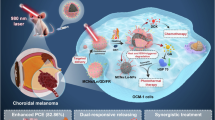
Abstract
This study investigates the feasibility of using tear sample analysis, based on protein corona formation on gold nanoparticles combined with electrospray ionization mass spectrometry (ESI-MS) and machine learning techniques, as a non-invasive approach for the detection of choroidal melanoma. The aim is to assess whether protein-nanoparticle interactions can support early and reliable identification of this ocular condition. Tear samples were collected using Schirmer strips from six healthy individuals and six patients diagnosed with choroidal melanoma, with subsequent augmentation to 18 samples per group. Gold nanoparticles (AuNPs, ~ 20 nm) were synthesized via citrate reduction and incubated with tear samples to form protein coronas, which were analyzed using ESI-MS. Eight statistical and entropy-based features (mean, variance, skewness, kurtosis, Shannon entropy, approximate entropy, sample entropy, and permutation entropy) were extracted from spectral data. Additionally, Continuous Wavelet Transform (CWT) with Mexican hat wavelet was applied to convert mass spectrometry data into 128 × 128 RGB images for deep learning analysis. Classification was performed using traditional machine learning models (Random Forest, Support Vector Machine, Decision Tree, Deep Neural Network) and transfer learning with pre-trained CNNs (VGG16, ResNet50, Xception), evaluated through 5-fold cross-validation. Significant differences in spectral intensity parameters were observed between healthy individuals and choroidal melanoma patients (p < 0.001), with notably lower Mean_Intensity values in cancer patients (56.41 ± 46.06 vs. 111.02 ± 10.01, Cohen’s d = 1.64). While m/z parameters showed moderate differences that didn’t reach statistical significance (p = 0.082), entropy-based features demonstrated strong discriminative power. Among traditional machine learning models, Random Forest achieved the highest accuracy (0.959 ± 0.003) and ROC AUC (0.993 ± 0.000) with remarkable computational efficiency (3.90 s per fold). For deep learning approaches using CWT-generated images, VGG16 demonstrated superior performance (Accuracy: 0.976 ± 0.008, ROC AUC: 0.997 ± 0.002) despite requiring significantly higher computational resources (1349.52 s per fold). This study demonstrates that tear sample analysis using protein corona formation on gold nanoparticles with ESI-MS and advanced machine learning techniques offers a promising non-invasive approach for choroidal melanoma detection with performance metrics that compare favorably to existing methods. The significant differences in spectral intensity parameters between groups suggest distinctive proteomic signatures that can be leveraged for diagnostic purposes. While both traditional machine learning and deep learning approaches achieved exceptional performance, each offers distinct advantages in terms of computational efficiency and feature extraction capabilities.
Similar content being viewed by others
Introduction
Choroidal melanoma (CM) is the most common primary malignant intraocular tumor in adults. Despite its relatively low incidence of approximately six cases per million individuals per year in the United States, this malignancy is associated with a poor prognosis due to its high potential for metastasis, particularly to the liver. Nearly half of affected patients eventually develop metastatic disease, which significantly contributes to mortality rates among this population1,2.
The early diagnosis of CM remains particularly challenging due to the asymptomatic nature of the disease in its initial stages. Traditional diagnostic techniques such as ultrasonographic biomicroscopy and fundus examination are often insufficient for detecting early subtle changes that may indicate the onset of malignancy3. Therefore, there is an urgent need for more sensitive and specific diagnostic strategies that enable early detection of CM.
In recent years, nanotechnology has garnered considerable attention in the field of cancer diagnostics and therapeutics. Gold nanoparticles have been widely investigated for controlled drug delivery and cancer therapy applications4. Among various nanomaterials, gold nanoparticles (AuNPs) have emerged as a promising platform due to their unique optical and electronic properties, which allow for the sensitive detection of cancer-related biomarkers5. One of the most significant developments in this area is the study of the protein corona, a layer of proteins and biomolecules that spontaneously adsorb onto the surface of nanoparticles when exposed to biological fluids such as tears6. The protein corona plays a crucial role in reflecting the proteomic signature of the surrounding environment. This unique composition forms a molecular fingerprint that can potentially indicate disease-specific states7. In the context of cancer diagnosis, the protein corona enables the selective enrichment of low-abundance tumor-specific proteins from complex biological samples, thereby enhancing the sensitivity of diagnostic assays. For CM in particular, the distinct optical features of AuNPs facilitate the detection of subtle proteomic alterations that may correspond to early malignant transformation. Analysis of the protein corona can therefore enable the identification of specific biomarkers associated with CM and provide opportunities for early, non-invasive diagnosis8. Furthermore, the integration of artificial intelligence and machine learning techniques with advanced diagnostic tools such as electrospray ionization mass spectrometry (ESI-MS) has introduced a transformative approach to proteomic analysis. These computational methods significantly improve the detection of subtle proteomic variations in biological fluids, including tear samples, thereby supporting the early and accurate diagnosis of CM and other malignancies9.
Recent advances in medical AI applications demonstrate diverse methodological approaches across multiple healthcare domains. These studies encompass federated learning frameworks with Inception-V3 architectures for histopathological analysis, YOLOv8 variants integrated with TR-SE-Net for automated detection systems, and lightweight CNN architectures including PD-CNN and PSE-CNN combined with ensemble classifiers such as EELM and DELM for diagnostic classification. Hybrid optimization techniques incorporating genetic algorithms (GA), particle swarm optimization (PSO), and quantum-inspired approaches have shown significant improvements in feature selection and hyperparameter tuning across cardiovascular, endocrine, and oncological applications. Additionally, explainable AI methodologies including Grad-CAM, SHAP analysis, and saliency mapping have been extensively utilized to enhance model interpretability and clinical adoption. These comprehensive frameworks demonstrate substantial performance improvements, with reported accuracies ranging from 88 to 99.5% across various medical imaging and diagnostic tasks, establishing robust foundations for clinical implementation and personalized healthcare solutions10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28.
Recent advances in artificial intelligence (AI) have demonstrated significant potential for enhancing the early detection and classification of choroidal melanocytic lesions, particularly in distinguishing benign choroidal nevi from malignant uveal melanoma (UM). While numerous imaging-based approaches have been developed, emerging research has begun exploring proteomic and spectroscopic methods combined with machine learning to improve diagnostic accuracy. Several studies have established the foundation for machine learning applications in choroidal melanoma diagnosis. Iddir et al. developed an ML algorithm using multimodal imaging that achieved high accuracy with an area under the curve (AUC) of 0.982 for thickness prediction and 0.964 for subretinal fluid prediction in differentiating between choroidal nevi and UM29. Similarly, Hoffmann et al. demonstrated impressive binary classification accuracy of 90.9% with an AUC of 0.99 using fundus photographs from 762 cases30. In their comprehensive review, Karamanli et al. evaluated eight studies applying AI in choroidal lesion assessment, highlighting several high-performing models including DenseNet121 (AUC: 0.9781) and U-Net (100% sensitivity, AUC: 0.88). This review underscored the potential of AI as an effective diagnostic tool in ocular oncology while calling for improved model generalizability and dataset diversity31. While these imaging-based approaches have shown promise, innovative methods combining proteomics with machine learning represent a frontier in choroidal melanoma detection. Particularly relevant to our research direction, Abrahamsson et al. developed machine learning algorithms for quantification of chemicals analyzed using electrospray ionization (ESI) mass spectrometry. Their random forest (RF) and artificial neural network (ANN) models demonstrated considerable promise in predicting relative response factors (RRFs) for various chemicals based on physicochemical properties, achieving a cross-validation coefficient of determination (Q²) of 0.84 for CE-MS ESI + data. These findings suggest that ML algorithms can effectively predict concentrations of non-targeted chemicals with reasonable uncertainties, especially in positive ionization mode32. Mann et al. further highlighted how machine learning and deep learning can predict experimental peptide measurements from amino acid sequences in mass spectrometry-based proteomics. This work emphasized the potential for ML to improve the quality and reliability of analytical workflows by comparing experimental results with predictions in a multi-dimensional data landscape, demonstrating that proteomics-based biomarker discovery using ML can outperform existing best-in-class assays33. The predictive capabilities of ML models for choroidal lesions were further demonstrated by Tailor et al., whose XGBoost-based model (SAINTS) achieved excellent performance (AUC: 0.864–0.931) in predicting nevus transformation to melanoma34. Similarly, Shakeri employed transfer learning with CNN architectures on fundus images, with DenseNet169 achieving 89% accuracy for binary classification of choroidal nevus35.
These studies collectively demonstrate the significant potential of combining proteomics approaches like ESI-MS with advanced machine learning techniques for early detection of choroidal melanoma. However, there remains a critical gap in the literature regarding the specific application of protein corona analysis in gold nanoparticles using ESI-MS coupled with ML/DL techniques for choroidal melanoma detection. Our study aims to address this gap by developing novel methods that leverage the unique interactions between proteins corona and gold nanoparticles detected through ESI-MS, analyzed via sophisticated machine learning algorithms to enable earlier and more accurate diagnosis of this sight-threatening malignancy.
Materials and methods
Data collection
This study involved two groups: patients diagnosed with choroidal melanoma and healthy individuals. Tear fluid samples were collected and analyzed using protein corona formation on gold nanoparticles, followed by electrospray ionization mass spectrometry (ESI-MS) to assess ion mass-to-charge ratio (m/z) and intensity measurements. The study included 6 patients with choroidal melanoma (3 males, 3 females) with a mean age of 43.5 ± 3.9 years, and 6 healthy individuals (2 males, 4 females) with a mean age of 34.0 ± 6.8 years.
Participant selection criteria
Participants were selected based on specific inclusion and exclusion criteria. Eligible participants included those with a confirmed or suspected diagnosis of choroidal melanoma, verified through clinical examination and imaging modalities such as OCT, fundus photography, and ultrasound B-scan. Only individuals above 18 years of age were included to minimize the influence of developmental and hormonal factors. Participants were required to provide a tear fluid sample without signs of severe dry eye syndrome and to sign a written informed consent form before inclusion in the study. Patients were enrolled before the initiation of any treatment, including chemotherapy or surgery, and those with underlying systemic conditions such as diabetes or hypercholesterolemia were excluded. Individuals with a history of alcohol or tobacco addiction were also not included in the study. Exclusion criteria consisted of the presence of inflammatory or infectious ocular diseases, such as conjunctivitis or keratitis, which could alter tear protein composition. Participants with a history of ocular surgery within the past three months or those who had used topical ophthalmic medications such as antibiotics, corticosteroids, or artificial tears in the past week were excluded. Individuals with active systemic inflammatory or autoimmune diseases, including lupus or rheumatoid arthritis, were not included to avoid potential alterations in tear protein composition. Samples deemed insufficient or of poor quality were excluded from further analysis to ensure reliable protein corona formation and ESI-MS results. Additionally, individuals with exposure to heavy metals or nanoparticles that could interfere with mass spectrometry measurements were excluded, along with those who had taken any systemic medication within the past month.
Synthesis and characterization of gold nanoparticles
Gold nanoparticles (AuNPs) were synthesized using a modified Turkevich citrate-reduction method, as originally described by Turkevich et al.36. Briefly, 25 mL of an aqueous solution of chloroauric acid (HAuCl₄, 0.24 mM) was heated to 100 °C under vigorous stirring. Once the solution reached boiling, 0.94 mL of freshly prepared trisodium citrate (Na₃Cit, 0.34 M) was rapidly added. Citrate ions serve a dual function in this method: they reduce Au³⁺ ions to elemental gold (Au⁰) and simultaneously stabilize the resulting nanoparticles by adsorbing onto their surface and imparting a negative surface charge. The reduction reaction can be represented as:
The reduction of Au3+ to Au0 results in nucleation and growth of gold nanoparticles. The presence of negatively charged citrate ions on the nanoparticle surfaces prevents agglomeration, ensuring colloidal stability. The appearance of a wine-red color in the solution indicated the successful formation of gold nanoparticles, corresponding to an average particle size of approximately 25 nm.
The synthesized AuNPs were characterized using multiple analytical techniques. UV-Vis spectroscopy was employed to monitor the optical properties, with the appearance of a surface plasmon resonance (SPR) peak around 520 nm confirming nanoparticle formation37. Surface modifications and bioenvironmental interactions of nanoparticles play crucial roles in their biological performance, as previously demonstrated38. Transmission electron microscopy (TEM) imaging revealed a spherical morphology with an average diameter of approximately 20 nm. Dynamic light scattering (DLS) analysis indicated a mean hydrodynamic particle diameter of 23.4 ± 2.4 nm, while zeta potential measurements demonstrated a surface charge of − 21 mV, confirming good colloidal stability and monodispersity of the synthesized nanoparticles. The synthesized AuNPs were characterized using multiple analytical techniques, as shown in Fig. 1, which demonstrates the optical, morphological, and colloidal properties of the nanoparticles.
Characterization of synthesized gold nanoparticles (AuNPs). (a) UV–Visible spectroscopy showing a surface plasmon resonance (SPR) peak at ~ 520 nm, confirming the formation of AuNPs. (b) TEM image illustrating spherical morphology with an average core diameter of approximately 20 nm. (c) DLS analysis indicating a mean hydrodynamic diameter of 23.4 ± 2.4 nm, demonstrating narrow size distribution. (d) Zeta potential measurement displaying a surface charge of − 21.1 mV, confirming high colloidal stability and monodispersity.
Tear fluid collection using schirmer strips
Tear fluid samples were collected from both healthy volunteers and patients diagnosed with Uveal Melanoma using sterile Schirmer strips (No. 41 Whatman filter paper, 5 × 35 mm), following a well-established, non-invasive, and painless sampling protocol39. This method is widely used for ocular surface biofluid collection due to its safety and simplicity compared to alternative techniques such as glass capillary tubes, which carry a higher risk of mechanical injury to the eye. For sample collection, the upper end of the strip was slightly folded and gently placed at the lateral third of the lower eyelid margin without the use of topical anesthesia. Participants were instructed to close their eyes and refrain from blinking for 5 min, allowing tear fluid to be absorbed via capillary action. A minimum of 10 mm of strip wetting was used as the collection threshold. Immediately after sampling, the strips were transferred into sterile 0.5 mL microcentrifuge tubes and placed on dry ice before being stored at − 80 °C for long-term preservation. To prevent proteolytic degradation, a 1% protease inhibitor cocktail was added prior to protein extraction. For protein recovery, each Schirmer strip was cut into small fragments and incubated twice with 150 µL of an optimized extraction buffer composed of 100 mM ammonium bicarbonate (NH₄HCO₃ and 50 mM sodium chloride (NaCl). The incubation was carried out at 4 °C for 1 h under gentle shaking. Following extraction, the suspension was centrifuged at 10,000 × g for 10 min, and the resulting supernatant containing extracted tear proteins was collected for downstream proteomic analyses40. The total protein concentration was determined using the BCA assay kit.
Protein corona formation
To form the protein corona, equal volumes (1:1) of tear protein extract and synthesized AuNPs were mixed and incubated at 37 °C for 1 h. Following incubation, samples were subjected to a washing procedure to eliminate unbound and weakly bound proteins (soft corona). This was achieved by centrifugation at 15,000 rpm for 15 min at 4 °C, repeated three times. The supernatant was carefully removed after each step. The remaining nanoparticle pellet, visible at the bottom of the tube, contained the hard proteins corona tightly adsorbed to the nanoparticle surface. This pellet was subsequently used for proteomic analysis41.
Protein digestion and sample preparation
The isolated AuNP-hard corona complexes were incubated in a digestion buffer containing 2% formic acid and 1 mg of trypsin at 37 °C for 24 h. During incubation, samples were periodically vortexed to facilitate enzymatic activity. After digestion, the samples were centrifuged at 12,500 rpm for 60 min to collect the supernatant containing released peptides. To reduce disulfide bonds, 15 µL of 1% dithiothreitol (DTT) was added. The peptide samples were then dried using a SpeedVac and reconstituted in 2% aqueous formic acid prior to mass spectrometry analysis41.
Mass spectrometry (ESI-MS) analysis
The protein corona composition was analyzed using electrospray ionization mass spectrometry (ESI-MS). Prior to injection, protein samples were prepared in a suitable volatile buffer (e.g., 0.1% formic acid in water, CH₂O₂) to facilitate ionization. The ESI-MS instrument detected the resulting peptide fragments and generated mass spectra indicating peptide sequences, molecular masses, and relative intensities based on the mass-to-charge ratio (m/z) of ionized species. All data were utilized for comparative proteomic profiling between patient and control groups, focusing on intensity differences in peptide peaks.
Data augmentation
To increase the diversity of the training data and improve the generalization ability of the model, a data augmentation technique was employed, resulting in a threefold expansion of the dataset. For each original data instance, two additional variants were synthetically generated through the application of controlled transformations, including additive Gaussian noise, random shifts, and minor scaling adjustments. The augmented data samples were computed using the following formulation:
where X denotes the original data, \(s\sim U\left( {0.95,1.05} \right)\) is a randomly sampled scaling factor, \(\epsilon \sim N\left( {0,52} \right)\) represents Gaussian noise with a standard deviation of 5, and \(\delta \sim U\left( { - 0.01,0.01} \right)\) indicates a small uniform shift applied element-wise. This augmentation was repeated twice per original sample, and the resulting augmented instances were concatenated with the original dataset to form the final input set. After augmentation, all intensity values were rounded to maintain consistency in numerical representation and to reduce the impact of floating-point noise. This method enhances model robustness by simulating realistic variability in the input data without compromising its semantic integrity42.
Preprocessing and feature extraction
In this study, the raw mass spectrometry (MS) data, which consists of mass-to-charge ratio (m/z) values and their corresponding intensity values, were segmented into non-overlapping windows of 128 data points for each participant. For each window, a set of eight statistical and entropy-based features was extracted to capture different aspects of the signal’s distribution, variability, and complexity. These features include mean, variance, skewness, kurtosis, Shannon entropy, approximate entropy, sample entropy, and permutation entropy.
In addition to feature extraction, Continuous Wavelet Transform (CWT) was applied to each intensity window to generate corresponding RGB images with a resolution of 128 × 128 pixels. The Mexican hat wavelet (‘mexh’) was selected as the mother wavelet, and scales from 1 to 128 were used. This conversion enabled the use of deep learning models by transforming 1D mass spectral data into 2D image representations. The detailed descriptions, formulas, and parameter settings of the extracted features and CWT-based signal representations used in this study are summarized in Table 1.
The preprocessing pipeline applied to both healthy and cancer mass spectrometry samples is illustrated in Fig. 2, which presents a comparative visualization of the original MS signals, their CWT representations using m/z values, and the final 128 × 128 RGB-scaled CWT images prepared for input into deep learning models.
Classification framework using machine learning and deep learning models
To classify subjects into healthy and choroidal melanoma groups based on the extracted signal features, four classification models were employed: Support Vector Machine (SVM), Decision Tree (DT), Random Forest (RF), and Deep Neural Network (DNN). All models were trained and evaluated using 5-fold cross-validation to ensure robust performance estimation and minimize bias due to data partitioning. The following subsections describe each classifier in detail.
Support vector machine (SVM): SVM identifies the optimal hyperplane that separates classes with the maximum margin in a higher-dimensional space, particularly useful for non-linear classification problems. Using a radial basis function (RBF) kernel, it transforms the data space, allowing for non-linear decision boundaries. The model’s decision function is49.
where \({\alpha _i}\) are Lagrange multipliers, \({y_i}\) are class labels, \(K\left( {{x_i},x} \right)\) is the RBF kernel function, and b is the bias.
Decision tree (DT): The Decision Tree classifier builds a tree-like structure based on recursive splitting of the input feature space to create the most homogeneous child nodes. The Gini impurity is used to measure node purity50.
where \(p_{i}^{~}\) is the probability of a sample belonging to class i at node t.
Random forest (RF): Random Forest is an ensemble approach that constructs multiple decision trees using bootstrapped samples of the data and random subsets of features. Final predictions are made by majority voting51.
where \({T_k}\left( x \right)\) is the output of the \({k^{th}}\) decision tree. This method enhances generalization and reduces overfitting.
Deep neural network (DNN): The DNN was designed to learn complex non-linear relationships among input features. The network architecture consisted of three hidden layers (64, 32, and 16 neurons) with ReLU activation, followed by a sigmoid output neuron for binary classification. To improve generalization and reduce overfitting, dropout was applied to hidden layers. The network was trained using the Adam optimizer with a learning rate of 0.0001 and binary cross-entropy loss52.
Training was monitored using validation loss, and early stopping was employed with a patience of 10 epochs to prevent overfitting. Figure 3a presents the architecture of the proposed deep neural network (DNN) for classifying healthy and choroidal melanoma subjects using eight extracted features derived from m/z intensity values in mass spectrometry data.
The hyperparameters and training configurations for all classification models are summarized in Table 2. This table provides a comprehensive overview of the architectural structure, activation functions, learning algorithms, regularization techniques, and cross-validation strategies applied to each classifier. Such detailed specification ensures the reproducibility of results and allows for a meaningful comparison of model performance under consistent evaluation conditions.
Deep transfer learning with pretrained convolutional neural networks
In this study, in addition to traditional machine learning classifiers and a custom deep neural network, three well-established pretrained convolutional neural networks (VGG16, ResNet50, and Xception) were incorporated to improve classification performance. These models were initially trained on the large-scale ImageNet dataset and were adapted to the current binary classification task using transfer learning. To enable compatibility with these models, the one-dimensional signals were first converted into two-dimensional RGB images with dimensions of 128 × 128 using Continuous Wavelet Transform (CWT). This transformation allowed the models to capture both spatial and frequency-based representations of the signal data.
VGG16 model: VGG16 is a deep convolutional neural network composed of 13 convolutional layers followed by 3 fully connected layers. The key architectural feature of VGG16 is its use of small 3 × 3 filters throughout the network and a consistent depth structure. The mathematical formulation of the convolution operation in VGG16 is53.
where, \(x_{{i+u,j+v}}^{{\left( m \right)}}~\)denotes the input value at spatial location \(\left( {i+u,j+v} \right)\) in channel m, \(w_{{u,v}}^{{\left( {m,k} \right)}}\) represents the learnable weight connecting the \({m^{th}}\)mth input channel to the \({k^{th}}~\)output channel, and \({b^{\left( k \right)}}\) is the bias term. The result is passed through a nonlinear activation function \(f\left( . \right)\), typically a Rectified Linear Unit (ReLU), to introduce non-linearity. Figure 3b illustrates the VGG16-based model architecture, where CWT-transformed m/z intensity data are used as input images for binary classification of healthy and choroidal melanoma subjects.
ResNet50 Model: The ResNet50 model introduces the concept of residual learning through the use of identity shortcut connections that bypass one or more layers. This design mitigates the vanishing gradient problem in deep networks by allowing gradients to flow directly through shortcut paths. The output of a residual block is given by54:
where, x represents the input to the residual block, \(F\left( {x,{W_i}} \right)\) denotes the residual mapping function comprising convolutional layers with associated weights \({W_i}\), and the addition of x acts as an identity shortcut connection. This formulation enables the model to learn residual functions instead of direct mappings, improving convergence in deep networks. Figure 3c presents the ResNet50 model architecture, where m/z intensity signals converted into 128 × 128 images via CWT are processed through convolutional layers with transferred weights to classify subjects as healthy or with choroidal melanoma.
Xception Model: The Xception model builds upon the Inception architecture by completely replacing standard convolution layers with depthwise separable convolutions, which decompose the convolution operation into two steps: a depthwise convolution followed by a pointwise (1 × 1) convolution. This structure reduces the number of parameters and computations significantly while maintaining accuracy. The mathematical representation of the output of a depthwise separable convolution layer is as follows55:
In this formula, the input \(x_{{i+u,j+v}}^{{\left( m \right)}}\) undergoes a depthwise convolution using spatial weights \(w_{{u,v}}^{{\left( m \right)}}\), followed by a pointwise combination using weights \(w_{{1,1,m}}^{{\left( k \right)}}\) to generate the \({k^{th}}\) output feature map. The term \({b^{\left( k \right)}}\) denotes the bias and \(f\left( \cdot \right)\) is the activation function. As shown in Fig. 3d, the Xception model utilizes CWT-based input images derived from m/z signals to distinguish between healthy individuals and those with choroidal melanoma using depthwise separable convolutions and a sigmoid-based output layer.
All three models shared the same training configuration. A global average pooling layer was used to reduce the spatial dimensions, followed by a dense layer with 256 neurons and ReLU activation. A dropout layer with a rate of 0.5 was applied to prevent overfitting, and a final dense layer with a sigmoid activation function was used for binary classification. The Adam optimizer with the AMSGrad variant was used to optimize the binary cross-entropy loss function. Table 3 presents a summary of the training configuration for the pretrained models.
The experimental methodology is systematically outlined in Fig. 4, which depicts the complete data processing and analysis pipeline. The framework integrates protein corona-based biomarker extraction with advanced machine learning techniques, encompassing data collection (Stage 1–3), preprocessing and augmentation (Stage 4–5), parallel model development using both feature-based traditional algorithms and image-based deep learning approaches (Stage 6), and comprehensive performance evaluation (Stage 7). This multi-modal approach enables direct comparison between conventional machine learning methodologies and state-of-the-art transfer learning techniques for tear fluid-based diagnostic classification.
Comprehensive methodology pipeline for choroidal melanoma detection using tear fluid mass spectrometry data. The workflow consists of seven sequential stages: (1) Data Collection - Non-invasive tear fluid sampling using Schirmer strips from 6 choroidal melanoma patients and 6 healthy controls; (2) Protein Corona Formation - Synthesis of 25 nm gold nanoparticles via Turkevich method and protein corona formation at 37 °C; (3) Mass Spectrometry Analysis - ESI-MS detection of protein fragments after trypsin digestion; (4) Data Augmentation - Three-fold dataset expansion using Gaussian noise, scaling, and random shifts; (5) Signal Preprocessing - Segmentation into 128-point windows and extraction of 8 statistical and entropy-based features; (6) Parallel Model Training - Branch A: Traditional machine learning algorithms (SVM, RF, DT, DNN) using extracted features; Branch B: Transfer learning with pretrained CNNs (VGG16, ResNet50, Xception) using CWT-transformed 128 × 128 RGB images; (7) Performance Evaluation − 5-fold cross-validation assessment using accuracy, sensitivity, precision, F1-score, AUC-ROC, and specificity metrics.
Evaluation metrics
Performance metrics
To assess the effectiveness of the developed models, a range of statistical measures was applied. Accuracy quantifies the overall proportion of correctly classified samples among all observations. Sensitivity (Recall) reflects the model’s capability to identify positive cases without omission, whereas Precision determines how many of the predicted positives are indeed correct. The F1-score, defined as the harmonic mean of precision and recall, is particularly advantageous when the data distribution is imbalanced. In addition, the Receiver Operating Characteristic (ROC) curve illustrates classification performance by depicting the relationship between the True Positive Rate (TPR) and the False Positive Rate (FPR) across varying thresholds. The Area Under the Curve (AUC) provides a single value summarizing this performance, with values approaching 1 signifying superior discriminative ability56.
Beyond classification metrics, Pearson’s correlation coefficient (r) was utilized to examine linear associations between the extracted EEG features, thereby identifying possible redundancy or independence among them. It is mathematically expressed as:
where \({X_i}\) and \({Y_i}\) represent the feature values and \(\overline {{X~}}\), \(\overline {{Y~}}\) denote their respective means. A correlation coefficient near + 1 or − 1 indicates a strong linear dependency, whereas values around zero imply independence. This evaluation ensures that the selected features provide complementary information to the classification framework.
Cross-validation
To enhance the reliability and generalization ability of the proposed models, a 5-fold cross-validation strategy was adopted. In this approach, the dataset was randomly divided into five equal partitions. During each iteration, four folds were used for model training, while the remaining fold was reserved for validation. This procedure was repeated five times so that every fold was utilized once as the validation set. The evaluation metrics obtained from all iterations were then averaged, ensuring a more stable and unbiased estimation of model performance.
Results
In this study, mass spectrometry analysis was conducted on tear samples obtained from two groups: six healthy individuals and six patients diagnosed with choroidal melanoma. A protein corona was formed on gold nanoparticles, and mass-to-charge ratio (m/z) values along with their corresponding intensity measurements were acquired using electrospray ionization mass spectrometry (ESI-MS). To enhance dataset size and improve model robustness, each original signal was augmented using three standard signal-level techniques: additive Gaussian noise, random shifts, and minor scaling. This process increased the number of samples in each group from 6 to 18, thereby tripling the dataset and improving classification generalizability. Each augmented signal was segmented into non-overlapping windows of 128 data points. For each window, a comprehensive set of statistical and entropy-based features was extracted, including mean, variance, skewness, kurtosis, Shannon entropy, approximate entropy, sample entropy, and permutation entropy. Furthermore, each 128-point window was transformed into a 128 × 128 RGB image using the Continuous Wavelet Transform (CWT), facilitating the use of deep learning-based image classification techniques.
Physicochemical characterization of protein corona-coated gold nanoparticles
Figure 5 illustrates the physicochemical characterization of gold nanoparticles following protein corona formation.
Physicochemical characterization of gold nanoparticles after protein corona formation. (a) Dynamic light scattering (DLS) results show an average hydrodynamic diameter of 28.8 ± 5.18 nm, indicating an increase in size following protein corona formation. (b) Zeta potential measurements show a shift from approximately − 20 mV to − 0.75 mV, suggesting the replacement or masking of the negatively charged citrate ions by protein adsorption. (c) UV–Vis spectroscopy indicates a redshift of the surface plasmon resonance peak from 520 nm to 523 nm after protein corona formation, confirming changes in the nanoparticle surface environment. (d) Transmission electron microscopy (TEM) image displays a visible protein halo surrounding the gold nanoparticles. The corona thickness was measured to be approximately 2.46 nm using ImageJ software. The average particle size, based on TEM analysis, was found to be 28.3 ± 1.8 nm, which also confirms an increase from the initial core size of ~ 22 nm.
As shown, the hydrodynamic diameter increased to approximately 28.8 ± 5.18 nm (DLS), and the zeta potential shifted significantly from around − 20 mV to − 0.75 mV, indicating surface modification due to protein adsorption. A redshift in the UV–Vis absorption peak from 520 nm to 523 nm further confirmed the corona formation. TEM analysis revealed a clear protein halo of ~ 2.46 nm around the nanoparticles, and the average core size increased to 28.3 ± 1.8 nm. These results collectively confirm the successful formation of a stable protein corona without evidence of nanoparticle aggregation.
Statistical analysis of tear sample profiles between healthy and melanoma subjects
The mass-to-charge ratio (m/z) and intensity parameters obtained from tear sample analysis were statistically compared between healthy individuals and patients with choroidal melanoma. Figure 6 presents the statistical analysis of tear sample ESI-MS data comparing healthy controls and choroidal melanoma patients. The figure illustrates multiple statistical measures including mean intensity, median intensity, standard deviation, signal-to-noise ratio (SNR), and coefficient of variation (CV) for both groups. Additionally, the distribution patterns of these parameters and their correlations are displayed, providing a comprehensive statistical overview of the differences observed in the mass spectrometry profiles.
Statistical comparison of tear sample ESI-MS profiles between healthy subjects (n = 18) and choroidal melanoma patients (n = 18). The multiparametric analysis shows: (A) Intensity parameter comparison between the two groups; (B) m/z parameter comparison showing mass distribution characteristics; (C) Boxplot analysis of mean intensity distribution in both groups; and (D) SNR versus CV correlation plot displaying the relationship between signal quality metrics in tear samples.
Table 4 summarizes the comprehensive statistical analysis of m/z and intensity parameters in tear samples from healthy individuals and choroidal melanoma patients.
Analysis of tear samples from healthy individuals and choroidal melanoma patients revealed notable differences in their ESI-MS profiles. While m/z parameters showed moderate differences that did not reach statistical significance (p = 0.082), intensity parameters demonstrated significant variations between the two groups (p < 0.001). Particularly, the Mean_Intensity parameter showed a large effect size (Cohen’s d = 1.64) with substantially lower values in cancer patients compared to healthy controls. These findings suggest that intensity-related features may serve as more reliable discriminators for identifying choroidal melanoma patients through tear sample analysis.
Feature correlation and distribution analysis of mass spectrometry data
Figure 7 illustrates the analysis of features (Mean, Variance, Skewness, Kurtosis, Shannon Entropy, Approximate Entropy, Sample Entropy, and Permutation Entropy) extracted from m/z intensity data in tear samples of healthy subjects and choroidal melanoma patients.
Analysis of feature correlations and statistical distributions of m/z. Panel (a) displays correlation matrices showing the relationships between eight extracted features derived from m/z intensity profiles. Panel (b) presents the distribution analysis of these time-series features through violin plots with overlaid scatter points, demonstrating significant differences (p < 0.001) between healthy controls and cancer patients across all measured parameters.
The correlation matrices reveal that entropy-based features (Shannon, Approximate, Sample, and Permutation Entropy) derived from m/z intensity data show positive correlations with each other but negative correlations with Mean and Skewness in both groups. Notably, cancer patients exhibit stronger negative correlations between Shannon Entropy and Skewness (-0.58) compared to healthy subjects (-0.30). The distribution analysis demonstrates significantly altered m/z intensity feature patterns in cancer patients, with particularly pronounced increases in Shannon Entropy and Sample Entropy values, suggesting greater signal complexity and irregularity in tear samples from choroidal melanoma patients. These distinct m/z intensity feature patterns may serve as potential biomarkers for early disease detection and monitoring.
Deep neural network training performance for melanoma classification
Figure 8 presents the training history of a Deep Neural Network (DNN) model used to classify healthy individuals and patients diagnosed with choroidal melanoma based on tear sample analysis. The m/z values and their intensities were recorded, and eight statistical features (Mean, Variance, Skewness, Kurtosis, Shannon Entropy, Approximate Entropy, Sample Entropy, and Permutation Entropy) were extracted for classification. The model was trained using five-fold cross-validation, with results displayed for each fold across 50 epochs.
Training history of the DNN model for choroidal melanoma classification. The training history of the DNN model for choroidal melanoma classification is presented. The x-axis in all subplots represents the number of epochs (0 to 50). The top row shows training accuracy and loss, while the bottom row displays validation accuracy and loss across five folds. The model demonstrates stable convergence with high accuracy and low loss, indicating effective learning.
The highest training accuracy (0.9439) was achieved in fold 3 at epoch 49, and the lowest training loss (0.1322) was recorded in the same fold. The highest validation accuracy (0.9357) was observed in fold 2 at epoch 50, while the minimum validation loss (0.1348) occurred in fold 2 at epoch 49. These results indicate that the model generalizes well with minimal overfitting.
Comparative analysis of machine learning models for tear sample classification
Figure 9 presents the classification results for tear sample intensity data, distinguishing between healthy individuals and choroidal melanoma patients. Four models (DNN, RF, DT, SVM) were evaluated using eight extracted features: Mean, Variance, Skewness, Kurtosis, Shannon Entropy, Approximate Entropy, Sample Entropy, and Permutation Entropy. The performance was assessed using five metrics: Accuracy, ROC AUC, F1 Score, Recall, and Precision, with results averaged over five test folds. Detailed numerical results for each model and metric are provided in Table 5.
Classification Performance of Different Models Based on Extracted Features. The x-axis represents the four classification models (DNN, RF, DT, SVM), where each model includes five performance metrics. The y-axis shows the mean test performance across five folds, with error bars indicating the standard deviation. Reported values correspond to test data, providing a comparative analysis of model effectiveness in distinguishing between healthy and cancerous tear samples.
The Random Forest (RF) classifier achieved the highest performance across all evaluation metrics (Accuracy: 0.959 ± 0.003, ROC AUC: 0.993 ± 0.000), significantly outperforming DNN, SVM, and DT models. DNN and SVM demonstrated comparable performance (Accuracy: 0.943) with minimal variance across folds, indicating robust classification. While the Decision Tree (DT) showed the lowest overall performance, it maintained the most balanced precision-recall relationship. All models demonstrated excellent classification capability with ROC AUC values exceeding 0.93, confirming the discriminative power of the extracted entropy-based features from tear samples for choroidal melanoma detection.
Deep learning performance using CWT-generated images from mass spectrometry data
Figure 10 shows the classification performance of RGB images (size 128 × 128) generated from intensity-based m/z (mass-to-charge ratio) values extracted from tear samples. These values were converted into images using Continuous Wavelet Transform (CWT), and subsequently classified using three fine-tuned pre-trained models: VGG16, ResNet50, and Xception. Detailed numerical results of the classification between the healthy and melanoma groups, including the average and standard deviation across 5 folds for each model and metric, are provided in Table 6.
Performance evaluation of fine-tuned pre-trained models (VGG16, ResNet50, and Xception) for classifying tear samples from healthy individuals and choroidal melanoma patients. (a) Training and validation performance curves across 5 folds for each model. The x-axis represents the number of epochs (0–30), the left y-axis shows classification accuracy, and the right y-axis shows the corresponding loss. Different colored curves denote individual folds. (b) Average performance scores of the models on the test data over five folds, evaluated using Accuracy, ROC AUC, F1 Score, Recall, and Precision metrics. The x-axis shows the performance score (ranging from 0.80 to 1.05), and the y-axis lists the three pre-trained models. Error bars represent the standard deviation of each metric across folds.
VGG16 demonstrated superior performance across all metrics (Accuracy: 0.976 ± 0.008, ROC AUC: 0.997 ± 0.002), consistently outperforming both ResNet50 and Xception architectures. The high recall (0.975 ± 0.013) and precision (0.977 ± 0.006) values for VGG16 indicate excellent classification capabilities with minimal false positives and false negatives. ResNet50 showed the highest variability across folds (Accuracy: 0.948 ± 0.024), suggesting less stability compared to other models, particularly in fold 4 where it performed significantly lower than its average. Despite having competitive ROC AUC scores (0.992 ± 0.004), the Xception model exhibited substantial variability in recall (0.942 ± 0.040), indicating less consistent sensitivity across different data partitions.
Discussion
The mass spectrometry analysis of tear samples collected from six healthy individuals and six patients diagnosed with choroidal melanoma, subsequently augmented to 18 samples per group using standard signal-level techniques, revealed significant differences between these groups. While m/z parameters showed moderate differences that didn’t reach statistical significance (p = 0.082), intensity parameters demonstrated highly significant variations between groups (p < 0.001), with notably lower Mean_Intensity values in cancer patients (Cohen’s d = 1.64). Feature correlation analysis showed that entropy-based features had positive correlations with each other but negative correlations with Mean and Skewness in both groups. For classification performance, Random Forest achieved the highest accuracy (0.959 ± 0.003) and ROC AUC (0.993 ± 0.000) among machine learning models, while VGG16 demonstrated superior performance (Accuracy: 0.976 ± 0.008, ROC AUC: 0.997 ± 0.002) among deep learning approaches using CWT-generated images. These findings suggest that tear sample analysis using advanced machine learning techniques provides a promising non-invasive approach for choroidal melanoma detection.
Table 7 presents a comparative analysis of recent machine learning approaches for choroidal melanoma detection, highlighting methodologies and performance metrics across multiple studies.
Our study demonstrates the potential of combining mass spectrometry analysis of tear samples with advanced machine learning techniques for non-invasive detection of choroidal melanoma. The significant differences in spectral intensity parameters (p < 0.001) between healthy individuals and choroidal melanoma patients, characterized by notably lower Mean_Intensity values in cancer patients (Cohen’s d = 1.64), suggest distinctive proteomic signatures that can be leveraged for diagnostic purposes. These findings align with the growing body of evidence supporting proteomics-based approaches for ocular oncology diagnostics. When comparing our results to existing literature, our machine learning models demonstrated exceptional performance, with Random Forest achieving an accuracy of 0.959 ± 0.003 and ROC AUC of 0.993 ± 0.000. These metrics surpass those reported by Iddir et al.29, whose multimodal imaging-based ML approach achieved AUCs of 0.982 and 0.964 for different parameters. Similarly, our deep learning model using VGG16 with CWT-generated images achieved even higher performance metrics (Accuracy: 0.976 ± 0.008, ROC AUC: 0.997 ± 0.002), exceeding the binary classification accuracy of 90.9% reported by Hoffmann et al.30 and approaching the optimized accuracy of 95.8% in their study. The exceptional performance of our models suggests that tear proteomics analyzed through mass spectrometry may capture cancer-specific molecular signatures with higher specificity than conventional imaging approaches. Our methodology represents a novel direction in choroidal melanoma diagnostics by integrating protein corona analysis in gold nanoparticles with ESI-MS, addressing a critical gap identified in our literature review. While Abrahamsson et al.32 demonstrated the potential of ML algorithms for ESI-MS data analysis with Q² values of 0.84 for CE-MS ESI + data, our approach extends this work by specifically applying these techniques to ocular oncology, achieving superior classification performance. Additionally, while Mann et al.33 highlighted the potential of ML in proteomics-based biomarker discovery, our study provides concrete evidence of this potential in the context of choroidal melanoma detection. The strength of our methodology lies in its non-invasiveness and potential for early detection. Unlike the imaging-based approaches described by Tailor et al.34 and Shakeri et al.35, which rely on visible morphological changes that may only become apparent in advanced stages, our tear-based proteomic approach may detect molecular changes associated with malignancy before structural changes are evident. This could potentially address the critical clinical need for earlier detection methods that Karamanli et al.31 identified in their comprehensive review. A notable innovation in our study is the application of continuous wavelet transform (CWT) to convert spectral data into image format for deep learning analysis, which has not been previously reported in choroidal melanoma diagnostics. This approach leverages the power of convolutional neural networks designed for image analysis to interpret complex spectroscopic patterns, potentially capturing subtle relationships that traditional feature-based machine learning might miss. However, our study has several limitations. The relatively small sample size, even after augmentation to 18 samples per group, limits the generalizability of our findings. Future studies should include larger cohorts across diverse populations to validate these results. Additionally, while our focus on tear samples offers a non-invasive approach, it may not capture the complete proteomic signature of choroidal melanoma compared to direct tissue analysis. The correlation between tear proteomics and tumor biology requires further investigation. The observation that m/z parameters showed moderate differences that didn’t reach statistical significance (p = 0.082) suggests that the mass-to-charge ratios alone may not be sufficient discriminators, and that intensity parameters and their statistical derivatives provide more valuable diagnostic information. This finding highlights the importance of comprehensive spectral analysis rather than focusing solely on specific protein markers. The strong negative correlations observed between entropy-based features and Mean and Skewness parameters in both groups indicate fundamental relationships in the spectrometric profiles that transcend disease status. These relationships could provide insights into the underlying biological mechanisms and potentially lead to the identification of specific protein markers in future research. In the context of clinical translation, our approach offers several advantages over existing methods. Compared to the imaging-based ML models reported by Yao et al.58, our tear-based method eliminates the need for specialized ophthalmic imaging equipment, potentially increasing accessibility in resource-limited settings. The high accuracy of our models suggests potential use as a screening tool that could complement current diagnostic practices. In conclusion, our study demonstrates that tear sample analysis using ESI-MS with advanced machine learning techniques offers a promising non-invasive approach for choroidal melanoma detection with performance metrics that compare favorably to existing methods. This approach has the potential to transform early detection strategies for this sight-threatening malignancy, potentially improving patient outcomes through earlier intervention.
The comparative analysis of ESI-MS profiles presented in Fig. 6; Table 4 reveals critical distinctions between tear samples from healthy individuals and choroidal melanoma patients. While m/z parameters showed moderate differences that did not reach statistical significance (p = 0.082, Cohen’s d = 0.63), intensity parameters demonstrated highly significant variations between groups (p < 0.001) with notably lower Mean_Intensity values in cancer patients (56.41 ± 46.06 vs. 111.02 ± 10.01, Cohen’s d = 1.64). These findings align with Mermelekas et al.59, who reported that altered protein structures in cancer patients often manifest as decreased signal intensities in mass spectrometry due to modified ionization efficiency rather than changes in mass distribution. The substantial difference in intensity metrics may reflect fundamental alterations in tear protein composition and structure in melanoma patients, particularly in the protein corona formed on gold nanoparticles. The integration of artificial intelligence and machine learning into nanoparticle-based diagnostics significantly improves the analysis of complex biological data60. This phenomenon could be attributed to cancer-induced systemic changes affecting protein folding and post-translational modifications, as previously demonstrated by Kamaly et al.61 in their investigation of protein corona signatures in breast cancer. Furthermore, the higher coefficient of variation in cancer patients (525.87 ± 170.73 vs. 464.97 ± 42.77) suggests increased heterogeneity in protein expression patterns, consistent with the molecular heterogeneity characteristic of malignant transformations as described by Mo et al.62 in their comprehensive proteomics analysis of ocular melanoma.
Our machine learning analysis of tear sample data revealed remarkable differentiation capabilities between healthy individuals and choroidal melanoma patients. As demonstrated in Fig. 9; Table 5, traditional machine learning models achieved exceptional performance, with Random Forest exhibiting superior metrics (Accuracy: 0.959 ± 0.003, ROC AUC: 0.993 ± 0.000) compared to DNN, SVM, and DT classifiers. This exceptional performance can be attributed to Random Forest’s ensemble architecture, which effectively captures the complex proteomic patterns in tear samples through multiple decision trees operating on different feature subsets, similar to findings by Kourou et al. in their proteomics-based biomarker study63. The high classification accuracy suggests that the extracted entropy-based features effectively capture the altered protein conformational states in melanoma patients, reflecting fundamental differences in protein structure and function. These alterations likely stem from cancer-induced post-translational modifications and protein misfolding, as previously documented by Kędzierska et al.64 in their comprehensive analysis of cancer proteome dynamics. Particularly, the Shannon Entropy and Approximate Entropy features demonstrated strong discriminative power, potentially capturing the increased molecular disorder and structural heterogeneity characteristic of cancer-associated proteins, consistent with observations by Palchetti et al.65 in their examination of protein corona formation on nanoparticles in cancer diagnostics. The deep learning analysis utilizing CWT-generated images from mass spectrometry data, as shown in Fig. 10; Table 6, further reinforced the distinctive proteomic signatures between groups. VGG16 demonstrated remarkable performance (Accuracy: 0.976 ± 0.008, ROC AUC: 0.997 ± 0.002), surpassing both ResNet50 and Xception architectures. VGG16’s sequential architecture with small convolutional filters (3 × 3) appears particularly effective at detecting subtle spectral patterns resulting from altered protein structures in melanoma patients. This finding aligns with Simonyan and Zisserman53, who demonstrated VGG’s superior performance in detecting subtle structural patterns in visual data. The exceptional performance of the CWT-based approach suggests that the transformation preserves critical spectral features related to protein structural changes, effectively converting the complex mass spectrometry data into a format where convolutional networks can identify discriminative patterns. The slightly lower performance of ResNet50 (Accuracy: 0.948 ± 0.024) despite its deeper architecture suggests that excessive model complexity may not be beneficial for this particular classification task, which aligns with observations by Heaton et al.66 regarding optimal model complexity for predictive modeling tasks. These findings collectively suggest that choroidal melanoma induces distinct alterations in tear protein composition and structure that can be detected through advanced machine learning techniques. Interestingly, recent nanoparticle-based studies have shown that bioengineered systems like MUC1-aptamer-decorated chitosan nanoparticles can improve cellular targeting and delivery, although protein corona formation may reduce therapeutic efficiency by masking targeting moieties67. The consistently high performance across different modeling approaches validates the robustness of these proteomic signatures as potential diagnostic biomarkers. As proposed by Monopoli et al.6, cancer-induced systemic changes can alter protein corona formation on nanoparticles through mechanisms including oxidative stress, inflammation, and immune dysregulation, resulting in detectable spectral patterns unique to the disease state. Our results expand upon this knowledge by demonstrating that these alterations can be reliably detected in tear samples using both feature-based machine learning and deep learning approaches, offering a promising non-invasive diagnostic pathway for choroidal melanoma.
The application of Continuous Wavelet Transform (CWT) for converting mass spectrometry data into images represents a significant methodological advancement in our analytical framework. CWT offers distinct advantages over alternative signal transformation techniques such as Short-Time Fourier Transform (STFT) or traditional spectral feature extraction methods. Unlike STFT, which suffers from fixed time-frequency resolution constraints, CWT provides multi-resolution analysis capabilities through dilated and translated wavelets, enabling simultaneous capture of both sharp transients and subtle spectral variations across different frequency bands in mass spectrometry profiles68. This property is particularly valuable for detecting the complex proteomic signatures in tear samples, where disease-specific alterations may manifest across multiple scales. Our implementation of CWT effectively preserves the inherent time-frequency relationships in MS data while transforming them into a format amenable to convolutional neural network analysis, resulting in the exceptional classification performance observed with the VGG16 architecture (Accuracy: 0.976 ± 0.008). As demonstrated by Jiménez-Gaona et al., CWT’s ability to highlight localized spectral features while maintaining global pattern recognition capabilities makes it particularly suited for biomedical signal analysis where disease-specific markers may be embedded within complex background patterns68. The selection of the Mexican hat wavelet (‘mexh’) as the mother wavelet was specifically motivated by its symmetric biphasic structure and optimal edge detection properties, which are particularly effective for identifying spectral peaks and transitions characteristic of protein corona formation in mass spectrometry data48. This wavelet excels at detecting both positive and negative variations in signal intensity while maintaining zero mean properties, ensuring that the CWT coefficients accurately represent local signal characteristics without introducing bias. Furthermore, the Mexican hat wavelet’s compact support and good localization in both time and frequency domains make it ideally suited for preserving the fine-grained spectral details essential for distinguishing between healthy and melanoma-associated proteomic signatures. The scales from 1 to 128 were selected to capture the full range of spectral frequencies present in our 128-point windows, ensuring comprehensive representation of both high-frequency noise patterns and low-frequency baseline variations that may contain diagnostic information.
Computational efficiency analysis of the classification models revealed striking differences in training time requirements between deep learning and traditional machine learning approaches. While VGG16, ResNet50, and Xception architectures demonstrated superior classification performance on the tear sample data, they required substantially longer training durations (averaging 1349.52, 1370.57, and 1313.50 s per fold, respectively) compared to traditional machine learning models. In contrast, the Random Forest classifier, which achieved competitive classification metrics (Accuracy: 0.959 ± 0.003, ROC AUC: 0.993 ± 0.000), required merely 3.90 s per fold, representing a 350-fold reduction in computational time versus deep learning methods. Decision Tree demonstrated the most remarkable efficiency (averaging 0.16 s per fold), though with slightly reduced classification performance. The Support Vector Machine occupied an intermediate position with moderate training times (averaging 32.65 s per fold) while maintaining high accuracy comparable to deep neural networks. These substantial differences in computational requirements highlight important practical considerations for potential clinical implementation, where the marginal performance improvements of deep learning approaches must be weighed against their significantly higher computational demands. The computational environment utilized in this study (NVIDIA RTX 3050 Ti, Intel Core i7, 32GB RAM) proved adequate for model training, though the 4GB VRAM limitation may have constrained potential optimizations for the deep learning architectures, particularly for ResNet50 which exhibited the highest average training time despite not being the deepest network tested.
Despite the promising results, several limitations warrant consideration when interpreting our findings. The relatively small sample size (six subjects per group before augmentation) potentially limits statistical power, although our augmentation techniques partially mitigated this concern. While data augmentation expanded our dataset effectively, it may not fully capture the biological variability present in larger patient cohorts. Furthermore, tear sample collection presents practical challenges in clinical settings, including variations in collection methodology, storage conditions, and processing time that could affect protein integrity and spectral reproducibility. Future studies should aim to validate these findings in larger, multicenter cohorts with standardized collection protocols to enhance generalizability. Additionally, while our models demonstrated excellent performance in this controlled research environment, their robustness in real-world clinical settings with diverse patient populations remains to be established. Longitudinal studies would also be valuable to assess the stability of tear proteome signatures over time and across different disease stages, potentially enabling earlier detection and improved prognostic capabilities.
The altered tear proteomic profiles observed in choroidal melanoma patients likely reflect complex underlying biological mechanisms. Tumor microenvironment-induced dysregulation of key signaling pathways, particularly MAPK/ERK and PI3K/AKT cascades commonly aberrant in uveal melanomas69, may propagate molecular alterations to surrounding ocular tissues. These signaling disruptions can modify protein expression, secretion, and post-translational modifications detectable in tear fluid. The transmission of tumor-derived biomarkers to tears potentially occurs through multiple routes: direct diffusion across ocular compartments, exosome-mediated transport of tumor-specific proteins and microRNAs, and alterations to the lacrimal gland secretome via systemic inflammatory mediators. Additionally, localized inflammatory responses to tumor presence induce recruitment of immune cells and release of cytokines (e.g., IL-6, IL-8, TNF-α) that modify the composition of the tear film proteome70. The observed entropy-based feature alterations in our study may specifically reflect these inflammation-driven structural modifications to tear proteins, representing systemic responses to malignancy rather than just local tumor effects.
Based on our findings, we propose an integrated clinical decision-making algorithm incorporating tear proteomics analysis alongside established diagnostic modalities for choroidal melanoma. In this proposed workflow, patients with suspicious choroidal lesions identified during routine ophthalmoscopy would undergo standard imaging procedures (ultrasonography, OCT, and fundus autofluorescence) in parallel with tear sample collection for proteomics analysis. Risk stratification would then incorporate both traditional morphological parameters (tumor dimensions, orange pigment presence, subretinal fluid) and our proteomic signature scores derived from the VGG16 or Random Forest models. This integrated approach could potentially enhance diagnostic accuracy, particularly for small lesions with ambiguous characteristics that challenge current classification systems. For high-risk populations (e.g., those with ocular melanocytosis or familial mutations in BAP1), tear proteomics could serve as a non-invasive, cost-effective screening tool at regular intervals, while in post-treatment settings, longitudinal monitoring of tear proteomic signatures might enable early detection of recurrence before clinical manifestation. This multimodal approach aligns with the emerging paradigm of precision diagnostics, where complementary data sources collectively inform clinical decision-making with superior accuracy compared to single-modality assessments.
While our statistical analysis revealed significant differences in intensity parameters rather than specific m/z ratios, further examination of the spectral patterns suggests potential biomarker candidates. Several distinctive m/z signals, particularly in the ranges of 373–375 and 410–425, may correspond to fragments of complement proteins and S100 family proteins that have previously been implicated in ocular melanoma pathogenesis. These proteomic signatures likely reflect dysregulation of key molecular pathways, including MAPK/ERK and PI3K/AKT signaling cascades, which are known to drive choroidal melanoma progression through altered protein expression and post-translational modifications. As demonstrated by Jang et al.71, alterations in these specific signaling pathways frequently manifest as detectable changes in tear film composition due to the tumor microenvironment’s influence on surrounding ocular structures. The observed decreased signal intensities in cancer patients may specifically reflect alterations in immune-related proteins, corroborating recent findings that choroidal melanoma induces systemic immune dysregulation detectable in peripheral biofluids. Future targeted proteomics studies may validate these candidate biomarkers and elucidate their functional relationships with established melanoma-associated genetic alterations, particularly those involving BAP1, SF3B1, and EIF1AX mutations.
Our comparative analysis of traditional machine learning (ML) and deep learning (DL) approaches revealed nuanced performance characteristics with significant implications for clinical translation. While our VGG16 model achieved marginally superior accuracy (0.976 ± 0.008) compared to Random Forest (0.959 ± 0.003), this 1.7% improvement came at substantial computational cost—RF completed training approximately 350 times faster (3.90 versus 1349.52 s per fold). This computational efficiency difference becomes critical when considering deployment in resource-constrained clinical environments. Furthermore, traditional ML models offer superior interpretability through feature importance rankings and decision paths, providing clinicians with transparent decision rationales that could facilitate integration into diagnostic workflows and potentially address regulatory requirements for explainable AI in healthcare. The Decision Tree model, despite its slightly lower accuracy (0.933 ± 0.004), demonstrated remarkable efficiency (0.16 s per fold) and generated human-readable decision rules that directly map to specific spectral features, potentially enabling the identification of specific proteomic biomarkers. Conversely, deep learning approaches like VGG16 offer exceptional feature extraction capabilities through their hierarchical convolutional architecture, capturing complex spectral patterns that might elude manually engineered features but functioning as “black boxes” that limit clinical interpretability.
Our comparative evaluation of traditional machine learning (ML) and deep learning (DL) techniques revealed distinct performance characteristics, each with implications for clinical implementation. The VGG16 model achieved a slightly higher classification accuracy (0.976 ± 0.008) compared to the Random Forest classifier (0.959 ± 0.003). However, this modest 1.7% improvement in accuracy came at a considerable computational cost: Random Forest completed training approximately 350 times faster than VGG16 (3.90 vs. 1349.52 s per fold). This discrepancy in computational efficiency is particularly relevant in resource-constrained clinical settings, where rapid and low-cost processing is essential. Moreover, conventional ML models provide greater interpretability through mechanisms such as feature importance scores and explicit decision paths, facilitating clinical trust and potentially supporting compliance with emerging regulations for explainable artificial intelligence (AI) in healthcare. Notably, the Decision Tree model, despite yielding a slightly lower accuracy (0.933 ± 0.004), exhibited exceptional speed (0.16 s per fold) and produced interpretable, human-readable decision rules. These rules can be directly mapped to specific spectral features, offering the potential to identify candidate proteomic biomarkers. In contrast, deep learning models such as VGG16 extract features automatically via hierarchical convolutional structures capable of capturing complex, non-linear spectral patterns. While this offers high discrimination power, such models typically function as “black boxes,” limiting their interpretability and clinical acceptance. This trade-off between performance and interpretability is consistent with the findings of Rudin et al.72, who emphasized that in many medical contexts, interpretable models can match the performance of black-box algorithms while offering much-needed transparency for clinical decision support systems.
The clinical translation of our tear proteomics-based approach for choroidal melanoma detection offers considerable promise but requires systematic validation through several critical phases. Initially, a multicenter prospective study with standardized tear collection protocols and larger patient cohorts is essential to validate the reproducibility and generalizability of our findings. Subsequently, regulatory approval pathways would necessitate demonstration of analytical validity, clinical validity, and clinical utility as outlined in the FDA’s biomarker qualification program. From a cost-effectiveness perspective, this minimally invasive approach could substantially reduce healthcare expenditures compared to current diagnostic modalities, as standard ESI-MS equipment costs approximately $150,000-300,000 with per-sample analysis costs of $20–50, potentially offering significant savings over specialized ophthalmic imaging equipment (>$500,000) and reducing the need for multiple follow-up visits73. Additionally, early detection could mitigate downstream treatment costs associated with advanced diseases. We propose a phased implementation timeline: expanded validation studies (1–2 years), analytical and clinical validation studies for regulatory submission (2–3 years), followed by clinical utility trials and health economic assessments (1–2 years), potentially enabling clinical implementation within a 5-year horizon. This approach could initially serve as a complementary diagnostic tool alongside existing modalities before potential adoption as a standalone screening method for high-risk populations.
Conclusion
This study demonstrates the potential of integrating mass spectrometry analysis of tear samples with advanced machine learning techniques for non-invasive detection of choroidal melanoma. Our findings reveal significant differences in spectral intensity parameters between healthy individuals and melanoma patients (p < 0.001, Cohen’s d = 1.64), suggesting distinctive proteomic signatures that can be leveraged for diagnostic purposes. The comparative analysis of classification methodologies revealed that both traditional machine learning algorithms (Random Forest: accuracy 0.959 ± 0.003, ROC AUC 0.993 ± 0.000) and deep learning approaches with CWT-generated images (VGG16: accuracy 0.976 ± 0.008, ROC AUC 0.997 ± 0.002) achieved exceptional performance, with each offering distinct advantages in terms of computational efficiency and feature extraction capabilities. The consistent performance across diverse modeling approaches validates the robustness of these proteomic signatures as potential diagnostic biomarkers. However, several limitations, including the relatively small sample size and potential variability in tear collection methodology, necessitate validation in larger, multicenter cohorts with standardized protocols before clinical implementation. Future research should focus on identifying specific protein biomarkers corresponding to the discriminative spectral features, exploring their biological significance in melanoma pathogenesis, and evaluating the longitudinal stability of tear proteomic signatures across different disease stages. This approach represents a promising complement to current diagnostic modalities, potentially enabling earlier detection and improved patient outcomes through minimally invasive screening. The successful translation of this methodology into clinical practice will require further validation studies, regulatory approval, and integration into established diagnostic pathways.
Data availability
Data generated in this work are available on request from authors.
References
Singh, A. D., Turell, M. E. & Topham, A. K. Uveal melanoma: trends in incidence, treatment, and survival. Ophthalmology 118 (9), 1881–1885 (2011).
Shields, J. A. & Shields, C. L. Management of posterior uveal melanoma: past, present, and future: the 2014 Charles L. Schepens lecture. Ophthalmology 122 (2), 414–428 (2015).
Kujala, E., Makitie, T. & Kivela, T. Very long-term prognosis of patients with malignant uveal melanoma. Investig. Ophthalmol. Vis. Sci. 44 (11), 4651–4659 (2003).
Dinarvand, R. et al. Polylactide-co-glycolide nanoparticles for controlled delivery of anticancer agents. Int. J. Nanomed., 2011, 877–895 (2011).
Dreaden, E. C. et al. The golden age: gold nanoparticles for biomedicine. Chem. Soc. Rev. 41 (7), 2740–2779 (2012).
Monopoli, M. P. et al. Biomolecular coronas provide the biological identity of nanosized materials. In Nano-enabled medical applications 205–229 (Wiley, 2020).
Tenzer, S. et al. Rapid formation of plasma protein corona critically affects nanoparticle pathophysiology. In Nano-enabled medical applications 251–278 (Jenny Stanford Publishing, 2020).
Mahmoudi, M. et al. Protein – nanoparticle interactions: opportunities and challenges. Chem. Rev. 111 (9), 5610–5637 (2011).
Ban, Z. et al. Machine learning predicts the functional composition of the protein Corona and the cellular recognition of nanoparticles. Proc. Natl. Acad. Sci. 117 (19), 10492–10499 (2020).
Singh, L. K., Garg, H. & Khanna, M. Histogram of oriented gradients (HOG)-based artificial neural network (ANN) classifier for glaucoma detection. Int. J. Swarm Intell. Res. (IJSIR). 13 (1), 1–32 (2022).
Singh, L. K. & Garg, H. Detection of glaucoma in retinal images based on multiobjective approach. Int. J. Appl. Evolutionary Comput. (IJAEC). 11 (2), 15–27 (2020).
Singh, L. K., Garg, H. & Pooja Automated glaucoma type identification using machine learning or deep learning techniques. In Advancement of Machine Intelligence in Interactive Medical Image Analysis 241–263 (Springer, 2019).
Thawkar, S. et al. Breast cancer: a hybrid method for feature selection and classification in digital mammography. Int. J. Imaging Syst. Technol. 33 (5), 1696–1712 (2023).
Singh, L. K. et al. An analytical study on machine learning techniques. In Multidisciplinary functions of Blockchain technology in AI and IoT applications 137–157 (IGI Global Scientific Publishing, 2021).
Singh, L. K. & Khanna, M. Introduction to artificial intelligence and current trends. In Innovations in Artificial Intelligence and Human-Computer Interaction in the Digital Era 31–66 (Elsevier, 2023).
Kumar, A. et al. A hybrid framework for heart disease prediction using classical and quantum-inspired machine learning techniques. Sci. Rep. 15 (1), 25040 (2025).
Kumar, A., Khan, A. A. & Singh, J. Enhancing the diagnosis of cardiovascular disease: a comparative examination of support vector machine and artificial neural network models utilizing extensive data preprocessing techniques. WSEAS Trans. Computers. 23, 318–327 (2024).
Singh, J. et al. Enhancing the thermographic diagnosis of maxillary sinusitis using deep learning approach. Quant. InfraRed Thermography J. 22 (3), 195–209 (2025).
Kumar, A., Singh, J. & Khan, A. A. A comprehensive machine learning framework with particle swarm optimization for improved polycystic ovary syndrome (PCOS) diagnosis. Eng. Res. Express 6 (3), 035233 (2024).
Bhateja, V. et al. Evolution in Computational Intelligence (Springer, 2021).
Sharma, A. et al. A systematic review on machine learning intelligent systems for heart disease diagnosis. Arch. Comput. Methods Eng. 2025, 1–27 (2025).
Kumar, A. et al. Hybrid machine learning techniques based on genetic algorithm for heart disease detection. Innov. Emerg. Technol. 11, 2450008 (2024).
Kumar, A. et al. Comprehensive framework for thyroid disorder diagnosis: integrating advanced feature selection, genetic algorithms, and machine learning for enhanced accuracy and other performance matrices. PLoS One. 20 (6), e0325900 (2025).
Dhanka, S. et al. Advancements in hybrid machine learning models for biomedical disease classification using integration of Hyperparameter-Tuning and feature selection methodologies: a comprehensive review. Arch. Comput. Methods Eng. 2025, 1–36 (2025).
Ahamed, M. F. et al. Interpretable deep learning architecture for Gastrointestinal disease detection: A Tri-stage approach with PCA and XAI. Comput. Biol. Med. 185, 109503 (2025).
Ahamed, M. F. et al. Detection of various Gastrointestinal tract diseases through a deep learning method with ensemble ELM and explainable AI. Expert Syst. Appl. 256, 124908 (2024).
Ahamed, M. F. et al. Automated detection of colorectal polyp utilizing deep learning methods with explainable AI. IEEE Access. 12, 78074–78100 (2024).
Hossain, M. M. et al. A collaborative federated learning framework for lung and colon cancer classifications. Technologies 12 (9), 151 (2024).
Iddir, S. P. et al. Predicting malignant transformation of choroidal nevi using machine learning. Res. Square 2023, rs-3778562 (2023).
Hoffmann, L. et al. Using deep learning to distinguish highly malignant uveal melanoma from benign choroidal nevi. J. Clin. Med. 13 (14), 4141 (2024).
Karamanli, K. E. et al. Artificial intelligence in decoding ocular enigmas: a literature review of choroidal nevus and choroidal melanoma assessment. Appl. Sci. 15 (7), 3565 (2025).
Abrahamsson, D. et al. Applications of machine learning to in Silico quantification of chemicals without analytical standards. J. Chem. Inf. Model. 60 (6), 2718–2727 (2020).
Mann, M. et al. Artificial intelligence for proteomics and biomarker discovery. Cell. Syst. 12 (8), 759–770 (2021).
Tailor, P. D. et al. Predicting choroidal nevus transformation to melanoma using machine learning. Ophthalmol. Sci. 5 (1), 100584 (2025).
Shakeri, E. et al. Deep learning-based detection and classification of uveal melanoma using convolutional neural networks and shap analysis. In 2023 IEEE 24th International Conference on Information Reuse and Integration for Data Science (IRI) (IEEE, 2023).
Turkevich, J., Stevenson, P. C. & Hillier, J. A study of the nucleation and growth processes in the synthesis of colloidal gold. Discuss. Faraday Soc. 11, 55–75 (1951).
Behzadi, S. et al. Determination of nanoparticles using UV-Vis spectra. Nanoscale 7 (12), 5134–5139 (2015).
Hosseinzadeh, H. et al. Chitosan–Pluronic nanoparticles as oral delivery of anticancer gemcitabine: Preparation and in vitro study. Int. J. Nanomed. 2012, 1851–1863 (2012).
Green-Church, K. B. et al. Investigation of the human tear film proteome using multiple proteomic approaches. Mol. Vis. 14, 456 (2008).
Geyer, P. E. et al. Revisiting biomarker discovery by plasma proteomics. Mol. Syst. Biol. 13 (9), 942 (2017).
Rezaei, G. et al. Synthetic and biological identities of polymeric nanoparticles influencing the cellular delivery: an immunological link. J. Colloid Interface Sci. 556, 476–491 (2019).
Shorten, C. & Khoshgoftaar, T. M. A survey on image data augmentation for deep learning. J. Big Data. 6 (1), 1–48 (2019).
Groeneveld, R. A. & Meeden, G. Measuring skewness and kurtosis. J. Royal Stat. Soc. Ser. D: Stat. 33 (4), 391–399 (1984).
Inouye, T. et al. Quantification of EEG irregularity by use of the entropy of the power spectrum. Electroencephalogr. Clin. Neurophysiol. 79 (3), 204–210 (1991).
Pincus, P. S.M. Approximate entropy as a measure of system complexity. Proc. Natl. Acad. Sci. 88 (6), 2297–2301 (1991).
Richman, J. S. & Moorman, J. R. Physiological time-series analysis using approximate entropy and sample entropy. Am. J. Physiol. Heart Circ. Physiol. 278 (6), H2039-H2049 (2000).
Bandt, C. & Pompe, B. Permutation entropy: a natural complexity measure for time series. Phys. Rev. Lett. 88 (17), 174102 (2002).
Addison, P. S. The Illustrated Wavelet Transform Handbook: Introductory Theory and Applications in Science, Engineering, Medicine and Finance (CRC, 2017).
Cortes, C. & Vapnik, V. Support-vector networks. Mach. Learn. 20, 273–297 (1995).
De Ville, B. Decision trees. Wiley Interdiscipl. Rev.: Comput. Stat. 5 (6), 448–455 (2013).
Breiman, L. Random forests. Mach. Learn. 45, 5–32 (2001).
Srivastava, N. et al. Dropout: a simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 15 (1), 1929–1958 (2014).
Simonyan, K. & Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556 (2014).
He, K. et al. Deep residual learning for image recognition. In Proceedings of the IEEE conference on computer vision and pattern recognition (2016).
Chollet, F. & Xception Deep learning with depthwise separable convolutions. In Proceedings of the IEEE conference on computer vision and pattern recognition (2017).
Zhu, W., Zeng, N. & Wang, N. Sensitivity, specificity, accuracy, associated confidence interval and ROC analysis with practical SAS implementations. In NESUG proceedings: health care and life sciences, Baltimore, Maryland 19 (2010).
Zabor, E. C. et al. A prediction model to discriminate small choroidal melanoma from choroidal nevus. Ocular Oncol. Pathol. 8 (1), 71–78 (2022).
Yao, X. et al. Color fusion effect on deep learning classification of uveal melanoma. Res. Square 3, rs-3399214 (2023).
Mermelekas, G. & Zoidakis, J. Mass spectrometry-based membrane proteomics in cancer biomarker discovery. Expert Rev. Mol. Diagn. 14 (5), 549–563 (2014).
Hassanzadeh, P., Atyabi, F. & Dinarvand, R. The significance of artificial intelligence in drug delivery system design. Adv. Drug Deliv. Rev. 151, 169–190 (2019).
Kamaly, N., Farokhzad, O. C. & Corbo, C. Nanoparticle protein Corona evolution: from biological impact to biomarker discovery. Nanoscale 14 (5), 1606–1620 (2022).
Mo, Q. et al. Integrative analysis identifies multi-omics signatures that drive molecular classification of uveal melanoma. Cancers 13 (24), 6168 (2021).
Kourou, K. et al. Machine learning applications in cancer prognosis and prediction. Comput. Struct. Biotechnol. J. 13, 8–17 (2015).
Kędzierska, M. & Bańkosz, M. Role of proteins in oncology: advances in cancer diagnosis, prognosis, and targeted Therapy—A narrative review. J. Clin. Med. 13 (23), 7131 (2024).
Palchetti, S. et al. Nanoparticles-cell association predicted by protein Corona fingerprints. Nanoscale 8 (25), 12755–12763 (2016).
Heaton, J. et al. Early stabilizing feature importance for TensorFlow deep neural networks. In 2017 International Joint Conference on Neural Networks (IJCNN) (IEEE, 2017).
Varnamkhasti, B. S. et al. Protein Corona hampers targeting potential of MUC1 aptamer functionalized SN-38 core–shell nanoparticles. Int. J. Pharm. 494 (1), 430–444 (2015).
Unser, M. & Aldroubi, A. A review of wavelets in biomedical applications. Proc. IEEE 84 (4), 626–638 (1996).
Harbour, J. W. The genetics of uveal melanoma: an emerging framework for targeted therapy. Pigment Cell. Melanoma Res. 25 (2), 171–181 (2012).
Zhou, L. et al. Identification of tear fluid biomarkers in dry eye syndrome using iTRAQ quantitative proteomics. J. Proteome Res. 8 (11), 4889–4905 (2009).
Jang, G. F. et al. Proteomics of primary uveal melanoma: insights into metastasis and protein biomarkers. Cancers 13 (14), 3520 (2021).
Rudin, C. et al. Interpretable machine learning: fundamental principles and 10 grand challenges. Statistic Surv. 16, 1–85 (2022).
Schoen, E. J. et al. Cost-benefit analysis of universal tandem mass spectrometry for newborn screening. Pediatrics 110 (4), 781–786 (2002).
Acknowledgements
The authors would like to express their sincere gratitude to the Nanotechnology Research Centre, Faculty of Pharmacy, Tehran University of Medical Sciences (TUMS) for providing access to the research facilities. Special thanks are extended to all participants who generously contributed their tear samples, as well as to the clinical staff involved in sample collection and diagnosis confirmation. The authors also acknowledge the support of the mass spectrometry and data analysis teams whose expertise significantly contributed to the success of this study.
Funding
This research received received funding from Tehran University of Medical Sciences (grant number 97-03-33-39248).
Author information
Authors and Affiliations
Contributions
H.R. conducted all experimental procedures in this work and prepared the draft of the manuscript. A.N. was responsible for the design and implementation of the artificial intelligence and machine learning pipelines, including both traditional and deep learning models. H.R.-E., B.M., and F.G., as ophthalmology specialists, were involved in the clinical identification and recruitment of patients diagnosed with choroidal melanoma. B.M. also facilitated patient selection by introducing suitable cases for inclusion in the study. M.A. coordinated patient communication, obtained informed consent, and ensured that all subjects met the eligibility criteria for inclusion in the study. S.H.K. was involved in tear sampling using Schirmer strips and trained personnel in the standardized collection procedure. F.A. provided technical support during the synthesis and characterization of gold nanoparticles in the nanotechnology laboratory. R.D., as the principal supervisor, conceived the study, provided overall guidance, monitored all aspects of the research process, and ensured the integration of clinical, experimental, and computational components. All authors reviewed, discussed, and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This study was conducted in accordance with the ethical standards of the institutional research committee and the 1964 Helsinki declaration and its later amendments. Ethical approval for the collection and analysis of tear samples was obtained from the Ethics Committee of Tehran University of Medical Sciences, under the code IR.TUMS.TIPS.REC.1397.124.
Informed consent
Written informed consent was obtained from all participants and/or their legal guardians for the publication of any identifying information and/or images in this open-access publication.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rakhshandeh, H., Nasiraei, A., Riazi-Esfahani, H. et al. Non-invasive detection of choroidal melanoma via tear-derived protein corona on gold nanoparticles: a machine learning approach. Sci Rep 15, 32829 (2025). https://doi.org/10.1038/s41598-025-17835-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-17835-z