Abstract
Peatland cloud forests, characterized by high altitude and humidity, are among the least-studied tropical ecosystems despite their significance for endemism and the bioavailable nitrogen (N) that can be emitted as N2O. While research has mainly focused on soil, the above-ground microbial N cycle remains largely unexplored. We quantified microbial N cycling genes across ecosystem compartments (soil, canopy soil, tree stems, and leaves) in relation to N2O and N2 fluxes and soil physicochemical properties in two peatland cloud forests and a wetland on Réunion Island. Complete denitrification minimized N2O emissions and increased N2 fluxes in wetland soils. In cloud forest soils, archaeal nitrification primarily produced nitrate (NO3–), while low pH potentially slowed denitrification, resulting in minimal N2O emissions. Soil N-fixers were more abundant in Erica reunionensis-dominated forests than in mixed forests. Tree stems varied between weak N2O sinks and sources, with fluxes unrelated to gene abundances in stems. High prokaryotic and fungal nirK gene abundance in forest canopy soil suggests potential for above-ground denitrification in wet conditions. nosZ-I genes found in forest canopy soil and leaves (E. reunionensis, Alsophila glaucifolia, and Typha domingensis) indicate that plants, including forest canopy, may play a significant role in the reduction of N2O.
Similar content being viewed by others
Introduction
In a terrestrial ecosystem, the primary productivity of plants relies on the nitrogen (N) supply. Microorganisms are key players in various N cycling processes. Starting from N fixation1 which is regulated by the nifH gene. Bacteria and archaea also perform ammonia oxidation (nitrification), including complete ammonia oxidation, and the bacterial and archaeal amoA genes regulate the process2,3. Denitrification is another crucial step of the microbial N cycle and is regulated by nitrite-reducing (nirS and nirK) genes and nitrous oxide-reducing (nosZ) gene, mainly4,5. Fungal denitrification is regulated by fungal nirK and p450nor6. Nitrification and denitrification processes can lead to the production of nitrous oxide (N2O), which is a potent greenhouse gas and stratospheric ozone-depleting substance7,8,9.
The organic soils with high nitrate content (NO3−) are hotspots for N2O emissions under drained conditions10. Similarly, tropical wetland soils during drought periods have been reported as significant sources of N2O11 and these emissions are correlated with the functional diversity of microbes and the processes dominated by ammonia-oxidizing archaea12. Tropical wetlands and forests are, thus, important components of global N2O budgets13,14. Even though accurate global estimation of N2O fluxes in ecosystems is still a major research challenge, tropical forests are considered a significant source of N2O compared to other ecosystems15,16,17 owing to their high organic N content18 and high microbial activity in varying soil moisture levels19. Field measurements and in situ analyses of various tropical forest ecosystems regarding the N cycle are valuable for developing process-based N cycle models for estimation and mapping of N2O emissions9,17,20.
The cloud forest ecosystem is one of the least studied tropical ecosystems, especially regarding the microbial N cycle. A cloud forest is a tropical or subtropical forest situated between 600 and 3200 m above sea level (m.a.s.l.), and it is defined by constant or frequent ground-level clouds passing through these forests21,22. While just covering 0.4% of the global land surface, these forests show relatively higher species richness and endemism than the rest of the forest ecosystems in the tropics23 and high levels of biologically fixed N in their soils24. In these forests, nitrogen fixation is associated with the ground cover of mosses and other bryophytes, as they host nitrogen-fixing bacteria25,26. The diversity and distribution of microbial communities that fix nitrogen heavily depend on soil moisture27. The clouds continuously feed these forests, and the moisture is thus retained; however, the dynamics of nitrogen fixation can be altered during the dry or warm periods. Soil moisture levels, temperature, and soil pH can influence nitrification28 and denitrification29,30 including N2O emissions31; however, little is known about the below- and above-ground N cycle processes and N2O emissions in tropical cloud forests.
The forest canopy (leaves) has recently been found to be an important area of study regarding N-cycling processes like nitrification32 as well as soil N2O mitigation33. Recent studies on the dynamics of N2O fluxes in forest canopy show significant absorption of N2O in beech (Fagus sylvatica) shoots34. Although a study32 has shown evidence of canopy nitrification through the presence of amoA genes, the above-ground N2O consumption process has still not been explained from a microbial perspective. Tree stems are shown to be the weak sources of N2O during the dry period in tropical upland forests35. During the wet period, tree stems in the tropical lowland rainforest are reported as weak sinks of N2O36. However, N2O reduction mechanisms in forest canopies and tree stems are newly reported phenomena and, thus, are largely unknown. In wetlands, it is shown that plants affect the soil microbiome that regulates N2O and N237 but the aboveground vegetation role is not yet explained. An integrated analysis of soil and phyllosphere N-cycle microbial communities could possibly shed light on N2O transformation in tree canopies38 and wetland vegetation.
The canopy soil, as a component of the canopy, has been considered in a few studies regarding the overall N cycling in tropical forest ecosystems39,40. Canopy soil is defined as the slowly decomposing material of dead epiphytes, cryptogams (bryophytes, algae, fungi, and lichens), mixed with dust, and plant litter, and harbors a variety of microbial and epiphytic communities that have significant importance in above-ground nutrient cycles41,42,43,44. In cloud forests, canopy soil is often related to above-ground N cycling, which is regulated differently from ground soil39. Globally, the cryptogams are reported as the sources of N2O45. Studies have shown differences in nutrient levels and microbial diversity in the canopy and ground soils in temperate forests46 and tropical rainforests42,47. Studies on nutrient cycles in tropical wetlands and peatland cloud forests, particularly integrating soil and vegetation, are scarce. This gap in knowledge hinders our ability to evaluate the sustainability and potential effects of wetland and forest management, climate change, and other disturbances on these ecosystems.
This study aimed to investigate the dynamics of the microbial N cycle and to measure N2O fluxes from peat soils and tree stems within tropical cloud forests, as well as from the wetland soils. The study assessed the abundance of functional genes related to the N cycle in two tropical peatland cloud forests (one featuring Erica reunionensis, and the other a mix of E. reunionensis and Alsophila glaucifolia) and a wetland on Réunion Island (Fig. 1). The main novelty of this study is investigating the microbial N cycle in all major forest compartments, including soil, canopy soil, leaves, and tree stem cores. We hypothesized that: (1) Due to different physicochemical conditions, the potential of microbial N cycle processes (nitrification, denitrification, and N fixation) are different in the belowground (peat soil), and above-ground (canopy soil, tree stems, and leaves) compartments of the cloud forests; (2) Archaeal nitrification dominates over denitrification in cloud forest peat soils; (3) The tropical cloud forest peat soils show higher N2O emissions and microbial potentials for producing N2O than the wetland.
Sampling sites details: (a) Plaine des Cafres, - Mixed forest: (i) dominated by endemic fern Alsophila glaucifolia (soil, n = 6), (ii) dominated by endemic shrub Erica reunionensis (soil, n = 6). (b) Plateau de Thym – Erica forest: featuring Erica reunionensis only (soil, n = 12). (c) Saint Paul wetland with Typha domingensis as a dominant plant species (soil, n = 6; sediment, n = 6). The areas marked in red on the maps indicate where all sampling took place. The embedded pictures of the sites show the soil N2O flux sampling using static chambers. (d) The locations of study sites on the Réunion Island. (e) Tree stem flux measurement with trace gas analyzers. Tree stem fluxes were measured only at cloud forest sites, in addition to the soil flux sampling. (f) Leaves of Erica reunionensis and (g) Alsophila glaucifolia were collected from the mixed forest, while only Erica reunionensis leaves were collected in the Erica forest (leaves, n = 17). (h) Canopy soil on the tree stems in the mixed forest comprising decomposing epiphytes (n = 19). Stem cores were also obtained from both forests (n = 8).
Results
Physical and chemical properties of soil
Welch’s ANOVA showed a significant effect of ecosystem type on soil temperature (F(2, 15.1) = 423.00, p < 0.001). The mean soil temperature at a 10 cm depth was 14.7 °C in mixed forest and 12.9 °C in Erica forest. The wetland mean soil temperature was 21.6 °C, significantly higher than both cloud forest sites (p < 0.001). The difference in soil temperature at the depths of 20 cm, 30 cm, and 40 cm followed the same pattern (Table S1). The SWC varied between 0.25 and 0.61 m3m− 3 in the mixed forest, while it ranged from 0.46 to 0.65 m3m− 3 in the Erica forest. In the wetland site, it was found to be between 0.87 and 0.90 m3m− 3. The Games-Howell tests revealed significant differences in SWC between Erica and mixed forests (p = 0.021), Erica forest and wetland (p < 0.001), and mixed forest and wetland (p < 0.001). The mean soil pH was 4.5 and 4.3 in the Erica and mixed forests, respectively. These significantly differed from the wetland samples’ mean pH (7.3, p < 0.001).
In the Erica forest, soil NH4+-N levels were significantly higher (p < 0.001) than in the mixed forest (Fig. 2a, Table S1). Meanwhile, the highest mean value for soil NH4+-N levels was found in the wetland’s sediment samples, significantly higher than the wetland soil (p < 0.05) and the mixed forest (p < 0.001). The soil NO3–-N levels were also significantly higher in Erica forest as compared to mixed forest (p < 0.001), and wetland samples (p < 0.001). Wetland soil as well as sediments showed the least NO3–-N levels (Fig. 2b).
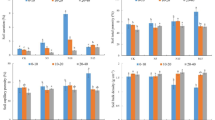
Boxplots of (a) soil NH4+-N levels, (b) soil NO3–-N levels, and (c) the soil N2O fluxes across the studied ecosystems (n = 36). (d) Fluxes of N2O (µg N m− 2 h− 1) from the stems of Erica reunionensis (n = 16, 10 in mixed forest and 6 in Erica forest) and Alsophila glaucifolia (n = 6) in cloud forests. The colors represent different species dominating the sampling points in a, b, and c. The box represents the interquartile range (IQR) containing the 25th and 75th percentiles of the data distribution. Lines extending from the box (whiskers) represent the range of data within 1.5 times the IQR. The bars inside the box represent the median, and the red dots represent the mean. The significance of the pair-wise relationships is indicated by * (p ≤ 0.05), ** (p ≤ 0.01), *** (p ≤ 0.001), and **** (p ≤ 0.0001). Insignificant relationships are not indicated.
Soil and tree stem fluxes
All sites exhibited low soil N2O emissions, with no significant differences between them (Fig. 2c). The mean N2O fluxes were 1.06 µg N m− 2 h− 1 in the mixed forest, 0.37 µg N m− 2 h− 1 in the Erica forest, and 3.84 µg N m− 2 h− 1 in the wetland soil. However, the wetland’s water surface was a weak N2O sink, and flux was measured at − 1.06 µg N m− 2 h− 1. We incubated intact soil cores to estimate potential N2 and N2O fluxes. The respective means of the soil potential N2 fluxes were 84.3, 76.4, and 240 µg N m− 2 h− 1 in the mixed forest, erica forest, and wetland sites (Fig. S1a), and potential N2 fluxes were significantly higher in the wetland than in mixed forest soils (p < 0.001). The means of the soil potential N2O fluxes were 13.1, 6.25, and 10.3 µg N m− 2 h− 1 in the mixed forest, Erica forest, and wetland sites, respectively, with no significant difference between them (Fig. S1b).
The mean N2O fluxes from the stems of Erica reunionensis in the Erica forest were − 0.267 µg N m− 2 h− 1, while in the mixed forest were 0.843 µg N m− 2 h− 1. The mean N2O fluxes from Alsophila glaucifolia stems in the mixed forest were − 0.016 µg N m− 2 h− 1. Overall, the E. reunionensis stems showed more variability in the stem N2O fluxes from weak sinks to weak sources of N2O (Fig. 2d).
Relationships between soil physicochemical properties
In the two cloud forest sites, the SWC had contrasting relationships with soil temperature: in the Erica-dominated forest, SWC positively correlated with soil temperature at a depth of 10 cm, while in the mixed forest, the correlation was negative (Fig. 3a). Both relationships were statistically significant (p < 0.05). In wetland soils, there was no significant correlation. Meanwhile, the low pH significantly correlated with the high NO3− values in the Erica forest (p < 0.05, Fig. 3b), while there was no significant correlation found between pH and NO3− values in wetland samples. Soil temperature had a positive yet statistically insignificant relationship with N2O flux in all ecosystems except the Erica forest (p = 0.05, Fig. 3c). SWC and soil N2O fluxes showed a positive relationship in both forests; however, the relationship was only statistically significant in the mixed forest (p < 0.05, Fig. 3d). In the case of the wetland, the relationship between SWC and N2O flux was negative (p = 0.05).
Relationships between (a) soil temperature at a depth of 10 cm and SWC (m3 m− 3) in forest soils, (b) pH and the soil NO3–-N levels in the forest and wetland soils, (c) soil temperature at a depth of 10 cm and soil N2O flux (µg N m− 2 h− 1), and (d) SWC (m3 m− 3) and soil N2O flux (µg N m− 2 h− 1) in the forest and wetland soils. Different colors represent the ecosystem type, and shades represent the 95% confidence intervals.
Gene abundances and proportions in soil
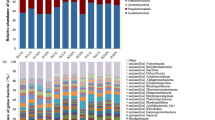
The post hoc tests conducted on the actual gene abundance values after the Welch’s ANOVA showed that the bacterial 16 S rRNA gene abundance was higher in the Erica forest than in the mixed forest (p < 0.001). In the Erica forest, the bacterial 16 S rRNA gene was also higher in proportion (in total prokaryotic abundance) compared to the wetland (p < 0.001). The difference in archaeal 16 S rRNA gene abundance was insignificant among all soils (Fig. 4, Table S2). However, the proportion of archaeal 16 S rRNA genes in the total microbial abundance was significantly higher (p < 0.001) in wetland samples than in the Erica forest (Table S3).
Boxplots of the abundance of different nitrogen-cycling functional genes in the peat soil of the Erica forest (n = 12), the Mixed forest (n = 12), and the wetland soils/sediments (n = 12). The colours of the boxes represent the dominant species found in the site where the soil was obtained. The red dots indicate means, while the bars represent medians. The whiskers show the 95% confidence intervals. The significance of the pairwise relationships is indicated by * (p ≤ 0.05), ** (p ≤ 0.01), *** (p ≤ 0.001), and **** (p ≤ 0.0001). Insignificant relationships are not shown.
The erica forest and the wetland soils had a higher abundance (Fig. 4) and proportion of the nifH genes than the mixed forest (p < 0.001), while the wetland showed the highest nifH proportion. The bacterial amoA gene abundance was low in forest soils compared to the wetland (p < 0.05), with no significant difference among the forest sites. The proportions of the bacterial amoA gene were too low for comparison (Table S3). The archaeal amoA gene abundance was found to be significantly higher in the Erica forest soil than in the mixed forest (p = 0.005) and the wetland (p < 0.001). The difference between the mixed forest and the wetland was also statistically significant (p = 0.05). The trend was followed by the proportion of archaeal amoA gene in total prokaryotic abundance, except that the cloud forest sites had no significant difference among them, but the difference was significant between wetland and cloud forests (p < 0.05). The proportion of archaeal amoA genes in the total archaeal abundance based on the archaeal 16 S rRNA gene was 5.4% in the Erica forest, 3% in the mixed forest, and 0.3% in the wetland. The mixed forest and the wetland had higher comammox amoA gene abundances than the Erica forest soils (p < 0.05). The trend was the same for the proportions of comammox amoA genes.
The abundance and proportion of the nirS gene were significantly higher (p < 0.05) in wetland samples compared to cloud forest soils (Fig. 4). The abundance of the nirK gene was higher in the Erica forest soil than in the mixed forest and wetland (p < 0.001). The proportion of nirK genes followed the same trend (Table S3); however, there was no significant difference between the cloud forests, but there was a significant difference between the wetland and the forests (p < 0.05). The fungal nirK abundance in the wetland samples was significantly higher than in the cloud forest soils (p < 0.05).
The nosZ-I abundance was significantly higher in the Erica forest soil (p < 0.01) than in the mixed forest and wetland (Fig. 4). The nosZ-I mean proportions followed the same trend. The soil of mixed forest dominated by A. glaucifolia showed the highest abundance and proportion of the nosZ-II, significantly different (p < 0.05) from the rest of the ecosystems. However, the other ecosystems showed no difference in abundance and proportionality of nosZ-II (Table S3).
The overall nir gene proportions were highest in the Erica forest soil (8.8%), followed by wetland (6.2%) and mixed forest (5.97%), and the difference was statistically insignificant. The ratio of nir to amoA genes showed the dominance of nir genes over amoA genes to be the strongest in the wetland, yet significantly different from the Erica forest only (p < 0.05). There was no significant difference in different ecosystems regarding nir: nosZ. Principal component analysis (PCA) suggested that the Erica and mixed forest show similarities in their microbial community composition compared to the wetland, where functional genes showed distinct patterns. The ratio of nir genes to amoA and nosZ genes was the highest in the wetland (Fig. 5a). The fungal nirK and prokaryotic nirS gene proportions also showed the same trend (Fig. 5b).
Principal component analysis (PCA) biplots of (a) gene abundances and (b) the gene proportions in the total prokaryotic abundances in different ecosystems as the groups. The x and y axes represent the first two principal components (PCs), capturing the most significant variation in the data. The length of vectors represents the importance of that gene in explaining the variance captured by the PCs. The ellipses in different colors represent the distribution of samples from different ecosystems (95% confidence intervals).
Gene abundances and proportions in canopy soil and plant samples
The mean bacterial and archaeal 16 S rRNA gene abundance in canopy soils and other plant samples is given in Table S4. The nifH, comammox amoA, and nosZ-II were not detected in any canopy samples. The bacterial amoA genes were only detected in the canopy soil samples. The archaeal amoA genes were detected in fern leaves, stems of E. reunionensis, and leaves and stalks of Typha domingensis. Since the proportions of these genes in total microbial abundance were too low, the difference between the abundances and the proportions in these samples was insignificant.
The denitrification genes were detected in most above-ground samples (Fig. 6). The nirS gene abundance was found to be significantly greater in the canopy soil as compared to other above-ground samples (p < 0.001). The nirS genes were not detected in the leaves of E. reunionensis and stalks of T. domingensis. Meanwhile, the nirK genes were abundant in all canopy samples, with canopy soil showing the highest mean value of 2 × 108 copies/g, which were significantly higher than leaves and stem cores (p < 0.01). Fungal nirK was only detected in the canopy soil. The nosZ-I was detected in all the samples taken from the canopy. The highest abundance was found in the canopy soil (p < 0.01).
The canopy soil from E. reunionensis had a higher abundance of nirS, nirK, and nosZ-I genes than A. glaucifolia’s canopy soil (Fig. 6a-c). However, the canopy soil of A. glaucifolia showed a higher mean abundance of fungal nirK as compared to the E. reunionensis canopy soil (p < 0.05). There was no significant difference between the nosZ-I abundances between leaf samples and typha stalks.
Boxplots of (a) nirS, (b) nirK, and (c) nosZ-I gene abundances in the above-ground samples. The colors represent different plant species. The line within the box shows the median. Dots outside the whiskers represent outliers. The whiskers show the 95% confidence intervals. The significance of the pairwise relationships is indicated by ns (not significant), * (p ≤ 0.05), ** (p ≤ 0.01), *** (p ≤ 0.001), and **** (p ≤ 0.0001).
The PCA analysis demonstrated that the canopy soil samples from E. reunionensis and A. glaucifolia (fern) formed distinct clusters along both principal components (Fig. 7). Despite this differentiation, the genes responsible for the variation in these components remained consistent, indicating a prevailing presence of denitrification genes (nirK and nosZ-I) across the various canopy soil samples. However, none of the denitrification gene was found correlating with stem N2O fluxes.
Principal component analyses of gene abundances in the canopy soil and plant leaf samples. The x and y axes represent the first two principal components (PC), capturing the most significant variation in the data. The length of vectors represents the importance of that gene’s abundance in explaining the variance captured by the PCs. The ellipses in different colors represent the sample type distribution (95% confidence intervals).
Relationships between gene parameters and soil chemical properties
The nifH gene abundance positively correlated with soil NH4+-N levels in the forest soil dominated by E. reunionensis (Fig. 8a). The relationship was weakly negative between nifH abundance and the ammonium in the forest soil dominated by A. glaucifolia. The multiple regression analysis showed a significant relationship between soil NO3–-N levels and the archaeal amoA in terms of both abundance and proportion (Fig. 8b). The estimate values of the regression between archaeal amoA and NO3–-N were significantly higher compared to the regressions with other nitrifying genes (P < 0.05). The ground coverage by Sphagnum mosses in the cloud forests positively correlated with nir:nosZ values (Fig. S1). Meanwhile, the gene abundance of the nosZ-I positively correlated with the ground percent cover of vascular plants. The mosses’ ground coverage and the soil N2O fluxes had a significant negative correlation.
(a) Relationship between the nifH abundance and the NH4+-N levels in the soil. (b) Multiple regression models between soil NO3– and nitrifiers’ abundance and the proportion in the total microbial abundance. Archaeal amoA shows the strongest positive relationship with the amount of soil NO3–-N among all nitrifiers. * represents the p < 0.001.
Discussion
The peat soils in tropical cloud forests, as well as the soil in the wetland ecosystem, were weak sources of N2O. Additionally, the surface of the wetland acted as a weak sink for N2O (Fig. 2c). Wetland soil showed the least N2O: (N2 + N2O) ratio (Fig. S1c). The tree stems in the tropical cloud forests varied from weak sources to weak sinks of N2O (Fig. 2d). There was no significant difference among the soil or tree stem flux values. Our study found high soil NH4+-N in the peatland cloud forest dominated by E. reunionensis (Fig. 2a). The levels of NH4+-N were significantly correlated with the nifH gene abundance in the peat soils dominated by E. reunionensis (Fig. 8a). This indicates a high potential for microbial N fixation in the peatland cloud forests. The nifH richness in arid and warm ecosystems is usually high48 and is related to the high nifH gene abundance and nitrogen fixation rates in tropical forest soils49. The relatively high nifH gene proportions in the Erica forest might suggest a symbiotic relationship between E. reunionensis and the nifH-containing microbes, as Ericaceae species have been associated with other N-fixing organisms previously50. No correlation was found between soil NH4+-N and nifH in soil dominated by A. glaucifolia despite possessing high abundance and proportions of nifH gene. A possible explanation could be rapid NH4+ consumption by these ferns; although this has not been previously reported specifically on A. glaucifolia, other fern species found in cloud forests have been shown to prefer the consumption of NH4+ over NO3–51. Furthermore, the symbiotic abilities, transport systems in the root’s plasma membranes, and affinities with the substrates52 might also promote the rapid NH4+ consumption by A. glaucifolia.
The high soil NO3–-N correlated with the archaeal amoA abundance and proportions (Fig. 8b). The bacterial amoA abundance was not prevalent in the studied ecosystems, and the results indicate that archaeal amoA dominated the overall nitrification process. Ammonia-oxidizing archaea (AOA) have been reported to majorly govern the nitrification process in tropical peatlands12. Among tropical soils, cloud forest soils have been shown to have the highest abundance of Archaeal amoA genes53; however, in our sites, this abundance was even higher than what has been reported before. The high soil NO3–-N also correlated with low soil pH values in both forests (Fig. 3b), which also suggests the active nitrification process, as the increased H+ ions generated during the ammonia oxidation result in the acidification of the soil54,55. Since AOA prefer and dominate the soils with low pH28,53,56,57and the nirS and nirK expression is reduced in soils with low pH29,58,59 we can concur that the efficient archaeal nitrification and a relatively slow nir-mediated denitrification process in these peatlands caused the accumulation of soil NO3–-N. The fungal nirK is unaffected by the low pH and can also perform denitrification, but the overall impact would not be substantial, as it constitutes a minor portion of nirK-type denitrifiers60. Our study found a small abundance of fungal nirK in the cloud forest soils, and hence, the role of fungal denitrification in N2O emission is minimized. Although the soil pH was low in both cloud forests, the peat soil dominated by E. reunionensis showed higher NO3–-N levels (Fig. 2). This indicates low NO3– consumption by E. reunionensis due to their restricted ability to utilize NO3[–61 and their preference for NH4+ as the major source of inorganic N62.
The nir:nosZ ratio in the soils from both (Erica and mixed) cloud forests was lower than in the soil from the wetland; meanwhile, the nosZ-I proportion was significantly higher in the Erica forest soil. The coexistence of nirS and nosZ-I in the same microbe can favor the complete denitrification63 and in tropical soils, N2O is often correlated with the nirK and fungal nirK abundances64. We observed very low N2O and high N2 emissions from the cloud forest peatland soil (Fig. 2c and Fig. S1a). The low pH is reported to inhibit the N2O-reducing microbial activity65. Therefore, multiple microbial processes can contribute to the high N2 flux from cloud forest soils, such as n-DAMO (nitrate-dependent anaerobic methane oxidation), which can yield N2 as a byproduct66. This can also be supported by the high abundance of NC-10 bacteria (involved in the n-DAMO process) in the peat soils of the same forests67. An RNA-based study in a similar ecosystem found the high expression of nosZ genes to be the reason behind low soil N2O fluxes68. In tropical soils, some acidophilic bacteria have been reported to reduce N2O at low pH levels69. Our N2O flux results align with a previous study at a similar elevation range in an Ecuadorian montane forest during the dry season70. The soil N2O fluxes were also negatively correlated with the ground coverage by mosses in the cloud forests (Fig. S2), which contrasts with the findings of previous studies71. The N2O fluxes from the soil correlated with the increase in SWC in both cloud forests, and the highest N2O flux was observed between the SWC range of 0.6–0.7 m3 m− 3 (Fig. 3d). This is the same range where many previous studies have observed their peak N2O flux in different ecosystems10,12,72.
Our study found the highest NH4+-N levels in the wetland sediments (Fig. 2a), which was positively correlated with the abundance of the nifH gene (Fig. 8a). Although nifH-containing microbes can perform nitrogen fixation in the sediments73 this can not be considered the sole reason behind high NH4+ in sediment. The prevailing anoxic conditions in the wetland also prevent active nitrification, which results in the accumulation of NH4+ in its sediments. Another reason can be the extensive use of NH4+-based fertilizers in the upslope regions of Réunion Island74 which can then become part of runoff and finally settle in the lowland wetland sediments. Meanwhile, we found very low NO3–-N levels in the wetland samples. Although we quantified a substantial abundance of archaeal amoA in the wetland, the highest comammox amoA found in our study was from the wetland. Typha and phragmites are known for the enormous oxygen release from their roots in the sediments75,76. This connective gas flow through the aerenchyma tissues of the T. domingensis can aerate the sediment enough to facilitate the comammox.
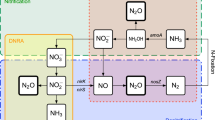
The low NO3–-N levels can be attributed to low nitrification as compared to the rapid denitrification since we found a high nir:amoA ratio in the wetland. Another reason could be the wetland’s fast-growing T. domingensis, which can efficiently uptake the NO3– from soil and sediment77. However, the highest N2 fluxes were from the wetland site (Fig. S1a). The dominance of the nirS gene and the high nir to amoA ratio (305 ± 132) suggest that the nir-based denitrification actively removed the NO3– from the wetland. The nirS-type denitrifiers usually dominate the wetland sites78 and can also contain the nosZ gene63. Consequently, in favorable conditions, these denitrifiers can successfully complete denitrification processes. The mean pH level observed in the wetland was recorded at 7.3, which supports the high expression of the nirS59norB, and nosZ genes30. The dominance of nir genes was coupled with a substantial abundance of nosZ-I and nosZ-II in the wetland. However, the nir: nosZ ratio (23.1 ± 3.05) was less than the nir: amoA ratio. The high SWC yields prevailing anaerobic conditions, which are conducive to the denitrification process. Typha has also been reported as supporting a high nosZ diversity in its rhizosphere79. The typha litter also enhances the growth of nosZ-containing bacteria in a wetland80. We concur that the presence of Typha in the wetland might be helpful in the wetland’s N2O-reducing efficiency. However, other factors such as n-DAMO can also contribute to the high N2 fluxes from the wetland. Our study also revealed a significant presence of nosZ-I on both typha leaves and stalks, indicating an additional potential for reducing N2O in the wetland vegetation (Fig. 6c). The hypoxic areas of the typha vegetation may contribute to the reduction of absorbed N2O from the sediment. The discovery of N2O reducers in the vegetative parts of wetlands is unprecedented and requires further research. A schematic N cycle in our studied ecosystems is shown in Fig. 9.
Schematic microbial N cycle in the cloud forest and the wetland site. The arrows represent the potential pathways of the processes in the light of N substrates, N fluxes, and the N cycling microbial genes. The red arrows represent the N2O fluxes. In the wetland, the N2O flux was positive in soils while it was negative in the wetland surface (hence two-sided arrow). However, the overall amounts were minimal. The large purple arrows represent the N2 flux from the soils of the forest and wetland. The difference in the arrows’ size represents the amount of the fluxes.
The absence of the nifH gene in the canopy samples indicated a lack of microbial N fixation potential in the canopy of cloud forests, contrasting with previous studies in a similar ecosystem, which estimated high N2 fixation in the canopy soil during the dry season81,82,83. However, the atmospheric NH3 converted to NH4+ in the presence of cloud droplets or precipitation can be deposited in the canopy84. This NH4+ becomes available for further microbial processes on the plants’ surfaces.
The presence of archaeal amoA in canopy samples shows a potential for nitrification in the canopy of cloud forests. The canopy’s microbial nitrification is a reported phenomenon in the forests of the global north, and the process claims 80% of the NO3– descending to the soil via throughfall32,85,86. Rainfall and cloud droplets might wash the NO3– from the canopy to the soil in our studied ecosystems, making the canopy another source of soil NO3–. The NO3– produced through canopy nitrification can also become available for uptake by the epiphytes growing on the canopy soil.
The presence of denitrifiers in the canopy reveals a potential for above-ground denitrification (Fig. 6, Table S4). Canopy soil showed a high abundance of nirS, nirK, fungal nirK, and nosZ-I genes. Our study also found a high abundance of nirK in stem cores and leaves. The NO3– from canopy nitrification32 assimilation, and foliar uptake from the atmosphere87 can further be utilized and reduced by the denitrifying microbes within the plant’s anoxic or hypoxic parts. This can also occur in the canopy soil when it becomes wet in case of precipitation, and anaerobic conditions on plant surfaces are thus realized. Metatranscriptomics evidence suggests that in such instances where denitrifiers are given anoxic conditions after long periods of oxic conditions, nirK-containing microbes exhibit faster rates of NO3– reduction. However, during a short-term anoxic situation (e.g., created by a brief spell of rain), only the nosZ-II can complete the denitrification88. However, our study did not find the nosZ-II in the cryptogamic canopy soil. Since our study was conducted in the dry season, where the only source of moisture was the clouds, complete anoxic conditions in the canopy soil were not realized. This can result in the N2O emission from the stems. We found stems of E. reunionensis as weak sources of N2O in the mixed forest where these trees possessed the canopy soil (Fig. 2d). The E. reunionensis stems in the Erica forest without any canopy soil were found to be the weak sinks of N2O. The absence of nosZ-II from the canopy soil can explain this difference. Canopy soil showed a high abundance of nosZ-I genes. However, the nosZ-I expression will only be highest during long-term anoxic conditions88 which was not observed in our study. Such conditions are realized during the rainy season or intense cloud formation in the cloud forests.
The nosZ-I was found in the leaf samples of all plant species under study. The N2O, which is produced in the soil and transported into different plant compartments89 can be reduced by the microbes containing nosZ-I found in the hypoxic parts of stems and leaves. The tree stems of the lowland tropical rainforest on the same island were found to be a weak N2O sink during the wet season36. Our study has confirmed the presence of nosZ-I denitrifiers in the cryptogamic canopy soil, providing further evidence that cryptogams can play a significant role in N2O uptake36. The presence of nosZ-I in the stem cores of the cloud forest trees can also explain the N2O uptake by E. reunionensis stems in our study. The nosZ-I microbes in the canopy during the wet season, when anoxic conditions prevail for longer periods on plant surfaces, can reduce the N2O emitted by soil and/or canopy soil.
Materials and methods
Site description
Two peatland cloud forests were studied on the tropical Réunion Island, France: Plaine des Cafres in Le Tampon municipality (21.145343° S, 55.569692° E) and Plateau de Thym, in Forêt de Bébour region of Saint-Benoît commune (21.097139° S, 55.548028° E) (Fig. 1). Both sites are peatland forests located between 1500 and 1650 m.a.s.l in the montane cloud forest vegetation band90.
The Plaine des Cafres’ peatland forest had a mix of endemic shrub species, E. reunionensis, and an endemic tree fern species, Alsophila glaucifolia. The epiphytic vegetation in this forest was dominated by Cordyline mauritiana and various fern species, such as Hymenophyllum inaequale, H. capillare, and Blechnum attenuatum. In the understory, Embelia angustifolia, Anthoxanthum odoratum, and Cynorkis ridleyi were the most frequent species. The bryophyte layer was patchy and dominated by Sphagnum species. The current study describes this forest as a mixed forest with two sub-sites based on the dominance of either E. reunionensis or A. glaucifolia (Fig. 1a).
The Plateau de Thym peatland forest had a dominance of E. reunionensis and Hubertia ambavilla. In the understory, the dominating species were Erica galioides and Juncus effusus, with patchy dominating Sphagnum species. The peatland in this forest is approximately 25,000 years old91. Peatland forest is described as the erica forest in the current study (Fig. 1b).
The wetland site was situated next to Saint Paul city (20.991416° S, 55.294264° E) at 4 m.a.s.l. The predominant vegetation at the site was Typha domingensis (Fig. 1c). Other commonly found species include Setaria geminata, aquatic Lemna sp, along with exotic Schinus terebinthifolius and Ipomoea cairica along the edges. In the current study, this site is referred to as a wetland.
All sampling, which included soil, gas fluxes, canopy soil, and plant material, was conducted in November 2022, which marks the early spring dry season on Réunion Island. At every site, 12 measurement points were selected, and soil collars were installed. In the Erica forest, all points were selected in the forest dominated by E. reunionensis, while in the mixed forest, 6 points were located in an area dominated by E. reunionensis and 6 points in an area dominated by A. glaucifolia. At the wetland site, 6 measurement points were located in the soil and 6 points in open water between Typha plants. The ground vegetation coverage within the defined area of soil collars was estimated in percentage for all species.
Soil, canopy soil, and plant sampling
Soil samples (n = 36) from all sites were collected from 0 to 10 cm depth for chemical and microbial analysis. Soil collection equipment was disinfected with ethanol between sampling points to prevent cross-contamination, and samples were packed in transparent plastic grip-seal bags. The soil samples for microbial analysis were stored at − 20 °C until DNA extraction. Canopy soil samples (n = 19) from the mixed forest were collected from the stem surfaces of E. reunionensis and A. glaucifolia for microbial analyses. Leaves (n = 17) were collected from both cloud forest sites from the different branches of E. reunionensis and A. glaucifolia. Stem core samples (n = 8) were also collected from the stems of E. reunionensis at both cloud forest sites. Tree stem samples were taken using a 400 mm length, 5.15 mm diameter, 3-threaded increment borer, and extractor (Haglöf Sweden AB, Langsle, Sweden). From the wetland site, Typha (Typha domingensis) leaves (n = 6) and stems/stalks (n = 6) were collected. The canopy soil and plant samples were placed in filter bags and packed inside sealed plastic bags with active silica gel to eliminate moisture. Silica gel was replaced until all moisture from the samples had been removed successfully. Drying was performed for the long-term preservation of these samples intended for DNA-based analyses92.
Physical and chemical properties of soil
At each measurement point, soil temperature was measured using probes (model CS 107, Campbell Scientific Inc., Logan, UT, USA). The soil water content (SWC) was determined using the ProCheck moisture sensor (Decagon Devices, Inc., USA). Soil chemical analyses were performed at the Estonian Environmental Research Centre in Tartu. The total nitrogen content, total carbon content, and pH level were measured. After extracting with 2 M KCl (1:10 ratio), NH4+-N and NO3−-N levels were determined from soil samples using flow-injection analysis according to standard methods93.
N2O sampling and flux calculations
To quantify the N2O exchange of the soil surface, 65 L polyvinyl chloride chambers (surface area 0.0196 m2 volume 0.065 m3and height 0.4 m) were placed to cover the soil surface within pre-installed chamber collars. At the wetland site, the chambers were buoyed in the open water using a foam noodle fixed underneath, and sampling was done without any vegetation inside the chamber. Gas samples were collected from the chambers and injected into 50 ml pre-vacuumed glass bottles during a one-hour measurement period at 20-minute intervals94. The N2O concentration of all gas samples was tested using two Shimadzu-2014 gas chromatographs equipped with an electron capture detector (GC-ECD), a thermal conductivity detector (GC-TCD), and a Loftfield-type autosampler95.
For tree stem N2O fluxes, static measurement chambers were installed on the stems of E. reunionensis (mixed forest n = 10, Erica forest n = 6) and A. glaucifolia (mixed forest n = 6) at one stem height per tree (approximately 20 cm above ground) in both cloud forest sites. The chambers were made from transparent rectangular plastic containers (Lock & Lock, Seoul, South Korea) with bottoms removed to expose the measured area of the tree stem. A neoprene band was glued to the bottom rim of the chamber to make the connection with the tree stem surfaces airtight. Each chamber was closed with an airtight removable lid connected to the gas analyzers. The gas concentrations were measured by circulating air in a closed loop between the chamber and a portable N2O/H2O trace gas analyzer (LI-7820, Li-Cor Biosciences, Lincoln, NE, USA) over 10 min. For soil and stem flux calculations, we determined the slope of the least-squares linear regression of the change in N2O concentrations in the chamber headspace over the measurement time. Detailed equations used for the calculations can be found in the study by Ranniku et al. 202496. The quality of each measurement session was validated using the adjusted R2 value of the linear regression for the CO2 measurements, also determined using the gas chromatograph, which certifies chamber closure quality. Flux values were accepted if the R2 value of the CO2 slope exceeded 0.9.
In addition, intact soil cores (diameter 6.8 cm) from the topsoil layer (0–10 cm) were obtained after the gas sampling was complete for each site. Potential N2 and N2O fluxes were measured from these soil cores in the laboratory using the helium atmosphere method for soil incubation97,98.
DNA extraction
Before DNA extraction, the canopy soil and the plant samples were crushed using a coffee grinder. The grinder parts were sterilized with 70% ethanol between every sample. The DNeasy PowerSoil Pro kit (Qiagen, Hilden, Germany) was used to extract DNA from 0.25 g of the soil and 0.12 g of canopy soil and plant material, following the instructions provided by the manufacturer. All samples (soil, canopy soil, stem, and leaves) were homogenized with lysis buffer using Precellys 24 Homogenizer (Berlin Technologies, Montigny-le-Bretonneux, France) at 5000 rpm for 20s. In the case of canopy soil and plant samples, the amount of lysis buffer (CD1) was increased up to 50–70% to maximize the amount of lysate. The concentration and quality of the extracted DNA were measured using a Tecan AG Infinite M200 spectrophotometer before storage at − 20 ºC.
Quantitative polymerase chain reaction (qPCR)
To determine the bacterial and archaeal abundance in all samples, the qPCR assay of 16 S ribosomal RNA (rRNA) genes was performed using RotorGene® Q equipment (Qiagen, Valencia, CA, USA). The abundance of N-fixing microbes was determined by quantifying the nifH gene, which encodes the nitrogenase enzyme, using qPCR. Similarly, nitrifier abundance was determined by quantifying bacterial and archaeal amoA and the comammox (complete ammonia oxidizers) amoA (encoding ammonia monooxygenase) gene. The abundance of nitrite-reducing denitrifiers was determined by quantifying the nirS (gene encoding cytochrome cd1 nitrite reductase) and the nirK (gene encoding the copper-containing nitrite reductase). The nitrous oxide reductase-encoding gene, nosZ (clade I and II), was quantified to determine the abundance of microbes involved in the final denitrification step. The primers and the qPCR program profiles are shown in Table S5. Negative controls were included in every qPCR run.
The qPCR data was analyzed using RotorGene Series Software (version 2.0.2, Qiagen, Hilden, Germany) and LinRegPCR program (version 2020.0). Gene copy numbers were determined from the samples’ threshold cycles and corrected by the dry weight%, expressed as gene copies per gram of dry weight of the sample (copies g-1 dw).
Statistical data analyses
Jamovi (version 2.4.8)99 was used for the basic descriptive analyses and was used to perform the normality tests on the data by employing the Shapiro-Wilks tests and drawing the histograms and quantile-quantile (Q-Q) plots. The variables were analyzed based on different ecosystem types and the dominant tree species in the ecosystem as factors. In RStudio (R version 2024.4.1.748)100a ggstatsplot package101 was used to perform one-way ANOVA (Welch) to compare the means of different variables in different ecosystems. Games-Howell tests as post hoc pairwise comparisons were employed to account for the violation of variance homogeneity. Principal component analysis (PCA) plots were created using the ggbiplot package in RStudio, while the rest of the plots were created using the ggplot2 package. The gene abundances and soil NH4+ values were log10 transformed to obtain normality before being used in different statistical analyses.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
Dixon, R. & Kahn, D. Genetic regulation of biological nitrogen fixation. Nat. Rev. Microbiol. 2, 621–631 (2004).
Rotthauwe, J. H., Witzel, K. P. & Liesack, W. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63, 4704–4712 (1997).
van Kessel, M. A. H. J. et al. Complete nitrification by a single microorganism. Nature 528, 555–559 (2015).
Zumft, W. G. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61, 533–616 (1997).
Yoon, S., Nissen, S., Park, D., Sanford, R. A. & Löffler, F. E. Nitrous oxide reduction kinetics distinguish Bacteria harboring clade I NosZ from those harboring clade II NosZ. Appl. Environ. Microbiol. 82, 3793–3800 (2016).
Aldossari, N. & Ishii, S. Fungal denitrification revisited – Recent advancements and future opportunities. Soil. Biol. Biochem. 157, 108250 (2021).
Conrad, R. Soil microorganisms as controllers of atmospheric trace gases (H2, CO, CH4, OCS, N2O, and NO). Microbiol. Rev. 60, 609–640 (1996).
Ravishankara, A. R., Daniel, J. S. & Portmann, R. W. Nitrous oxide (N2O): the dominant Ozone-Depleting substance emitted in the 21st century. Science 326, 123–125 (2009).
Butterbach-Bahl, K., Baggs, E. M., Dannenmann, M., Kiese, R. & Zechmeister-Boltenstern, S. Nitrous oxide emissions from soils: how well do we understand the processes and their controls? Philos. Trans. R Soc. B Biol. Sci. 368, 20130122 (2013).
Pärn, J. et al. Nitrogen-rich organic soils under warm well-drained conditions are global nitrous oxide emission hotspots. Nat. Commun. 9, 1135 (2018).
Liengaard, L. et al. Extreme emission of N2O from tropical wetland soil (Pantanal, South America). Front Microbiol 3 (2013).
Bahram, M. et al. Structure and function of the soil Microbiome underlying N2O emissions from global wetlands. Nat. Commun. 13, 1430 (2022).
Dalal, R. C. & Allen, D. E. Greenhouse gas fluxes from natural ecosystems. Aust J. Bot. 56, 369–407 (2008).
Tian, H. et al. A comprehensive quantification of global nitrous oxide sources and sinks. Nature 586, 248–256 (2020).
Werner, C., Butterbach-Bahl, K., Haas, E., Hickler, T. & Kiese, R. A global inventory of N2O emissions from tropical rainforest soils using a detailed biogeochemical model. Glob Biogeochem. Cycles 21 (2007).
Townsend, A. R., Cleveland, C. C., Houlton, B. Z., Alden, C. B. & White, J. W. Multi-element regulation of the tropical forest carbon cycle. Front. Ecol. Environ. 9, 9–17 (2011).
Yu, L. et al. Global variations and drivers of nitrous oxide emissions from forests and grasslands. Front Soil. Sci 2 (2022).
Taylor, P. G. et al. Organic forms dominate hydrologic nitrogen export from a lowland tropical watershed. Ecology 96, 1229–1241 (2015).
Pajares, S., Campo, J., Bohannan, B. J. M. & Etchevers, J. D. Environmental controls on soil microbial communities in a seasonally dry tropical forest. Appl. Environ. Microbiol. 84, e00342–e00318 (2018).
Swails, E., Hergoualc’h, K., Deng, J., Frolking, S. & Novita, N. How can process-based modeling improve peat CO2 and N2O emission factors for oil palm plantations? Sci. Total Environ. 839, 156153 (2022).
Hamilton, L. S. Mountain cloud forest conservation and research: A synopsis. Mt. Res. Dev. 15, 259–266 (1995).
Foster, P. The potential negative impacts of global climate change on tropical montane cloud forests. Earth-Sci. Rev. 55, 73–106 (2001).
Karger, D. N., Kessler, M., Lehnert, M. & Jetz, W. Limited protection and ongoing loss of tropical cloud forest biodiversity and ecosystems worldwide. Nat. Ecol. Evol. 5, 854–862 (2021).
Brookshire, E. N. J., Hedin, L. O., Newbold, J. D., Sigman, D. M. & Jackson, J. K. Sustained losses of bioavailable nitrogen from montane tropical forests. Nat. Geosci. 5, 123–126 (2012).
Markham, J. & Fernández Otárola, M. Bryophyte and lichen biomass and nitrogen fixation in a high elevation cloud forest in Cerro de La muerte, Costa Rica. Oecologia 195, 489–497 (2021).
Permin, A., Horwath, A. B., Metcalfe, D. B., Priemé, A. & Rousk, K. High nitrogen-fixing rates associated with ground-covering mosses in a tropical mountain cloud forest will decrease drastically in a future climate. Funct. Ecol. 36, 1772–1781 (2022).
Cusack, D. F., Silver, W. & McDowell, W. H. Biological nitrogen fixation in two tropical forests: Ecosystem-Level patterns and effects of nitrogen fertilization. Ecosystems 12, 1299–1315 (2009).
Nicol, G. W., Leininger, S., Schleper, C. & Prosser, J. I. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia oxidizing archaea and bacteria. Environ. Microbiol. 10, 2966–2978 (2008).
Albina, P. et al. Influence of hydrogen Electron donor, alkaline pH, and high nitrate concentrations on microbial denitrification: A review. Int. J. Mol. Sci. 20, 5163 (2019).
Bakken, L. R., Bergaust, L., Liu, B. & Frostegård, Å. Regulation of denitrification at the cellular level: a clue to the Understanding of N2O emissions from soils. Philos. Trans. R Soc. B Biol. Sci. 367, 1226–1234 (2012).
Zhang, J. et al. Temperature sensitivity of soil carbon dioxide and nitrous oxide emissions in mountain forest and meadow ecosystems in China. Atmos. Environ. 142, 340–350 (2016).
Guerrieri, R. et al. Substantial contribution of tree canopy nitrifiers to nitrogen fluxes in European forests. Nat. Geosci. 17, 130–136 (2024).
Mander, Ü. et al. Forest canopy mitigates soil N2O emission during hot moments. Npj Clim. Atmospheric Sci. 4, 1–9 (2021).
Machacova, K., Schindler, T., Bréchet, L., Mander, Ü. & Grams, T. E. E. Substantial uptake of nitrous oxide (N2O) by shoots of mature European Beech. Sci. Total Environ. 934, 173122 (2024).
Welch, B., Gauci, V. & Sayer, E. J. Tree stem bases are sources of CH4 and N2O in a tropical forest on upland soil during the dry to wet season transition. Glob Change Biol. 25, 361–372 (2019).
Machacova, K. et al. Trees as net sinks for methane (CH4) and nitrous oxide (N2O) in the lowland tropical rain forest on volcanic Réunion Island. New. Phytol. 229, 1983–1994 (2021).
Espenberg, M. et al. Towards an integrated view on microbial CH4, N2O and N2 cycles in brackish coastal marsh soils: A comparative analysis of two sites. Sci. Total Environ. 918, 170641 (2024).
Siegenthaler, A. et al. Temperate tree microbiomes: divergent soil and phyllosphere microbial communities share few but dominant taxa. Plant. Soil. 496, 319–340 (2024).
Hietz, P., Wanek, W., Wania, R. & Nadkarni, N. M. Nitrogen-15 natural abundance in a montane cloud forest canopy as an indicator of nitrogen cycling and epiphyte nutrition. Oecologia 131, 350–355 (2002).
Matson, A. L., Corre, M. D. & Veldkamp, E. Nitrogen cycling in canopy soils of tropical montane forests responds rapidly to indirect N and P fertilization. Glob Change Biol. 20, 3802–3813 (2014).
Leary, J. J. K., Singleton, P. W. & Borthakur, D. Canopy nodulation of the endemic tree legume Acacia Koa in the mesic forests of Hawaii. Ecology 85, 3151–3157 (2004).
Cardelús, C. L., Mack, M. C., Woods, C., DeMarco, J. & Treseder, K. K. The influence of tree species on canopy soil nutrient status in a tropical lowland wet forest in Costa Rica. Plant. Soil. 318, 47–61 (2009).
Victoriano-Romero, E. et al. Interactions between epiphytes during canopy soil formation: an experiment in a lower montane cloud forest of Southeast Mexico. Plant. Biol. 25, 468–477 (2023).
Matson, A. L., Corre, M. D. & Veldkamp, E. Canopy soil greenhouse gas dynamics in response to indirect fertilization across an elevation gradient of tropical montane forests. Biotropica 49, 153–159 (2017).
Lenhart, K. et al. Nitrous oxide and methane emissions from cryptogamic covers. Glob Change Biol. 21, 3889–3900 (2015).
Dangerfield, C. R., Nadkarni, N. M. & Brazelton, W. J. Canopy soil bacterial communities altered by severing host tree limbs. PeerJ 5, e3773 (2017).
Pittl, E., Innerebner, G., Wanek, W. & Insam, H. Microbial communities of arboreal and ground soils in the esquinas rainforest, Costa Rica. Plant. Soil. 329, 65–74 (2010).
Sepp, S. K. et al. Global diversity and distribution of nitrogen-fixing bacteria in the soil. Front Plant. Sci 14 (2023).
Reed, S. C., Townsend, A. R., Cleveland, C. C. & Nemergut, D. R. Microbial community shifts influence patterns in tropical forest nitrogen fixation. Oecologia 164, 521–531 (2010).
Rayner, M. C. Nitrogen fixation in Ericaceae. Bot. Gaz. 73, 226–235 (1922).
He, X. et al. Cooperation of earthworm and arbuscular mycorrhizae enhanced plant N uptake by balancing absorption and supply of ammonia. Soil. Biol. Biochem. 116, 351–359 (2018).
von Wirén, N., Gojon, A., Chaillou, S. & Raper, D. Mechanisms and Regulation of Ammonium Uptake in Higher Plants. in Plant Nitrogen (eds. Lea, P. J. & Morot-Gaudry, J.-F.) 61–77 (Springer, Berlin, Heidelberg, 2001).
Pett-Ridge, J., Petersen, D. G., Nuccio, E. & Firestone, M. K. Influence of oxic/anoxic fluctuations on ammonia oxidizers and nitrification potential in a wet tropical soil. FEMS Microbiol. Ecol. 85, 179–194 (2013).
Li, Y., Chapman, S. J., Nicol, G. W. & Yao, H. Nitrification and nitrifiers in acidic soils. Soil. Biol. Biochem. 116, 290–301 (2018).
Tao, J. et al. Nitrification-induced acidity controls CO2 emission from soil carbonates. Soil. Biol. Biochem. 192, 109398 (2024).
Zhalnina, K., de Dörr, P., Camargo, A. O. & Triplett, E. W. F. Drivers of archaeal ammonia-oxidizing communities in soil. Front Microbiol 3 (2012).
Zhou, Z. F. et al. A comparative study of ammonia-oxidizing archaea and bacteria in acidic and alkaline purple soils. Ann. Microbiol. 66, 615–623 (2016).
Saleh-Lakha, S. et al. Effect of pH and temperature on denitrification gene expression and activity in Pseudomonas mandelii. Appl. Environ. Microbiol. 75, 3903–3911 (2009).
Brenzinger, K., Dörsch, P. & Braker, G. pH-driven shifts in overall and transcriptionally active denitrifiers control gaseous product stoichiometry in growth experiments with extracted bacteria from soil. Front Microbiol 6 (2015).
Bösch, Y. et al. Distribution and environmental drivers of fungal denitrifiers in global soils. Microbiol Spectr. 11:e00061-23 (2022).
Havill, D. C., Lee, J. A. & Stewart, G. R. Nitrate utilization by species from acidic and calcareous soils. New. Phytol. 73, 1221–1231 (1974).
Pornon, A., Escaravage, N. & Lamaze, T. Complementarity in mineral nitrogen use among dominant plant species in a subalpine community. Am. J. Bot. 94, 1778–1785 (2007).
Graf, D. R. H., Jones, C. M. & Hallin, S. Intergenomic comparisons highlight modularity of the denitrification pathway and underpin the importance of community structure for N2O emissions. PLoS ONE. 9, e114118 (2014).
Lourenço, K. S., Costa, O. Y., de Cantarella, A., Kuramae, E. E. & H. & Ammonia-oxidizing bacteria and fungal denitrifier diversity are associated with N2O production in tropical soils. Soil. Biol. Biochem. 166, 108563 (2022).
Liu, B., Frostegård, Å. & Bakken, L. R. Impaired Reduction of N2O to N2 in Acid Soils Is Due to a Posttranscriptional Interference with the Expression of nosZ. mBio 5, e01383-14 (2014).
Ettwig, K. F., van Alen, T., van de Pas-Schoonen, K. T., Jetten, M. S. M. & Strous, M. Enrichment and molecular detection of denitrifying methanotrophic Bacteria of the NC10 phylum. Appl. Environ. Microbiol. 75, 3656–3662 (2009).
Kazmi, F. A. et al. Distinct microbial communities drive methane cycling in below- and above-ground compartments of tropical cloud forests growing on peat. Environ. Microbiome. 20, 54 (2025).
Gallarotti, N. et al. In-depth analysis of N2O fluxes in tropical forest soils of the congo basin combining isotope and functional gene analysis. ISME J. 15, 3357–3374 (2021).
He, G. et al. Sustained bacterial N2O reduction at acidic pH. Nat. Commun. 15, 4092 (2024).
Lamprea Pineda, P. A. et al. Ideas and perspectives: patterns of soil CO2, CH4, and N2O fluxes along an altitudinal gradient – a pilot study from an Ecuadorian Neotropical montane forest. Biogeosciences 18, 413–421 (2021).
Li, A., DeLuca, T. H., Sun, S., Zhang, J. & Wang, G. Bryophytes impact the fluxes of soil non-carbon dioxide greenhouse gases in a subalpine coniferous forest. Biol. Fertil. Soils. 56, 1151–1163 (2020).
Kazmi, F. A. et al. Meltwater of freeze-thaw cycles drives N2O-governing microbial communities in a drained peatland forest soil. Biol. Fertil. Soils. 61, 667–680 (2025).
Caton, I. R., Caton, T. M. & Schneegurt, M. A. Nitrogen-fixation activity and the abundance and taxonomy of NifH genes in agricultural, pristine, and urban prairie stream sediments chronically exposed to different levels of nitrogen loading. Arch. Microbiol. 200, 623–633 (2018).
Billen, G., Garnier, J., Pomet, A. & Bonnet, B. Is food self-sufficiency possible for reunion island?? Reg. Environ. Change. 24, 58 (2024).
Armstrong, W., Armstrong, J., & Beckett, P. M. Measurement and Modelling of Oxygen Release from Roots of Phragmites australis. in Constructed Wetlands in Water Pollution Control (eds. Cooper, P. F. & Findlater, B. C.) 41–51 (Pergamon, 1990).
Konnerup, D., Sorrell, B. K. & Brix, H. Do tropical wetland plants possess convective gas flow mechanisms? New. Phytol. 190, 379–386 (2011).
Ghamary, E. & Mohajeri, J. Efficiency of Cyperus alternifolius, Typha latifolia, and Juncus inflexus in the removal of nitrate from surface water. AQUA - Water Infrastruct. Ecosyst. Soc. 70, 654–664 (2021).
Ligi, T. et al. Effects of soil chemical characteristics and water regime on denitrification genes (nirS, nirK, and nosZ) abundances in a created riverine wetland complex. Ecol. Eng. 72, 47–55 (2014).
Ruiz-Rueda, O., Hallin, S. & Bañeras, L. Structure and function of denitrifying and nitrifying bacterial communities in relation to the plant species in a constructed wetland. FEMS Microbiol. Ecol. 67, 308–319 (2009).
Chen, Y., Wen, Y., Zhou, Q. & Vymazal, J. Effects of plant biomass on denitrifying genes in subsurface-flow constructed wetlands. Bioresour Technol. 157, 341–345 (2014).
Matson, A. L., Corre, M. D., Burneo, J. I. & Veldkamp, E. Free-living nitrogen fixation responds to elevated nutrient inputs in tropical montane forest floor and canopy soils of Southern Ecuador. Biogeochemistry 122, 281–294 (2015).
Shi, X., Wang, L., Sun, J., Lucas-Borja, M. E. & Wang, J. Nitrogen cycling-related functional genes exhibit higher sensibility in soil than leaf phyllosphere of different tree species in the subtropical forests. Plant. Soil. 493, 173–185 (2023).
Stanton, D. E., Batterman, S. A., Von Fischer, J. C. & Hedin, L. O. Rapid nitrogen fixation by canopy Microbiome in tropical forest determined by both phosphorus and molybdenum. Ecology 100, e02795 (2019).
Behera, S. N., Sharma, M., Aneja, V. P. & Balasubramanian, R. Ammonia in the atmosphere: a review on emission sources, atmospheric chemistry and deposition on terrestrial bodies. Environ. Sci. Pollut Res. 20, 8092–8131 (2013).
Guerrieri, R., Vanguelova, E. I., Michalski, G., Heaton, T. H. E. & Mencuccini, M. Isotopic evidence for the occurrence of biological nitrification and nitrogen deposition processing in forest canopies. Glob Change Biol. 21, 4613–4626 (2015).
Watanabe, K. et al. Microbial nitrification in throughfall of a Japanese Cedar associated with archaea from the tree canopy. SpringerPlus 5, 1596 (2016).
Bourgeois, I., Clément, J. C., Caillon, N. & Savarino, J. Foliar uptake of atmospheric nitrate by two dominant subalpine plants: insights from in situ triple-isotope analysis. New. Phytol. 223, 1784–1794 (2019).
Sennett, L. B. et al. Determining how oxygen legacy affects trajectories of soil denitrifier community dynamics and N2O emissions. Nat. Commun. 15, 7298 (2024).
Machacova, K. et al. Pinus sylvestris as a missing source of nitrous oxide and methane in boreal forest. Sci. Rep. 6, 23410 (2016).
Heymans, L. et al. Do native and alien species differ in their ecological strategies?? A test with Woody plants in tropical rainforests on réunion Island (Mascarene archipelago, Indian Ocean). Plants 12, 3990 (2023).
Margenat, H. & Le Roux, G. POST EXPEDITION REPORT Field Expedition La Réunion Island, France ATMO-PLASTIC Project. https://doi.org/10.5281/zenodo.7643599 (2023).
Smenderovac, E. et al. Drying as an effective method to store soil samples for DNA-based microbial community analyses: a comparative study. Sci. Rep. 14, 1725 (2024).
Standard Methods for the Examination of Water and Wastewater. (American Public Health Association, Washington, DC, (2005).
Soosaar, K. et al. Dynamics of gaseous nitrogen and carbon fluxes in riparian alder forests. Ecol. Eng. 37, 40–53 (2011).
Loftfield, N., Flessa, H., Augustin, J. & Beese, F. Automated gas chromatographic system for rapid analysis of the atmospheric trace gases methane, carbon dioxide, and nitrous oxide. J. Environ. Qual. 26, 560–564 (1997).
Ranniku, R. et al. Dry and wet periods determine stem and soil greenhouse gas fluxes in a Northern drained peatland forest. Sci. Total Environ. 928, 172452 (2024).
Butterbach-Bahl, K., Willibald, G. & Papen, H. Soil core method for direct simultaneous determination of N2 and N2O emissions from forest soils. Plant. Soil. 240, 105–116 (2002).
Mander, Ü. et al. Isotopologue ratios of N2O and N2 measurements underpin the importance of denitrification in differently N-Loaded riparian alder forests. Environ. Sci. Technol. 48, 11910–11918 (2014).
The jamovi project. jamovi. The jamovi project. (2023).
Posit team. RStudio: Integrated Development Environment for R. Posit software, PBC. (2024).
Patil, I. Visualizations with statistical details: the ‘ggstatsplot’ approach. J. Open. Source Softw. 6, 3167 (2021).
Acknowledgements
We acknowledge the Doctoral School of Earth Sciences and Ecology of the University of Tartu for arranging the research expedition to the Réunion Island. This study was supported by the European Research Council (ERC; ÜM) under the grant agreement No 101096403 (MLTOM23415R), and the European Union HORIZON-CSA project No 101079192 “Living Labs for Wetland Forest Research (LiWeFoR)”. KK was supported by the Estonian Research Council (grant no PSG714) and The Estonian Ministry of Education and Research, Centre of Excellence for Sustainable Land Use (FutureScapes, TK232). MÖ and ÜM are supported by the Estonian Research Council (grants no PRG1789 and PRG2032, respectively). The authors would like to thank the Observatory of the Science of the Universe - Réunion (OSU-R) for the use of the facilities at the Mare Longue Research field station during the fieldwork campaign and acknowledge the National Park of La Réunion for the collecting permit DIR-I-2022-281 (for sample collection). We acknowledge Dr. Alar Teemusk and master’s student Timothy Ukah for their contributions to the lab analysis of gas and canopy soil samples, respectively. We also thank Dr. Salla A.M. Tenhovirta and Dr. Monica M. Palta for their guidance during the manuscript preparation.
Author information
Authors and Affiliations
Contributions
F. Kazmi conceptualized the study, performed fieldwork, formal analysis, and data visualization, and prepared the original draft. Ü. Mander conceptualized the study, acquired funding, supervised the fieldwork, and reviewed the manuscript. R. Ranniku performed field work and formal analyses and reviewed the manuscript. M. Öpik performed the fieldwork and reviewed the manuscript. K. Püssa performed the fieldwork. K. Soosaar performed the fieldwork. K. Kasak performed the fieldwork and reviewed the manuscript. M. Masta reviewed the manuscript. C. Ah-Peng performed the fieldwork and reviewed the manuscript. M. Espenberg conceptualized the study, performed the fieldwork, provided resources for lab work, and supervised and reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kazmi, F.A., Mander, Ü., Ranniku, R. et al. Nitrogen cycling genes abundance in soil and aboveground compartments of tropical peatland cloud forests and a wetland on Réunion Island. Sci Rep 15, 27155 (2025). https://doi.org/10.1038/s41598-025-12367-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-12367-y