Abstract
Cancer of bone marrow is classified as Acute Lymphoblastic Leukemia (ALL), an abnormal growth of lymphoid progenitor cells. It affects both children and adults and is the most predominant form of infantile cancer. Currently, there has been significant growth in the identification and therapy of acute lymphoblastic leukemia. Therefore, a method is required that is capable to accurately assessing risk by an appropriate treatment strategy that takes into account all relevant clinical, morphological, cytogenetic, and molecular aspects. However, to enhance survival and quality of life for those afflicted by this aggressive haematological malignancy, more research and clinical trials are required to address the issues associated with resistance, relapse, and long-term toxicity. Consequently, a deep optimized Convolutional Neural Network (CNN) has been proposed for the early diagnosis and detection of ALL. The design of the deep optimized CNN model consisted of five convolutional blocks with thirteen convolutional layers and five max pool layers. The proposed deep optimized CNN model is tuned using the hyperparameters such as 30 epochs, batch size 32 and optimizers, namely Adam and Adamax. Out of the two optimizers, the proposed deep optimized CNN model has outperformed using Adam optimizer with the points of accuracy and precision as 0.96 and 0.95, respectively.
Similar content being viewed by others
Introduction
Acute lymphoblastic leukemia (ALL) is a type of cancer that affects the blood and bone marrow, fundamentally focusing on white blood cells called lymphocytes. Excess production of lymphocyte precursors in the bone marrow causes ALL. Lymphoblasts are the name given to these immature lymphocytes. Anemia, infection, and bleeding are only some of the consequences of lymphoblasts crowding out healthy blood cells1,2. Although it can happen to adults as well, it is the most prevalent form of cancer in children. Abnormal lymphoblasts, or immature white blood cells with improper function, proliferate rapidly in ALL patients. If the leukemia cells spread to the brain and spinal cord, patients may also experience headaches, vomiting, and difficulty with balance and coordination. A bone marrow biopsy may be performed to affirm the conclusion and decide the degree of the infection. Treatment for ALL more often not includes a combination of chemotherapy, focused on treatment, radiation treatment, and stem cell transplantation. In general survival rate for children with ALL is presently around 90%, whereas the survival rate for grown-ups is lower, around 40–50%. Several factors can influence the outlook of a patient, such as their age, the specific type of acute lymphoblastic leukemia (ALL), the presence of particular genetic mutations, and the effectiveness of the treatment response. But there are some gaps and limitations in current methodologies. There are performance limitations such as many existing CNN-based models suffer from overfitting due to small and imbalanced datasets. Additionally, the computational complexity of deep networks can lead to inefficiencies in real-world applications. Traditional CNN models may not effectively capture intricate morphological variations in leukemic cells, leading to misclassification. Some methods demonstrate high accuracy on benchmark datasets but fail to generalize well to real clinical blood smear images due to variations in staining techniques and imaging conditions. Many deep learning models operate as black boxes, making it difficult for medical professionals to interpret the decision-making process.
Scientific Contributions of proposed work are as follow: (1) Development of a CNN for ALL Detection: A novel deep convolutional neural network architecture is proposed, specifically designed to diagnose and detect Acute Lymphoblastic Leukemia (ALL) early. The design incorporates five convolutional blocks, comprising thirteen convolutional layers and five max-pooling layers to effectively capture spatial hierarchies and morphological patterns in medical imaging data. (2) Comprehensive Hyperparameter Tuning for Enhanced Performance: The CNN model was fine-tuned using various hyperparameters such as a batch size of 32, 30 training epochs, and two different optimizers—Adam and Adamax—to enhance model performance. This rigorous tuning helped the model generalize better and avoid overfitting, which is crucial when dealing with complex and high-dimensional biomedical data like ALL blood smear images. (3) Impact of Optimizer Selection in ALL Diagnosis: The study highlights the critical role of optimizer choice in the training dynamics and convergence of deep learning models for ALL detection. The Adam optimizer demonstrated superior performance over Adamax in terms of both accuracy and precision, achieving 96% accuracy and 95% precision. This finding emphasizes the significance of selecting appropriate optimization techniques in medical image classification tasks. (4) Advancement Toward AI-Assisted Hematological Diagnostics: By integrating deep learning with optimized training strategies, the research contributes to the growing field of AI-driven hematology diagnostics. It lays the groundwork for future work that may incorporate additional patient-specific clinical, cytogenetic, and molecular features to improve risk stratification and treatment planning for ALL.
The rest of the article is summarized in a way such as Sect. Related work depicts the literature review, a description of the dataset is given in Sect. Dataset description, methodology is given in Sect. Proposed methodology, followed by results in Sect. Results, Discussion in Sect. Discussion and Sect. Conclusion shows the conclusion.
Related work
In this, a review of the literature is performed using the research of different researchers on different datasets3. Atteia et al.4 presented Bayesian CNN model for the diagnosis of ALL in blood images. They had worked using a hybrid dataset by combining two datasets and a total of 368 images. Ullah et al.5 proposed a technique to address morphological similarities by utilizing an Efficient Channel Attention (ECA) module. Their method, applied to both normal and malignant cells, achieved an accuracy rate of 91.1%. Similarly, Sampathila et al.6 developed a CNN-based model for the diagnosis of leukemic cells. Using an Nvidia Tesla GPU for training, their model attained an accuracy of 95.54%. Rezayi et al.7 presented ResNet50 and VGG16 networks for the diagnosis of ALL by CNN model with obtained value of accuracy as 82.10%. Baig et al.8 presented the different CNN models for the diagnosis using 4150 images and five different classification algorithms with an accuracy of 97.04%. Karar et al.9 presented an Internet of Medical Things model for the detection of ALL. They performed using two datasets and had obtained the values of accuracy as 98.67% and 95.5% on binary and multi-classification respectively. Bhute et al.10 presented ensemble learning based approach for the leukemia detection. They had obtained the value of accuracy as 90% on 1800 microscopic images. Huang et al.11 presented ensemble ALL model for the classification of ALL. Bayesian optimization algorithm had used for the enhancement of model accuracy and achieved accuracy as 96.26%. ResNet50’s deep architecture and residual connections improve feature extraction and mitigate vanishing gradient issues, making it effective for leukemia detection. Additionally, Bayesian optimization was used in hyperparameter tuning, explaining how it efficiently selected the optimal parameters compared to traditional methods, leading to better model performance. Mondal et al.12 worked using two disease classes and had obtained the value of accuracy as 88.80% on ensemble model using WEN-kappa. Duggal et al.13 worked on binary classification and had obtained the value of accuracy as 90.78% for CNN model. Medical image classification (IC) played a key role in computer-aided diagnosis (CAD) systems, aiding radiologists in detecting tumors and disorders. The proposed method utilized pre-trained CNN models, such as DenseNet-121 and VGG-16, as feature extractors, along with bidirectional LSTM layers for temporal feature extraction. For classification, Support Vector Machine (SVM) and Random Forest (RF) algorithms are employed. Hyperparameters are optimized using a modified grey wolf optimization method. Experimental results on the Mammographic Image Analysis Society (MIAS) dataset shown impressive performance, with the VGG-16 model achieving an overall accuracy of 99.86%, and similar high results on the INbreast dataset14. Author15 addressed the global challenge of kidney diseases by developing an AI-driven diagnostic system for classifying kidney stones, cysts, and tumors. Using a dataset of 12,446 CT images, the model combined AlexNet’s feature extraction with ConvNeXt’s advanced attention mechanisms, achieving a high classification accuracy of 99.85%. The approach incorporated hierarchical spatial information and self-attention mechanisms for improved performance. A custom optimization technique inspired by Adam facilitated faster convergence. The model demonstrated outstanding metrics, including 99.89% precision, 99.95% recall, and 99.83% specificity. Emphasizing interpretability, the system shown promise for clinical deployment in kidney disease diagnosis. Author16 focused on improving early breast cancer (BC) detection using deep learning (DL) and machine learning (ML) techniques. Given the challenges posed by sparse disease-related data, the proposed method utilized ensemble pre-trained models, including DenseNet-121 and EfficientNet-B5, as feature extractors. A support vector machine is used for classification, with hyperparameters optimized via a modified meta-heuristic optimizer. Experimental results on the INbreast dataset demonstrated the effectiveness of the EfficientNet-B5 model for BC classification, achieving outstanding performance metrics: 99.9% accuracy, 99.9% sensitivity, 99.8% specificity, 99.1% precision, and 1.0 AUC, showcasing its potential for early cancer detection17. presented a novel deep learning approach for classifying poultry audio signals, incorporating a custom Burn Layer to enhance robustness. The model integrated audio signal processing, convolutional neural networks (CNNs), and the Burn Layer, which injected controlled noise during training to improve resilience. The architecture reduced trainable parameters to 191,235, significantly fewer than traditional models. The model used an Adamax optimizer and performed exceptionally well with sensitivity (96.77%), specificity (100%), precision (100%), accuracy (98.55%), and other metrics. This approach advanced animal health monitoring by utilizing robust deep learning for accurate poultry disease detection through audio signal analysis.
Dataset description
The images for this data collection were generated at the bone marrow lab located in Taleqani Hospital. Blood samples from 89 patients were expertly processed and stained, yielding 3256 PBS pictures for analysis. The data set is classified as either benign or malignant18. First, there are hematogones, and second, there are the three subtypes of malignant lymphoblasts that make up the Acute Lymphoblastic Leukemia (ALL) family.
The data in Table 1 indicates that there are 504 instances categorized as benign conditions and 985 instances identified as early-stage diseases. Furthermore, the table reveals that there are 963 cases classified as pre-diseases, alongside 804 cases designated as pro-diseases. Overall, the table provides an overview of the distribution of diseases across different classes. It highlights the presence of benign diseases, early diseases, pre-diseases, and pro-diseases, indicating the varied stages and severity of the conditions observed in the given dataset. The total count of diseases listed in the table is 3,256. Figure 1 shows the input image samples of the dataset. Figure 1(a) shows the Benign (Class 0), 1 (b) shows the Early (Class 1) malignant, (c) depicts Pre (Class 2) and (d) shows Pro (Class 3).
Data augmentation technique is applied as the dataset samples are very less for training. In the field of image processing and deep learning, enhancing a model’s generalization and robustness is often achieved through data augmentation. This technique involves applying various random transformations to images, including rotation, flipping, scaling, and translation. By doing so, the augmented dataset offers a broader range of examples for the model to learn from, ultimately improving its performance. This helps the model to become more invariant to variations in the input data, such as changes in lighting conditions, orientation, or size. In addition to geometric transformations, data augmentation can also involve changes to the color space, such as adjusting brightness, contrast, saturation, and hue. These modifications assist the model to learn features that are more robust to variations in color and illumination. Here the data augmentation techniques are applied in such a manner that all the four disease class images reach to 1500. The different techniques that are applied to increase image count are vertical and horizontal flip, 90 degrees right rotation as illustrated in Fig. 2.
Table 2 presents a summary of disease classes in a dataset before and after data augmentation techniques were applied. Data augmentation is a critical method in deep learning, especially when working with imbalanced datasets. By increasing the number of instances in the minority classes (Benign, Pre, and Pro), the augmented dataset becomes more balanced to improve the deep learning models’ performance that are trained on this data. To mitigate class awkwardness within the unique dataset and avoid one-sided demonstration execution, information enlargement methods such as rotation, flipping, and zooming were connected to produce extra images, guaranteeing an equal number of samples for both ALL and healthy classes. This balance empowered the model to viably learn features from both categories. A total of 1500 images per class was chosen to supply adequate variability whereas keeping up computational effectiveness, upgrading demonstrates generalization and decreasing the chance of overfitting. Observational assessments with changing test sizes (1000, 1200, 1500, 1800) uncovered that execution advancements plateaued beyond 1500 images, whereas preparing time and asset utilization expanded. Subsequently, 1500 images per class were considered ideal for this research. Out of these 6000 images, 80% of the data is used for training and 20% of the data is used for testing purposes.
Proposed methodology
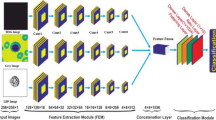
The proposed methodology is presented in Fig. 3. It depicts the dataset collection showing the image samples of the four disease classes. These data samples are given as input to the pre-processing stage for the data augmentation. In the data augmentation, two different techniques, such as flipping and rotation, are used to increase the image count of the data samples and to remove unbalancing in the dataset. With the help of augmentation techniques, the image count is increased to 6000. This dataset of 6000 image samples is then split into 80% as training and 20% as testing. Now the data is fed into the proposed CNN model. This convolutional neural network (CNN) architecture tackles blood cell classification by progressively extracting discriminative features from the input image. The initial Convolutional Block 1 starts with two 3 × 3 convolutional layers, each using 16 filters. ReLU activations and batch normalization follow each layer, stabilizing training and extracting low-level visual features like edges and textures. Subsequent max pooling reduces dimensionality and introduces some translation invariance. Building upon this foundation, each subsequent block doubles the number of filters (32 in Block 2, 64 in Block 3, and so on) while maintaining the 3 × 3 filter size. This gradual increase in complexity allows the network to capture progressively more intricate details, from shapes and patterns in Block 2 to specific cell morphologies and subtle inter-class variations in Block 5. Each block repeats the pattern of ReLU activation, batch normalization, and max pooling. After feature extraction, the flattened output from the final block feeds into two fully connected layers. The first layer with ReLU activation and 512 neurons, further processes the extracted information. Dropout helps prevent overfitting by randomly dropping units during training. In essence, this architecture leverages a cascade of convolutional blocks to refine feature representation, starting with basic visual cues and culminating in highly discriminative characteristics unique to different blood cell types. This allows the network to achieve accurate classification and distinguish between various cell morphologies. The training and testing of the models is done in the next step. The performance comparison on two optimizers, namely Adam and Adamax19, is performed with Adam as the best-performing optimizer. After this, the analysis of the proposed model is performed with different transfer learning models such as ResNet50 and YoloV8. In the end the proposed model classifies the dataset samples into four different classes. In this study deep model is used. Shallower networks exhibited faster training times but suffered from lower accuracy and poor feature extraction, while significantly deeper models led to overfitting and increased computational cost without substantial performance gains. The final architecture was selected based on a balance between performance metrics (accuracy, precision, and validation loss) and computational efficiency. Empirically, the chosen depth allowed the model to extract sufficient hierarchical features for effective ALL classification while maintaining generalization.
Results
Results were achieved using two distinct optimizers: Adam and Adamax. The analysis on the basis of epochs, training and validation curves, confusion matrix parameters etc. is performed in the results section.
Adam optimizer analysis
The Adam optimizer analysis are shown in this section. Here’s a summary of the Table 3, focusing on the changes in accuracy and loss values across different epochs. From the Epoch 5 to Epoch 10, accuracy increased from 0.705 to 0.816 (an improvement of 0.111). Loss decreased from 0.739 to 0.470 (a decrease of 0.269). From the epoch 10 to Epoch 15, validation accuracy amplified from 0.797 to 0.891. Validation loss decreased from 0.506 to 0.291. From epoch 15 to Epoch 20, accuracy increased from 0.901 to 0.940 (an improvement of 0.039). Loss decreased from 0.267 to 0.164 (a decrease of 0.103). However, after Epoch 20, there is a slight fluctuation in the accuracy and loss values. The validation accuracy also demonstrates consistent improvement, while the validation loss generally decreases over the epochs. It’s worth noting that the model has achieved high accuracy and low loss values after Epoch 30, suggesting that the training process has been effective.
Figure 4 illustrates the accuracy and loss metrics over the course of the training process, which was conducted over 30 epochs. Notably, between epoch 25 and epoch 30, the accuracy rose from 0.939 to 0.970, reflecting an enhancement of 0.031, as demonstrated in Fig. 4(a). Concurrently, the loss declined from 0.171 to 0.081, indicating a reduction of 0.090, as detailed in Fig. 4(b). Additionally, Fig. 4(c) presents the confusion matrix generated after utilizing the Adam optimizer in the analysis.
Adamax optimizer analysis
In Table 4, the model demonstrates a consistent enhancement in accuracy with the progression of epochs. The accuracy starts at 0.613 at Epoch 5 and consistently increases to 0.929 at Epoch 30. The validation accuracy follows a similar trend, starting at 0.623 at Epoch 5 and reaching 0.919 at Epoch 30. This signifies that the model’s performance generalizes well to unseen data. The loss value steadily decreases as the number of epochs increases. The loss starts at 0.984 at Epoch 5 and consistently decreases to 0.210 at Epoch 30. The validation loss also decreases over time, starting at 0.999 at Epoch 5 and reaching 0.231 at Epoch 30. In summary, both accuracy and loss values show consistent improvements as the number of epochs increases. This suggests that the model’s performance becomes better with additional training, as indicated by higher accuracy and lower loss on validation and training data.
Figure 5(a) depicts the accuracy curve for 30 epochs. The validation loss also decreases over time, starting at 0.999 at Epoch 5 and reaching 0.231 at Epoch 30 as shown in Fig. 5(b). In summary, both accuracy and loss values show consistent improvements as the number of epochs increases. Figure 5(c) depicts the confusion matrix after the analysis is performed on Adam optimizer. According to the confusion matrix analysis, the classification model demonstrates an accuracy rate of 93.8%. This indicates that 93.8% of the instances in the dataset were accurately classified. For class 0, the model achieved a precision of 99.2%, meaning that 99.2% of the instances predicted to be in class 0 were indeed part of that class. Furthermore, the recall for class 0 stands at 99.6%, which implies that 99.6% of the actual instances belonging to class 0 were correctly identified. In summary, the confusion matrix illustrates that the classification model is effectively predicting product arrival times.
Comparison of optimizer
The Table 5 shows the comparison results of both optimizers. Using the Adam optimizer, Disease Class 3 achieves a precision rate of 0.97 and an F1 score of 0.99, resulting in an overall accuracy of 0.96. On the other hand, when employing the Adamax optimizer, Disease Class 0 exhibits a precision of 0.96, with an overall accuracy of 0.91. In terms of precision, the Adam optimizer generally outperforms the Adamax optimizer for all disease classes. However, the Adamax optimizer achieves higher recall for Disease Class 0 compared to the Adam optimizer. But the overall accuracy of 0.96 is achieved with Adam optimizer.
Adam typically results in more aggressive updates to the model’s weights because of its use of the squared gradients. This can help in faster convergence, especially when the gradients are stable and not prone to large outliers. Adam includes bias correction terms for both the first and second moments (gradients and squared gradients), which helps make the learning process more stable in the early stages. Adam is generally known to work better with smaller batch sizes, which is typical in many deep learning tasks, because it adapts faster based on the gradient estimates.
Comparison with other transfer learning models
From the previous section, it is observed that the proposed model has outperformed with Adam optimizer with the accuracy rate as 0.96. Now, the results are calculated using different transfer learning models such as ResNet50 and YoloV8 model. The proposed Adam optimized CNN model is compared with these two transfer learning models. Table 6 shows the comparative analysis of Adam optimized proposed CNN model and transfer learning models.
The proposed model utilizes the Adam optimizer, which is known for its efficiency in terms of both speed and accuracy. The use of Deep CNN is another contributing factor to the performance. CNNs are highly effective in extracting hierarchical features from image data, which is crucial for tasks like image classification in medical datasets such as ALL20,21. The deep layers in CNNs help the model to capture complex patterns and structures in the data, which might be missed by shallower models or non-CNN-based approaches. Adam optimizes the model by adjusting the learning rate dynamically based on the momentums of the gradients. This leads to more stable and efficient training compared to static learning rate methods or methods that require frequent manual adjustments.
The Table 6and Fig. 6 presents a comparative analysis of three deep learning models—Proposed CNN, ResNet50, and YoloV8—based on four performance metrics: Precision, Recall, F1-score, and Accuracy. The Proposed CNN outperforms the other models in most metrics. It achieves the highest accuracy (0.96). This suggests that the Proposed CNN is highly effective in identifying true positives while minimizing false positives and false negatives.
YoloV8 follows closely, with a Precision of 0.938, and the highest F1-score of 0.939 among the three models. YoloV8’s high F1-score indicates a balanced trade-off between Precision and Recall. ResNet50 ranks third across all metrics, with an accuracy of 0.93. While it performs well, it is slightly behind the Proposed CNN and YoloV8, especially in F1-score and Accuracy. In comparison, the Proposed CNN demonstrates the best overall performance, particularly excelling in Precision, Recall, and Accuracy.
Visualization of results
The Fig. 7 depicts the visual results on the proposed deep optimized convolution neural network model. Here the categorization of a disease into four distinct classes based on its severity or stage is done. “Benign” (class 0) represents a non-threatening condition, while “Early” (class 1) indicates the initial stage of the disease. “Pre” (class 2) suggests a more advanced but not yet critical stage, and “Pro” (class 3) signifies the most severe form of the disease. These classifications help in diagnosing and determining appropriate treatment.
State-of-art techniques
The Table 7 shows a comparison of different techniques and their corresponding accuracies in identifying Acute Lymphoblastic Leukemia (ALL) in various datasets. Recent studies have investigated different deep learning and machine learning methods for classifying Acute Lymphoblastic Leukemia (ALL) utilizing diverse datasets and image volumes. The proposed Adam-optimized deep convolutional neural network (CNN) accomplished an accuracy of 96.00%, which illustrates strong execution, particularly considering the complexity of leukemia discovery tasks. When compared with existing strategies, a few models have reported somewhat higher accuracies, such as the ResNet50 model accomplishing 99.38% on the ALL-PBS dataset22, and the DarkNet19 ESA model achieving 98.52% employing an open dataset of 3256 pictures23. However, it is vital to note that numerous of the strategies with higher detailed accuracies utilized moderately smaller or different datasets. For instance, the IoT-based model and SVM approach reported accuracies of 95.5% and 97.4% separately, but were trained on only 179 and 260 images from the ALL-IDB dataset9,24.
In contrast, models prepared on bigger datasets, such as ALLNET and Ensemble-ALL, utilizing the C-NMC2019 dataset with 7,272 images, reported accuracies of 95.54% and 96.26%, individually6,25, which are comparable to the proposed model. Traditional models like VGG16 and basic CNNs too showed better than average results, such as 84.62% on the CodaLab dataset and 93.9% on C-NMC20197,26. Also, a Bayesian-based CNN accomplished 93.5% accuracy on the ALL-IDB dataset comprising 368 images4, whereas a 19-layer CNN reported 93.18% accuracy on only 293 open images8.
Generally, whereas a few strategies outperformed the proposed model in accuracy, they frequently used less images or different datasets, which may not generalize well to real-world clinical information. The proposed model, with its vigorous execution and adjusted dataset, offers a reliable and versatile solution for ALL classification (Fig. 8).
Discussion
This study has created a deep optimized convolutional neural network (CNN) for the early determination of Acute Lymphoblastic Leukemia (ALL), and the results illustrate that the proposed approach beats existing state-of-the-art approaches in terms of precision and accuracy. This discussion explains the implications of these discoveries, the comparative performance, and the limitations and future scope of this research. The precision and accuracy of the proposed model recommend that it can be an important device for pathologists in early-stage ALL discovery, possibly lessening diagnostic mistakes and assisting treatment planning. The structural plan of the CNN, joining five convolutional blocks and an addition of thirteen layers, permitted the model to extricate both low-level and high-level features successfully. Furthermore, tuning with numerous optimizers empowered the model to achieve high generalization ability. The model’s execution is assessed on a particular dataset, and its generalizability to other clinical datasets with distinctive imaging conditions, recoloring methods, or statistic varieties needs encourage approval. Whereas the model shows promise for early conclusion, its integration into real-world clinical workflows has not however been tried and will require further collaboration with medical experts and approval through clinical trials. Future work will investigate integrating multimodal data—combining cytogenetic, molecular, and clinical features—with image-based inputs to improve diagnostic robustness. In addition, deployment in clinical settings and approval over numerous institutions will be basic to ensure viable utility. In conclusion, this study presents a vigorous and precise deep learning model for ALL discovery, contributing to the developing field of AI-assisted hematological diagnostics. With further approval and integration, the proposed approach holds potential to become a valuable decision-support device in clinical oncology.
Conclusion
Acute lymphoblastic leukemia (ALL) presents a noteworthy experiment in healthcare, particularly affecting young individuals but also impacting adults. The diagnosis of Acute Lymphoblastic Leukemia (ALL) involves a comprehensive approach that combines blood analyses, physical check-ups, and additional laboratory assessments, such as bone marrow aspiration and biopsy. The developed model has undergone optimization and testing with two different optimizers: Adam and Adamax. The findings indicate impressive accuracy rates of 0.96 for Adam and 0.91 for Adamax. This advancement has the potential to transform how ALL is diagnosed and treated. By utilizing deep learning methods, the identification process can be made more efficient, facilitating earlier detection and prompt intervention. This, in turn, can improve patient outcomes and reduce the burden of this disease on individuals and healthcare systems. The success of our model underscores the importance of ongoing research and development in medical science and therapeutic techniques.
Looking ahead, the future holds great promise for individuals affected by ALL. With continued innovation and collaboration within the medical community, one can strive towards even better results and an improved quality of life for patients. By harnessing the power of deep learning and other cutting-edge technologies, one can continue to make meaningful strides in the fight against ALL and other forms of cancer.
Funding.
Manipal University Jaipur.
Data availability
The dataset [17] used in this study is publicly available on the Kaggle repository at https://www.kaggle.com/datasets/mehradaria/leukemia.
References
Liu, B. et al. Developing a new sepsis screening tool based on lymphocyte count, international normalized ratio and procalcitonin (LIP score). Sci. Rep. 12(1), 20002. https://doi.org/10.1038/s41598-022-16744-9 (2022).
Sheykhhasan, M., Manoochehri, H. & Dama, P. Use of CAR T-cell for acute lymphoblastic leukemia (ALL) treatment: A review study. Cancer Gene Ther. 29, 1080–1096 (2022).
Atteia, G., Alhussan, A. A. & Samee, N. A. BO-ALLCNN: Bayesian-Based Optimized CNN for Acute Lymphoblastic Leukemia Detection in Microscopic Blood Smear Images. Sensors 22, 5520 (2022).
Zakir Ullah, M. et al. An Attention-Based Convolutional Neural Network for Acute Lymphoblastic Leukemia Classification. Appl. Sci. 11, 10662 (2021).
Sampathila, N. et al. Customized deep learning classifier for detection of acute lymphoblastic leukemia using blood smear images. Healthcare 2022, 10 (1812).
Rezayi, S., Mohammadzadeh, N., Bouraghi, H., Saeedi, S. & Mohammadpour, A. Timely diagnosis of acute lymphoblastic leukemia using artificial intelligence-oriented deep learning methods. Comput. Intell. Neurosci. 2021, 5478157 (2021).
Baig, R., Rehman, A., Almuhaimeed, A., Alzahrani, A. & Rauf, H. T. Detecting malignant leukemia cells using microscopic blood smear images: A deep learning approach. Appl. Sci. 12, 6317 (2022).
Karar, M. E., Alotaibi, B. & Alotaibi, M. Intelligent medical IoT-enabled automated microscopic image diagnosis of acute blood cancers. Sensors 22, 2348 (2022).
Bhute, A. et al. Acute lymphoblastic leukemia detection and classification using an ensemble of classifiers and pre-trained convolutional neural networks. Int. J. Intell. Syst. Appl. Eng. 12(1), 571–580 (2024).
Huang, M. L. & Huang, Z. B. An ensemble-acute lymphoblastic leukemia model for acute lymphoblastic leukemia image classification. Math. Biosci. Eng. 21(2), 1959–1978 (2024).
Mondal, C. et al. Ensemble of convolutional neural networks to diagnose acute lymphoblastic leukemia from microscopic images. Inf. Med. Unlocked 27, 100794 (2021).
Duggal, R., Gupta, A., Gupta, R. and Mallick, P., 2017. SD-layer: stain deconvolutional layer for CNNs in medical microscopic imaging. In Medical Image Computing and Computer Assisted Intervention− MICCAI 2017: 20th International Conference, Quebec City, QC, Canada, September 11–13, 2017, Proceedings, Part III 20 (pp. 435–443). Springer International Publishing.
Alnowaiser, K., Saber, A., Hassan, E. & Awad, W. A. An optimized model based on adaptive convolutional neural network and grey wolf algorithm for breast cancer diagnosis. PLoS ONE 19(8), e0304868 (2024).
Elbedwehy, S., Hassan, E., Saber, A. & Elmonier, R. Integrating neural networks with advanced optimization techniques for accurate kidney disease diagnosis. Sci. Rep. 14(1), 21740 (2024).
Saber, A., Elbedwehy, S., Awad, W.A. and Hassan, E., 2024. An optimized ensemble model based on meta-heuristic algorithms for effective detection and classification of breast tumors. Neural Computing and Applications, pp.1–14.
Hassan, E., Elbedwehy, S., Shams, M. Y., Abd El-Hafeez, T. & El-Rashidy, N. Optimizing poultry audio signal classification with deep learning and burn layer fusion. J. Big Data 11(1), 135 (2024).
Aria, M. et al. Acute Lymphoblastic Leukemia (ALL) image dataset. Kaggle https://doi.org/10.34740/KAGGLE/DSV/2175623 (2021).
Anand, V., Koundal, D., Alghamdi, W. Y. & Alsharbi, B. M. Smart grading of diabetic retinopathy: An intelligent recommendation-based fine-tuned EfficientNetB0 framework. Front. Artif. Intell. 7, 1396160 (2024).
Zhang, T. & Xue, G. Fuzzy attention-based deep neural networks for acute lymphoblastic leukemia diagnosis. Appl. Soft Comput. 171, 112810 (2025).
Reza, H. et al. Acute lymphoblastic leukemia diagnosis and subtype segmentation in blood smears using CNN and U-Net. Indones. J. Electr. Eng. Comput. Sci. 38(2), 950–959 (2025).
Muduli, D. et al. Deep learning-based detection and classification of acute lymphoblastic leukemia with explainable AI techniques. Array 26, 100397 (2025).
Başaran, E., Çelik, G. and Toğaçar, M., 2025. Regionally focused neural-coder model designed for the diagnosis of acute lymphoblastic leukemia disease. Measurement, p.118176
Almahdawi, H., Akbas, A. and Rahebi, J., 2025. Deep learning neural network based on PSO for leukemia cell disease diagnosis from microscope images. J. Imag. Inf. Med., pp.1–10.
Alim, M. S. et al. Integrating convolutional neural networks for microscopic image analysis in acute lymphoblastic leukemia classification: A deep learning approach for enhanced diagnostic precision. Syst. Soft Comput. 6, 200121 (2024).
Khan Tusar, M. T. H., Islam, M. T., Sakil, A. H., Khandaker, M. N. & Hossain, M. M. An Intelligent telediagnosis of acute lymphoblastic leukemia using histopathological deep learning. J. Comput. Theor. Appl. 2(1), 1–12 (2024).
Papaioannou, D., Christou, I., Anagnou, N. and Chatziioannou, A., 2024. Deep Learning Algorithms for Early Diagnosis of Acute Lymphoblastic Leukemia. arXiv preprint arXiv:2407.10251.
Huang, M. L. & Huang, Z. B. An ensemble-acute lymphoblastic leukemia model for acute lymphoblastic leukemia image classification. Math. Biosci. Eng. 21(2024), 1959–1978 (2024).
Jawahar, M., Anbarasi, L. J., Narayanan, S. & Gandomi, A. H. An attention-based deep learning for acute lymphoblastic leukemia classification. Sci. Rep. 14(1), 17447 (2024).
Luan, S. et al. Deep learning for fast super-resolution ultrasound microvessel imaging. Phys. Med. Biol. 68(24), 245023. https://doi.org/10.1088/1361-6560/ad0a5a (2023).
Funding
Open access funding provided by Manipal University Jaipur.
Author information
Authors and Affiliations
Contributions
Conceptualization, V.A., P.B.; methodology, V.A., P.B., D.K.; software, V.A., P.B., D.K.; validation, D.K., and A.D.; formal analysis, D.K., and A.D.;investigation, D.K., and A.D.; resources, D.K., and A.D.; data curation, D.K., and A.D.; writing—original draft preparation, V.A., P.B., D.K.; writing—review and editing, V.A., P.B., D.K.; visualization, P.B., D.K., and A.D.; project administration, D.K., and A.D.; Funding, A.D.All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Anand, V., Bachhal, P., Koundal, D. et al. Deep learning model for early acute lymphoblastic leukemia detection using microscopic images. Sci Rep 15, 29147 (2025). https://doi.org/10.1038/s41598-025-13080-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-13080-6