anesthesiol. 22(1), 366 (2022).
Liu, H., Peng, Y. & Weng, C. How good is chatgpt for medication evidence synthesis?. Stud. Health Technol. Inform. 302, 1062–1066. https://doi.org/10.3233/shti230347 (2023).
Truhn, D., Reis-Filho, J. S. & Kather, J. N. Large language models should be used as scientific reasoning engines, not knowledge databases. Nat. Med. 29(12), 2983–2984. https://doi.org/10.1038/s41591-023-02594-z (2023).
Wang, C. et al. keqing: knowledge-based question answering is a nature chain-of-thought mentor of LLM (2023)
Zhang, G. et al. Leveraging generative ai for clinical evidence summarization needs to achieve trustworthiness (2023) https://doi.org/10.48550/arXiv.2311.11211 [cs.AI]
Wong, C. et al. Scaling clinical trial matching using large language models: A case study in oncology (2023)
Zhang, R. et al. Drug repurposing for covid-19 via knowledge graph completion. J. Biomed. Inform. 115, 103696. https://doi.org/10.1016/j.jbi.2021.103696 (2021).
Liu, A., Huang, Z., Lu, H., Wang, X. & Yuan, C. Bb-kbqa: Bert-based knowledge base question answering. In: Chinese Computational Linguistics: 18th China National Conference, CCL 2019, Kunming, China, October 18–20, 2019, Proceedings, pp. 81–92. Springer, Berlin, Heidelberg (2019). https://doi.org/10.1007/978-3-030-32381-3_7
Han, X. et al. Overview of the ccks 2019 knowledge graph evaluation track: Entity, relation, event and qa (2020) https://doi.org/10.48550/arXiv.2003.03875
Yuan, J., Tang, R., Jiang, X. & Hu, X. Large language models for healthcare data augmentation: An example on patient-trial matching. AMIA Annu. Symp. Proc. 2023, 1324–1333 (2023).
Yuan, M. et al. Large language models illuminate a progressive pathway to artificial healthcare assistant: A review (2023)
Ortner, V. K. et al. Accelerating patient recruitment using social media: Early adopter experience from a good clinical practice-monitored randomized controlled phase i/iia clinical trial on actinic keratosis. Contemp. Clin. Trials Commun. 37, 101245. https://doi.org/10.1016/j.conctc.2023.101245 (2024).
Guan, Z. et al. CohortGPT: An enhanced GPT for participant recruitment in clinical study (2023)
Yang, Z. et al. Talk2Care: Facilitating asynchronous patient-provider communication with large-language-model (2023)
Soumen, P., Manojit, B., Aminul, I. M. & Chiranjib, C. Chatgpt or llm in next-generation drug discovery and development: pharmaceutical and biotechnology companies can make use of the artificial intelligence-based device for a faster way of drug discovery and development. Int. J. Surg. 109(12), 4382–4384. https://doi.org/10.1097/JS9.0000000000000719 (2023).
Sawant, S., Madathil, K. C., Molloseau, M. & Obeid, J. Overcoming recruitment hurdles in clinical trials: An investigation of remote consenting modalities and their impact on workload, workflow, and usability. Appl. Ergon. 114, 104135. https://doi.org/10.1016/j.apergo.2023.104135 (2024).
GLM, T. et al. Chatglm: A family of large language models from glm-130b to glm-4 all tools. arXiv preprint arXiv:2406.12793 (2024).
Siriwardhana, S., McQuade, M., Gauthier, T., Atkins, L., Neto, F.F., Meyers, L., Vij, A., Odenthal, T., Goddard, C. & MacCarthy, M., et al. Domain adaptation of llama3-70b-instruct through continual pre-training and model merging: A comprehensive evaluation. arXiv preprint arXiv:2406.14971 (2024).
Yang, A. et al. Qwen2. 5 technical report. arXiv preprint arXiv:2412.15115 (2024).
Corredor Casares, J.: Large language model evaluation. B.S. thesis, Universitat politècnica de catalunya (2025).
Briman, M.K.H. & Yildiz, B. Beyond rouge: A comprehensive evaluation metric for abstractive summarization leveraging similarity, entailment, and acceptability. Int. J. Artif. Intell. Tools. 33 (05), 2450017 https://doi.org/10.1142/S0218213024500179(2024).
Acknowledgements
We sincerely thank the Science and Technology Innovation 2030 - Major Project of 1“New Generation Artistic Intelligence” (2020AAAA0109300) and the Shanghai Municipal Administration of Traditional Chinese Medicine Clinical Project (2020LP018) for their support and encouragement. This paper has greatly benefited from the insights provided by these funding sources. Additionally, we extend our gratitude to all colleagues who participated in this project.
Funding
This work was supported by Science and Technology Innovation 2030 - Major Project of “New Generation Artistic Intelligence (2020AAAA0109300)” and Shanghai Municipal Administration of Traditional Chinese Medicine Clinical Project (2020LP018).
Author information
Authors and Affiliations
Contributions
ZiHang Chen:Conceptualization,Methodology,Writing- Original Draft. Liang Liu:Writing - Review. QianMin Su: Supervision,Funding acquisition. GaoYi Cheng: Writing - Review & Editing,Investigation. JiHan Huang: Resources,Supervision. Ying Li:Resources.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable. The data used in this framework consists of open-source data and patient data simulated by large language models.
Consent to participate
All authors disclosed no relevant relationships.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zihang, C., Liang, L., Qianmin, S. et al. Enhanced pre-recruitment framework for clinical trial questionnaires through the integration of large language models and knowledge graphs. Sci Rep 15, 27398 (2025). https://doi.org/10.1038/s41598-025-11876-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-11876-0