Abstract
The Poisson-Inverse Gaussian regression model is a widely used method for analyzing count data, particularly in over-dispersion. However, the reliability of parameter estimates obtained through maximum likelihood estimation in this model can be compromised when multicollinearity exists among the explanatory variables. Multicollinearity means that high correlations between explanatory variables inflate the variance of the maximum likelihood estimates and increase the mean squared error. To handle this problem, the Poisson-Inverse Gaussian ridge regression estimator has been proposed as a viable alternative. This paper introduces a generalized ridge estimator to estimate regression coefficients in the Poisson-Inverse Gaussian regression model under multicollinearity. The performance of the proposed estimator is evaluated through a comprehensive simulation study, covering various scenarios and employing the mean squared error as the evaluation criterion. Furthermore, the practical applicability of the estimator is demonstrated using two real-life datasets, with its performance again assessed based on mean squared error. Theoretical analyses, supported by simulation and empirical findings, suggest that the proposed estimator outperforms existing methods, offering a more reliable solution in multicollinearity.
Similar content being viewed by others
Introduction
Count data characterized by non-negative integer values frequently arises in various disciplines, necessitating the use of specialized regression models to explore relationships between the response variable and its predictors. These models facilitate the identification of significant explanatory variables and their impact on the response1,2. Among the most commonly employed models is the Poisson regression model (PRM), which assumes that the mean and variance are equal (i.e., equi-dispersion). However, this assumption is often unrealistic in empirical datasets, where the variance either exceeds (over-dispersion) or is less than (under-dispersion) the mean. In such situations, applying the Poisson model may lead to biased estimates and unreliable conclusions3. To address the limitations of equi-dispersion, alternative count regression models, including the negative binomial (NBRM)4, Bell (BRM)5, quasi-Poisson (QPRM)6, generalized Poisson Lindley (GPLRM)7, Conway-Maxwell Poisson (CMPRM)8, and Poisson-Inverse Gaussian (PIGRM) models9, have been proposed. These models extend the Poisson framework to accommodate over-dispersed and under-dispersed data, thereby enhancing model accuracy and interpretability.
The PIGRM is highly effective for datasets with heavy tails, where a small number of extreme values differ significantly from most data near zero. This type of data is common in fields like actuarial science, biology, engineering, and medical research. The PIGRM is preferred over the NBRM for its ability to handle greater skewness and kurtosis, making it ideal for heavy-tailed count data9. Willmot3 highlighted the PIGRM as a better alternative to the NBRM for highly skewed and heterogeneous datasets. Putri et al.10 found PIGRM effective in managing overdispersion when compared to NBRM. Similarly, Saraiva et al.11 used PIGRM for overdispersed dengue data, and Husna and Azizah12 applied it to study dengue hemorrhagic fever in Central Java, showcasing its versatility for complex count data.
One of the fundamental assumptions of the generalized linear regression model is the absence of correlation among explanatory variables. However, in empirical settings, explanatory variables frequently exhibit strong or near-strong linear relationships, thereby violating this assumption and giving rise to the problem of multicollinearity13. This issue also manifests in the context of the PIGRM, where multicollinearity complicates parameter estimation, inflated variance, and increases mean squared error (MSE). The maximum likelihood estimator (MLE), commonly employed for estimating regression coefficients in the PIGRM, is particularly sensitive to multicollinearity, as it results in inflated variances and unreliable parameter estimates14. To tackle multicollinearity in regression models, ridge regression and the Liu estimator are two commonly studied techniques. Ridge regression, introduced by Hoerl and Kennard15, has been extensively explored, with notable studies focusing on optimal shrinkage parameters by Golub et al.16, and Alkhamisi et al.17. Segerstedt18 extended ridge regression to the generalized linear model (GLM), with further advancements by researchers like Månsson19, Saleh et al.20, Amin et al.21, Sami et al.22, and Shahzad et al.23. On anther hand Kejian24 introduced the Liu estimator, which offers a linear shrinkage parameter \(d\) and has been applied to different models. Kibria25 and Alheety and Kibria26 developed the Liu estimator for linear regression models (LRMs) and later Kurtoǧlu27 defined the Liu estimator for GLMs. Then, inspiring further applications by Qasim et al.28 and Bulut29.
Several studies have focused on developing generalized versions of the ridge estimator to handle multicollinearity in different types of regression models. Rashad et al.30 proposed a generalized ridge estimator for the NBRM, while Akram et al.31 extended this idea to both generalized ridge and generalized Liu estimators in the gamma regression. Fayose and Ayinde32 explored how to define an appropriate biasing parameter for the generalized ridge regression estimator, and Bhat and Raju33 introduced a class of such estimators. Gómez et al.34 discussed biased estimation using generalized ridge regression for multiple LRMs. Abdulazeez and Algamal35 combined shrinkage estimation with particle swarm optimization to define a generalized ridge estimator. In addition, van Wieringen36 applied the generalized ridge approach to estimate the inverse covariance matrix, and Mohammadi37 suggested a test for detecting harmful multicollinearity based on this method. Bello and Ayinde38 also proposed feasible generalized ridge estimators for models affected by both multicollinearity and autocorrelation. Recent works30,35,39,40 have shown that the performance of biased estimators can be improved by allowing the biasing parameters to vary across observations (such as \(k_j\)) rather than using a fixed value (such as \(k\)). Based on this, the main goal of the present paper is to develop a more general form of the ridge estimator to better address multicollinearity in the PIGRM.
This paper discusses the advantages of the generalized ridge estimator in the PIGRM. We also present new methods for determining the optimal values of the shrinkage coefficients (kj), based on the methodologies of Rashad30 and Dawood et al.39 The careful selection of these coefficients is crucial to improving the performance of the biased generalized ridge estimation method. Monte Carlo simulations use the MSE to evaluate the performance of the proposed estimator compared to existing estimators. To validate the simulation results, we use two real-world data studies that confirm the effectiveness of the estimator, highlighting its improved accuracy and robustness in addressing problems related to multicollinearity.
The structure of this paper is as follows: Section 2 provides a summary of the PIGRM model and examines biased estimation approaches, including PIGRRE and the proposed estimator. It also discusses theoretical comparisons of the proposed estimator and methods for selecting biasing parameters. Section 3 describes the study and results of the Monte Carlo simulation used to evaluate the proposed estimator. Section 4 highlights the application of the estimator through two real-world datasets to confirm the simulation results. Finally, Section 5 summarizes the results and discusses their broader significance.
Methodology
The PIG distribution combines two probability distributions: the Poisson and the Inverse Gaussian. Let \(Y\) represent a random variable following a Poisson distribution with a mean of \(\mu \nu\), where \(\nu\) itself follows an Inverse Gaussian distribution with mean 1 and dispersion parameter \(\phi\). The probability mass function (PMF) of \(Y\) is expressed as1,41:
where \(\nu \sim \text {IG}(1, \phi )\), and the probability density function of \(\nu\) is given by42:
The marginal PMF of \(Y\), representing the PIG distribution, is derived by integrating over \(\nu\). This results in the following expression:
where \(s = y - 0.5\), \(\alpha ^2 = \phi ^2 \left( 1 + 2\frac{\mu }{\phi } \right)\), and \(\textbf{R}_s(\alpha )\) denotes the modified Bessel function of the third kind43. The mean and variance of the distribution are \(E(Y) = \mu\) and \(V(Y) = \mu + \frac{\mu ^3}{\phi }\), respectively.
In the context of PIGRM, the linear predictor \(g(\mu ) = \eta _i\) is used, which is equivalent to \(\mu = \exp {x_i^T \beta }\), where \(\eta _i\) is the linear combination of the explanatory variables, defined as \(\eta _i = x_i^T \beta\), where \(\beta = (\beta _0, \beta _1, \dots , \beta _{p-1})^T\) is the vector of regression coefficients, and \(x_i = (1, x_{i1}, \dots , x_{i(p-1)})^T\) is the vector of explanatory variables for the \(i\)-th observation1,43.
To estimate the parameters of the PIGRM, the MLE method is employed. The log-likelihood function for the model is as follows:
The log-likelihood function with respect to the regression coefficients \(\beta\) is given by:
To obtain the MLE of the parameters, we differentiate the log-likelihood function concerning the parameters and set the derivatives equal to zero. The first derivative with respect to \(\beta _j\) is10:
Similarly, thetove with respect to \(\phi\) is:
To estimate the parameters \(\beta\) and \(\phi\) through MLE, numerical methods such as the Newton-Raphson algorithm or the iteratively reweighted least squares (IRLS) algorithm are utilized, given the non-linearity of the equations (Eqs. (6) and (7)). Upon completion of the iterative procedure, the MLE estimate for \(\hat{\beta }\) is derived from the following expression14:
where \(\hat{W} = \text {diag}(\mu _i + \alpha \mu _i^3)\) and \(\hat{u}_i\), the adjusted response variable, is computed as \(\hat{u}_i = \log (\mu ) + \frac{y_i - \mu _i}{\mu _i + \mu _i^3 / \phi }\). To evaluate the accuracy of the estimator, the mean squared error matrix (MSEM) and MSE of \(\hat{\beta }_{\text {MLE}}\) are determined through spectral decomposition of the matrix \(X^T\hat{W} X= \mathcal {Q} G \mathcal {Q}^T\). where \(G = \text {diag}(g_1, g_2, \dots , g_p)\) is the diagonal matrix of the eigenvalues of \(X^T \hat{W} X\), and \(\mathcal {Q}\) is the orthogonal matrix with columns corresponding to the eigenvectors of \(X^T \hat{W} X\).
The MSEM for the estimator is given by:
The MSE is calculated as:
where \(\hat{\phi }\) is the estimated value of \(\phi\) and computed as \(\hat{\phi } = \frac{\sum _{i=1}^n \frac{(y_i - \hat{\mu }_i)^2}{V(\hat{\mu }_i)}}{n - p}\).
Poisson-Inverse Gaussian ridge regression estimator
Segerstedt18 proposed a ridge regression estimator to handle multicollinearity in GLM as an alternative to the MLE. Accordingly, Månsson19, Månsson and Shukur44, Kibria et al.45, and Ashraf46 apply it to many GLMs. Following this research, Batool et al.14 proposed a PIGRRE to address multicollinearity in PIGRM. The PIGRRE is given by:
where \(k~ (k>0)\) represents a PIGRRE parameter, and \(I\) is the identity matrix of \(p \times p\). In particular, when \(k = 0\), the PIGRRE reduces to MLE. The bias and covariance matrix associated with the PIGRRE are as follows:
where \(G_k = \text {diag}(g_1 + k, g_2 + k, \dots , g_p + k)\) is a diagonal matrix consisting of the eigenvalues of the covariance matrix, adjusted by the ridge parameter.
The MSEM of the PIGRRE estimator combines both the covariance and the squared bias:
Expanding the above expression:
where \(b_k = \text {Bias}(\hat{\beta }_k) = -k \mathcal {Q} G_k^{-1} \alpha\). The MSE is calculated as the trace of the MSEM:
where \(\alpha _j\) denotes the \(j\)-th component of the vector \(\alpha = \mathcal {Q}^T \hat{\beta }_{\text {MLE}}\), representing the MLE-adjusted coefficient values.
Proposed estimator
Building on the work of Bhat and Raju33 and extending the contributions of Rashad et al.30 and Akram et al.31, we propose the generalized ridge estimator for the PIGRM. The Poisson-Inverse Gaussian generalized ridge estimator (PIGGRE) enhances standard ridge regression by assigning a unique shrinkage parameter to each regression coefficient, improving its ability to address multicollinearity.
In the PIGGRE method, the ridge parameter matrix is written as \(K = \text {diag}(k_1, k_2, \ldots , k_p)\), where each \(k_j\) controls how much shrinkage is applied to the \(j\)-th regression coefficient. This setup gives more flexibility and improves prediction accuracy and addresses multicollinearity than the standard PIGRRE approach.
Key scenarios in the PIGGRE framework include:
-
1.
\(k_j = 0\) for all \(j\): the estimator reduces to the MLE.
-
2.
\(k_j = k\) (constant) for all \(j\): the estimator corresponds to PIGRRE.
-
3.
\(k_j\) varying across coefficients: the estimator is referred to as PIGGRE
The PIGGRE is defined as:
The bias and covariance matrix associated with the PIGGRE are as follows:
The MSEM of the PIGGRE estimator is defined by:
where \(G_K = \text {diag}(g_1 + k_1, g_2 + k_2, \dots , g_p + k_p)\) and \(b_K = \text {Bias}(\hat{\beta }_K) = -K \mathcal {Q} G_K^{-1} \alpha\). The MSE is calculated as the trace of the MSEM:
Superiority of PIGGRE
In this part, we compare the proposed PIGGRE method with other known estimators, such as MLE and PIGRRE. To support this comparison, we make use of the following lemma:
Lemma 1
Trenkler and Toutenburg47. Consider two linear estimators of \(\alpha\) , denoted by \(\hat{\alpha }_i = U_i w\) for \(i = 1, 2.\) Let \(E = \text {Cov}(\hat{\alpha }_1) - \text {Cov}(\hat{\alpha }_2) ,\) where \(\text {Cov}(\hat{\alpha }_i)\) is the covariance matrix of \(\hat{\alpha }_i ,\) and assume \(E > 0.\) Additionally, define the bias of \(\hat{\alpha }_i\) as \(b_i = (U_i X - I) \alpha .\) Then, the difference in MSEM between the two estimators can be expressed as:
where the MSEM of \(\hat{\alpha }_i\) is given by:
This inequality is satisfied if and only if:
Theorem 2.1
The PIGGRE estimator, \(\hat{\beta }_{K} ,\) is superior to an alternative estimator \(\hat{\beta }_{\text {MLE}}\) if and only if:
Proof
The difference (D) between the covariance matrices of the two estimators is given by:
This can be expressed as:
The matrix \(G^{-1} - G_K^{-1} G G_K^{-1}\) is positive definite if and only if \((g_j + k_j)^2 - g_j^2 > 0\) or \(k_j^2 +2k_j g_j > 0\). It is evident that for \(k_j > 0\) (\(i = 1, 2, \ldots , p\)), the term \(k_j^2 +2k_j g_j^2 > 0\) is positive, ensuring that \(G^{-1} - G_K^{-1} G G_K^{-1}\) is positive definite. Thus, by Lemma 3, the superiority condition of \(\hat{\beta }_{K}\) over \(\hat{\beta }_{\text {MLE}}\) is satisfied.\(\square\)
Theorem 2.2
The PIGGRE estimator, \(\hat{\beta }_{K}\), is superior to an alternative estimator \(\hat{\beta }_{k}\) if and only if:
Proof
The difference (D) between the covariance matrices of the two estimators is given by:
This can be expressed as:
The matrix \(G_k^{-1} G G_k^{-1} - G_K^{-1} G G_K^{-1}\) is positive definite if and only if \((g_j + k_j)^2 -( g_j+k)^2 > 0\) or \(k_j^2-k^2 +2 g_j(k_j-k) > 0\). It is evident that for \(k_j > 0\) (\(i = 1, 2, \ldots , p\)), the term \(k_j^2-k^2 +2 g_j(k_j-k) > 0\) is positive, ensuring that \(G_k^{-1} G G_k^{-1} - G_K^{-1} G G_K^{-1}\) is positive definite. Thus, by Lemma 3, the superiority condition of \(\hat{\beta }_{K}\) over \(\hat{\beta }_{k}\) is satisfied.\(\square\)
The biasing parameters of the PIGGRE
To estimate the optimal values of \(k_j\) for the proposed PIGRRE, the approach minimizes the MSE of the estimator, defined as:
To find the optimal values of \(k_j\), differentiate \(T(k_1, k_2, \ldots , k_p)\) with respect to \(k_j\) and set the derivative to zero \(\frac{\partial T(k_1, k_2, \ldots , k_p)}{\partial k_j} = 0.\) Solving for \(k_j\), we obtain:
where \(\hat{\phi }\) and \(\hat{\alpha }\) represent the estimated values of \(\phi\) and \(\alpha\), respectively.
Following Bhat and Raju33 and Rashad et al.30, we considered the following values of \(K\):
Monte Carlo simulation
Simulation design
The simulation procedure for evaluating the performance of the proposed estimator involves several steps, considering key factors such as sample size (\(n\)), the number of explanatory variables (\(p\)), levels of multicollinearity (\(\rho ^2\)), and dispersion parameters (\(\phi\)), as detailed in Table 1. The simulation process is outlined as follows:
-
1.
Choose the regression coefficients \(\beta = (\beta _1, \beta _2, \dots , \beta _{(p)})\) such that the sum of their squared values equals 1, i.e., \(\sum _{j=1}^p \beta _j^2 = 1\). This normalization is a common assumption in regression modeling.
-
2.
Use PIG(\(\mu , \phi\)) distribution to generate the response variable \(y_i\) for the PIGRM, where \(\mu = \exp (x_i^T \beta )\). Here, \(x_i^T\) represents the explanatory variables for observation \(i\), and \(\beta\) are the regression coefficients.
-
3.
Simulate the correlated explanatory variables \(x_{ij}\) using the formula:
$$\begin{aligned} x_{ij} = \sqrt{1 - \rho ^2} \, F_{ij} + \rho \, F_{i(j+1)}, \end{aligned}$$where \(F_{ij}\) are independent standard normal random variables. This ensures the correlation structure between the explanatory variables, with \(\rho ^2\) controlling the degree of correlation. This step is repeated for \(i = 1, \ldots , n\) and \(j = 1, \ldots , p+1\).
-
4.
Use the ‘gamlss‘ package in R to estimate the regression parameters based on the simulated data, selecting the PIG family for the model.
-
5.
Repeat the generation of the entire data and estimation process for different combinations of \(n\),\(\rho\), \(p\), and \(\phi\) for 1000 replications to ensure the robustness of the results.
-
6.
Evaluate the performance of the proposed estimator using the MSE criterion:
$$\begin{aligned} \text {MSE}(\hat{\beta }) = \frac{\sum _{i=1}^R (\hat{\beta }_i - \beta )^T (\hat{\beta }_i - \beta )}{R}, \end{aligned}$$(27)where \(\hat{\beta }_i\) represents the estimated parameters from the \(i\)-th replication, \(\beta\) is the true parameter vector, and \(R\) is the total number of replications (1000 in this case). This criterion quantifies the deviation between the true parameters and the estimated ones, helping to assess the accuracy of the estimator.
Following Segerstedt18, Månsson and Shukur44, and Batool et al.14, we use the following values of \(k\) for PIGRRE:
$$k_1 = \min \left( \frac{\hat{\phi }}{2 \hat{\alpha }_j^2 + \frac{\hat{\phi }}{g_j}}\right) , \quad \hat{k}_2 = \min \left( \frac{\hat{\phi }}{\hat{\alpha }_j^2}\right) , \quad \hat{k}_3 = \left( \frac{\hat{\phi }}{\sum _j^p \hat{\alpha }_j^2}\right) , \quad k_4 = \prod _{j=1}^p \left( \frac{\hat{\phi }}{2 \hat{\alpha }_j^2 + \frac{\hat{\phi }}{g_j}}\right) ^{\frac{1}{p}}, \quad k_5 = p\sum _{j=1}^p\left( \frac{\hat{\phi }}{2 \hat{\alpha }_j^2 + \frac{\hat{\phi }}{g_j}}\right) .$$
Simulation results
Tables 2, 3, 4, 5, 6, 7, 8, 9 and 10 introduced the simulated MSE values for the MLE, PIGRRE, and PIGGRE, based on the results of the simulation analysis:
-
1.
An increase in the degree of multicollinearity (\(\rho\)), the dispersion parameter (\(\phi\)), or the number of explanatory variables (\(p\)) results in higher MSE values for all estimators, indicating a degradation in performance under these conditions.
-
2.
The MSE values decrease as the sample size (\(n\)) increases, emphasizing the role of larger sample sizes in improving estimator reliability and precision.
-
3.
The PIGRRE demonstrates superior performance compared to the MLE across all scenarios, consistently yielding lower MSE values irrespective of variations in \(\phi\), \(\rho\), \(p\), and \(n\).
-
4.
The PIGGRE outperforms the PIGRRE, achieving the lowest MSE values among the evaluated estimators across all parameters irrespective of variations in \(\phi\), \(\rho\), \(p\), and \(n\).
-
5.
The results show that the proposed PIGGRE estimator works efficiently, especially when the dispersion parameter (\(\phi\)) is high and the explanatory variables are highly correlated. This indicates that the estimator is reliable even in complex situations where traditional methods may struggle.
-
6.
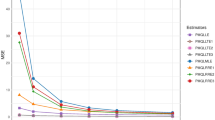
Figures 1a–d show the simulation results by presenting the mean squared errors (MSEs) for different estimators under various settings, such as different sample sizes (\(n\)), levels of multicollinearity (\(\rho ^2\)), numbers of predictors (\(p\)), and values of the dispersion parameter (\(\phi\)). The results clearly show that PIGGRE performs better than both PIGRRE and MLE by consistently achieving lower MSE values. This improvement is especially clear when using the ridge parameters \(\hat{K}_1\) and \(\hat{K}_4\), which help reduce estimation errors and improve the reliability of the method.
Applications
In this section, we compare our proposed estimator with existing estimators such as MLE and PIGRRE, using two real-world datasets to demonstrate the advantages and superiority of the proposed estimator and highlight its effectiveness in improving estimation accuracy, especially in the presence of multicollinearity.
Number of equations and citations
The performance of the proposed estimator is assessed using a real dataset from Fawcett and Higginson48, which is available in R through the AER package under the name EquationCitations. This data investigates the relationship between the number of citations received by evolutionary biology publications and the number of equations per page in the cited papers. This dataset comprises 649 observations, with the response variable representing the total number of equations (y). The explanatory variables include the total number of citations received (\(x_1\)), the number of equations present in appendix (\(x_2\)), the number of equations present in the main text (\(x_3\)), the number of citations made by the authors of the paper (\(x_4\)), the number of citations from theoretical papers (\(x_5\)), and the number of citations from non-theoretical papers (\(x_6\)).
The response variable in this study is count data, which requires specialized models. We compared the PRM, NBRM, COMPRM, and PIGRM to identify the best fit for the relationship between the response variable and explanatory variables in the number of equations and citation data. The selection criteria were the log-likelihood (LL), the Akaike Information Criterion (AIC), and the Bayesian Information Criterion (BIC), where a lower AIC, BIC, and higher LL indicate a better model. Results in Table 11 show that the PIGRM outperforms the other models with the lowest AIC, BIC, and highest LL.
With six explanatory variables in the dataset, the eigenvalues of the matrix \(X^T \hat{W} X\) are: 1754069.240, 76832.270, 59927.316, 17295.812, 4131.207, 129.039, and 63.036. Multicollinearity was assessed using the condition number (CN) and variance inflation factors (VIFs). The CN, defined as \(\text {CN} = \sqrt{\frac{\lambda _{\max }}{\lambda _{\min }}},\) where \(\lambda _{\max }\) and \(\lambda _{\min }\) are the largest and smallest eigenvalues of \(X^T\hat{W} X\), respectively, was calculated to be 166.81, indicating a high level of multicollinearity. In addition, the VIF for each explanatory variable, computed as: \(\text {VIF}_j = \frac{1}{1 - R_j^2},\) where \(R_j^2\) is the coefficient of determination from regressing the \(j\)-th variable on all other predictors, yielded values of 1201.15, 1.68, 1.57, 23.77, 255.37, and 463.52. These values confirm the presence of severe multicollinearity among several variables. This is further supported by the correlation matrix shown in Fig. 2, emphasizing the need for caution in interpreting parameter estimates and potentially adopting bias-reducing methods to improve model stability.
The coefficients of the PIGRM are estimated using three methods: MLE, PIGRRE, and PIGGRE, as specified in Eqs. 8, 11, and 17, respectively. The corresponding MSEs for each estimator are computed using Eqs. 10, 16, and 21. The results indicate that MLE performs less favorably than PIGRRE across all ridge parameter estimators, as reflected by its higher MSE. Conversely, PIGGRE demonstrates improved performance compared to both MLE and PIGRRE, yielding lower MSE values. A comparison of the ridge parameter estimators used in PIGGRE, presented in Table 12, highlights that \(\hat{K}_1\) through \(\hat{K}_3\) outperform other existing estimators. These findings are consistent with the results observed in the simulation studies, supporting the efficacy of the proposed estimators.
Australian institute of sports
To empirically evaluate the proposed estimator’s performance, we utilize the Australian Institute of Sport dataset, as presented by Telford and Cunningham49 and is available in the R software through the GLMsData package under the name AIS. This dataset encompasses physical and blood measurements of high-performance athletes from various sports, including basketball (BBall), field events, gymnastics (Gym), netball, rowing, swimming (Swim), track events over 400 meters (T400m), tennis, track sprints (TPSprnt), and water polo (WPolo). The dataset contains information for 202 athletes, comprising 102 males and 100 females. The response variable in this dataset is the plasma ferritin (in ng per decilitre) (y). In addition, the dataset includes 10 explanatory variables lean body mass (in kg) (\(x_1\)), height (in cm) (\(x_2\)), weight (in kg) (\(x_3\)), body mass index (in \(\textrm{kg}/\textrm{m}^{2}\)) (\(x_4\)), the sum of skin folds (\(x_5\)), percentage body fat (\(x_6\)), red blood cell count (in \(10^{12}\) per liter) (\(x_7\)), white blood cell count (in \(10^{12}\) per liter) (\(x_8\)), hematocrit (in percent) (\(x_9\)), and hemoglobin concentration (in grams per deciliter) (\(x_{10}\))
The dependent variable in this analysis is count data, which necessitates appropriate models. To determine the best model for the relationship between the dependent variable and the explanatory variables in the context of the number of equations and citation data, we compared four models: the PRM, NBRM, COMPRM, and PIGRM. We evaluated the models based on three criteria: LL, AIC, and BIC. A lower AIC, BIC, and higher LL indicate a more suitable model. As shown in Table 13, the PIGRM provides the best fit, with the lowest AIC and BIC values and the highest LL.
The dataset includes ten explanatory variables, the eigenvalues of the matrix \(X^T \hat{W} X\) are: 30689803.431, 702460.991, 114401.412, 8572.263, 1996.989, 1704.116, 1019.529, 123.657, 94.052, 19.553, and 0.037. Multicollinearity was assessed using the CN and VIFs. The observed CN value of 28699.25, alongside VIF values of 442.07, 56.21, 516.80, 79.96, 22.75, 7.27, 1.09, 15.95, 12.23, and 62.66, indicates substantial multicollinearity among the variables. These findings are further corroborated by the correlation matrix shown in Fig. 3. Such evidence highlights the necessity for careful interpretation and potential adjustments in subsequent analyses to mitigate the impact of multicollinearity.
The coefficients for the PIGRM were estimated using three methods: MLE, PIGRRE, and PIGGRE, as outlined in equations 8, 11, and 17. The corresponding MSEs were calculated based on equations 10, 16, and 21. The results indicate that MLE performs less effectively than PIGRRE, as reflected by its higher MSE values. In contrast, PIGGRE shows superior performance, with consistently lower MSEs compared to both MLE and PIGRRE. Additionally, a comparison of the ridge parameter estimators in PIGGRE, shown in Table 14, highlights that estimators \(\hat{K}_1\) and \(\hat{K}_4\) outperform \(\hat{K}_2\), \(\hat{K}_3\), and \(\hat{K}_5\) in PIGGRE. This can be attributed to the nature of the data, including the degree of multicollinearity among the predictors, the level of dispersion, and the sample size. These factors collectively influence the performance of the estimators and the sensitivity of each to the choice of the biasing parameter K. Overall, the PIGGRE estimator demonstrates superior performance, which is consistent with both the simulation results and the findings of the first application.
Conclusion
The PIGRM is one of the most widely used models for analyzing overdispersed count data. In the PIGRM, MLE is used to estimate regression coefficients. However, when the explanatory variables are highly correlated, this leads to multicollinearity, a problem that reduces the reliability of the regression coefficients and inflates the variance. To address this, we introduced in this paper a new biased estimation method called the PIGGRE, which uses shrinkage parameter techniques to reduce the effect of multicollinearity. We evaluated the performance of the proposed PIGGRE through simulation studies with different scenarios, such as sample sizes, levels of dispersion, multicollinearity, and number of predictors. The results showed that PIGGRE outperformed both MLE and PIGRRE. In particular, using shrinkage parameters \(K_1\) and \(K_3\) resulted in the lowest MSE, demonstrating higher accuracy. To confirm the simulation study results, we also applied the proposed PIGGRE to two real datasets. The results of these applications provided significant support and confirmation of the simulation results, demonstrating the advantages of using PIGGRE. Therefore, we recommend using PIGGRE with values of \(K_1\) and \(K_3\) to address multicollinearity in PIGRM. This approach could be extended in future work to other models, such as the zero-inflated negative binomial model, the zero-inflated Poisson model, and the Conway-Maxwell-Poisson model. Although PIGGRE outperforms in the context of PIGRM under multicollinearity, there is one important limitation worth mentioning. The estimator’s performance depends mainly on the K values, which balance bias and variance, and suboptimal K values can affect the estimator’s performance, making the estimator sensitive to the choice of parameters. Further improvements could be achieved by incorporating accurate estimation techniques, such as those proposed by Omara50 and Lukman et al.51.
Data availability
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
Dean, C., Lawless, J. F. & Willmot, G. E. A mixed Poisson-inverse-Gaussian regression model. Can. J. Stat. 17(2), 171–181 (1989).
Shoukri, M. M., Asyali, M. H., VanDorp, R. & Kelton, D. The Poisson inverse gaussian regression model in the analysis of clustered counts data. J. Data Sci. 2(1), 17–32 (2004).
Willmot, G. E. The Poisson-inverse gaussian distribution as an alternative to the negative binomial. Scand. Actuar. J. 1987(3–4), 113–127 (1987).
Hilbe, J. M. Negative Binomial Regression (Cambridge University Press, Cambridge, 2011).
Castellares, F., Ferrari, S. L. P. & Lemonte, A. J. On the bell distribution and its associated regression model for count data. Appl. Math. Model. 56, 172–185 (2018).
Ver Hoef, J. M. & Boveng, P. L. Quasi-Poisson versus negative binomial regression: how should we model overdispersed count data?. Ecology 88(11), 2766–2772 (2007).
Wongrin, W. & Bodhisuwan, W. Generalized Poisson–Lindley linear model for count data. J. Appl. Stat. 44(15), 2659–2671 (2017).
Sellers, K. F. & Premeaux, B. Conway–Maxwell–Poisson regression models for dispersed count data. Wiley Interdiscip. Rev. Comput. Stat. 13(6), e1533 (2021).
Zha, L., Lord, D. & Zou, Y. The Poisson inverse gaussian (pig) generalized linear regression model for analyzing motor vehicle crash data. J. Transp. Saf. Secur. 8(1), 18–35 (2016).
Putri, G. N., Nurrohmah, S. & Fithriani, I. Comparing Poisson-inverse gaussian model and negative binomial model on case study: Horseshoe crabs data. J. Phys: Conf. Ser. 1442(1), 012028 (2020).
Saraiva, E. F., Vigas, V. P., Flesch, M. V., Gannon, M. & de Bragança Pereira, C. A. Modeling overdispersed dengue data via Poisson inverse gaussian regression model: A case study in the city of Campo Grande, MS, Brazil. Entropy 24(9), 1256 (2022).
Husna, F. R. & Azizah, A. The Poisson inverse gaussian regression model in the analysis of dengue hemorrhagic fever case in central java. In AIP Conference Proceedings, vol. 3049(1) (2024).
Farrar, D. E. & Glauber, R. R. Multicollinearity in regression analysis: The problem revisited. Rev. Econ. Stat. 49, 92–107 (1967).
Batool, A., Amin, M. & Elhassanein, A. On the performance of some new ridge parameter estimators in the Poisson-inverse Gaussian ridge regression. Alex. Eng. J. 70, 231–245 (2023).
Hoerl, A. E. & Kennard, R. W. Ridge regression: Applications to nonorthogonal problems. Technometrics 12(1), 69–82 (1970).
Golub, G. H., Heath, M. & Wahba, G. Generalized cross-validation as a method for choosing a good ridge parameter. Technometrics 21(2), 215–223 (1979).
Alkhamisi, M., Khalaf, G. & Shukur, G. Some modifications for choosing ridge parameters. Commun. Stat. Theory Methods 35(11), 2005–2020 (2006).
Segerstedt, B. On ordinary ridge regression in generalized linear models. Commun. Stat. Theory Methods 21(8), 2227–2246 (1992).
Månsson, K. On ridge estimators for the negative binomial regression model. Econ. Model. 29(2), 178–184 (2012).
Saleh, A. M. E., Kibria, B. M. G. & Geroge, F. Comparative study of lasso, ridge regression, preliminary test and stein-type estimators for the sparse gaussian regression model. Stat. Optim. Inf. Comput. 7(4), 626–641 (2019).
Amin, M., Akram, M. N. & Majid, A. On the estimation of bell regression model using ridge estimator. Commun. Stat. Simul. Comput. 52(3), 854–867 (2023).
Sami, F., Amin, M. & Butt, M. M. On the ridge estimation of the Conway–Maxwell–Poisson regression model with multicollinearity: Methods and applications. Concurr. Comput. Pract. Exp. 34(1), e6477 (2022).
Shahzad, A., Amin, M., Emam, W. & Faisal, M. New ridge parameter estimators for the quasi-Poisson ridge regression model. Sci. Rep. 14(1), 8489 (2024).
Liu, K. A new class of Blased estimate in linear regression. Commun. Stat. Theory Methods 22(2), 393–402 (1993).
Kibria, B. G. Some Liu and ridge-type estimators and their properties under the ill-conditioned gaussian linear regression model. J. Stat. Comput. Simul. 82(1), 1–17 (2012).
Alheety, M. I. & Kibria, B. G. On the Liu and almost unbiased Liu estimators in the presence of multicollinearity with heteroscedastic or correlated errors. Surv. Math. Appl. 4, 155–167 (2009).
Kurtoğlu, F. & Özkale, M. R. Liu estimation in generalized linear models: Application on gamma distributed response variable. Stat. Pap. 57, 911–928 (2016).
Qasim, M., Kibria, B. G., Månsson, K. & Sjölander, P. A new Poisson Liu regression estimator: Method and application. J. Appl. Stat. 47(12), 2258–2271 (2020).
Bulut, Y. M. Inverse gaussian Liu-type estimator. Commun. Stat. Simul. Comput. 52(10), 4864–4879 (2023).
Rashad, N. K., Hammood, N. M. & Algamal, Z. Y. Generalized ridge estimator in negative binomial regression model. J. Phys: Conf. Ser. 1897(1), 012019 (2021).
Akram, M. N., Amin, M. & Faisal, M. On the generalized biased estimators for the gamma regression model: Methods and applications. Commun. Stat. Simul. Comput. 52(9), 4087–4100 (2023).
Fayose, T. S. & Ayinde, K. Different forms biasing parameter for generalized ridge regression estimator. Int. J. Comput. Appl. 181(37), 2–29 (2019).
Bhat, S. & Raju, V. A class of generalized ridge estimators. Commun. Stat. Simul. Comput. 46(7), 5105–5112 (2017).
Gómez, R. S., García, C. G. & Reina, G. H. Generalized ridge regression: Biased estimation for multiple linear regression models. arXiv preprint arXiv:2407.02583 (2024).
Abdulazeez, Q. A. & Algamal, Z. Y. Generalized ridge estimator shrinkage estimation based on particle swarm optimization algorithm. Electron. J. Appl. Stat. Anal. 14(1), 254–265 (2021).
van Wieringen, W. N. The generalized ridge estimator of the inverse covariance matrix. J. Comput. Graph. Stat. 28(4), 932–942 (2019).
Mohammadi, S. A test of harmful multicollinearity: A generalized ridge regression approach. Commun. Stat. Theory Methods 51(3), 724–743 (2022).
Bello, H. A. & Ayinde, K. Feasible ordinary and generalized ridge estimators for handling multicollinearity and autocorrelation in linear regression model. J. Sustain. Technol. 12(1), 24–35 (2023).
Dawoud, I., Abonazel, M. R. & Awwad, F. A. Generalized Kibria–Lukman estimator: Method, simulation, and application. Front. Appl. Math. Stat. 8, 880086 (2022).
Qian, F., Chen, R. & Wang, L. Generalized ridge shrinkage estimation in restricted linear model. Comput. Stat. 39(3), 1403–1416 (2024).
Shaban, S. A. Computation of the Poisson-inverse Gaussian distribution. Commun. Stat. Theory Methods 10(14), 1389–1399 (1981).
Folks, J. L. & Chhikara, R. S. The inverse Gaussian distribution and its statistical application—A review. J. R. Stat. Soc. Ser. B Stat Methodol. 40(3), 263–275 (1978).
Gómez-Déniz, E., Ghitany, M. E. & Gupta, R. C. Poisson-mixed inverse gaussian regression model and its application. Commun. Stat. Simul. Comput. 45(8), 2767–2781 (2016).
Månsson, K. & Shukur, G. A Poisson ridge regression estimator. Econ. Model. 28(4), 1475–1481 (2011).
Kibria, B. G., Månsson, K. & Shukur, G. Performance of some logistic ridge regression estimators. Comput. Econ. 40, 401–414 (2012).
Ashraf, B., Amin, M. & Akram, M. N. New ridge parameter estimators for the zero-inflated Conway–Maxwell–Poisson ridge regression model. J. Stat. Comput. Simul., 1–27 (2024).
Trenkler, G. & Toutenburg, H. Mean squared error matrix comparisons between biased estimators–an overview of recent results. Stat. Pap. 31(1), 165–179 (1990).
Fawcett, T. W. & Higginson, A. D. Heavy use of equations impedes communication among biologists. Proc. Natl. Acad. Sci. 109(29), 11735–11739 (2012).
Telford, R. D. & Cunningham, R. B. Sex, sport, and body-size dependency of hematology in highly trained athletes. Med. Sci. Sports Exerc. 23(7), 788–794 (1991).
Omara, T. Robust Liu-type estimator for sur model. Stat. Optim. Inf. Comput. 9(3), 607–617 (2021).
Lukman, A. F., Arashi, M. & Prokaj, V. Robust biased estimators for Poisson regression model: Simulation and applications. Concurr. Comput. Pract. Exp. 35(7), e7594 (2023).
Acknowledgements
Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2025R515), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
All authors have worked equally to write and review the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Almulhim, F.A., Hammad, A.T., Bakr, M.E. et al. Development of the generalized ridge estimator for the Poisson-Inverse Gaussian regression model with multicollinearity. Sci Rep 15, 31162 (2025). https://doi.org/10.1038/s41598-025-15334-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-15334-9