Abstract
The Andes arguably contains the most important biodiversity hot spot (Tropical Andes) in the world and is home to the only extant bear (spectacled bear, Tremarctos ornatus), in South America. Two molecular population genetics studies of the spectacled bear were recently carried out in Ecuador. However, these studies had contradictory results, which could hinder the development of an adequate conservation program for this species. To resolve this issue, we analyzed three molecular data sets representing a broad geographical sampling effort. The sets included six mitochondrial (mt) genes of 127 specimens, complete mitogenomes of 43 specimens, and seven nuclear DNA microsatellites of 88 specimens. The main results were as follows. (1) The mt genetic diversity levels obtained for the six mt gene data set were high and identical to the estimates of the two previous studies regardless of whether specific mt primers for this species were used. (2) The analysis of the six mt genes and mitogenome data revealed genetic heterogeneity among the spectacled bear samples of different Ecuadorian provinces, including significant heterogeneity between the northernmost and southernmost Ecuadorian areas. However, this heterogeneity is relatively small. (3) The genetic diversity levels with heterologous microsatellites were undifferentiable from the genetic levels reported with homologous microsatellites in a previous study. (4) Analysis of the microsatellite data set indicated a more significant spatial structure than previously reported. Up to seven different genetic clusters were detected but mixed in different areas of Ecuador. However, when they were analyzed at the province level only one unique genetic cluster was supported. (5) Based on these data, we suggest a single management plan for the entire spectacled bear population in Ecuador rather than a different management plan for each small spectacled bear population as suggested elsewhere.
Similar content being viewed by others
Introduction
The Andes is the longest mountain range (cordillera) in the world, nearly 8,900 km in length and up to 750 km wide. It has an average height of about 4,000 m above sea level (masl), and crosses parts of Venezuela, Colombia, Ecuador, Peru, Bolivia, Chile and Argentina. It occupies an approximate area of 2,870,000 km2 and it is home to some of the tallest volcanoes in the world including Aconcagua, which reaches 6,962 masl. The Andes has an extraordinary biological diversity. Out of 36 world biodiversity hot spots, the Andean cordillera contains three: The Tumbes-Chocó-Magdalena area in Peru, Ecuador, and Colombia; the Tropical Andes in Venezuela, Colombia, Ecuador, Peru, Bolivia, and some residual areas in northern Argentina and Chile; and the Chilean Winter Rainfall Valdivian Forests. The Tropical Andes (1,258,000 km2) is arguably the most important biodiversity hot spot in the world. It contains 45,000 plant species (about 1/6 of all plant species in the world) of which 20,000 are endemic1. For a relative comparison, the second most biodiverse hot spot has 25,000 plant species. Additionally, the Tropical Andes contains a diverse assemblage of animals (980 amphibian species [670 endemic], 1,700 bird species [900 endemic], and 570 mammal species [around 80–100 endemic])2,3,4.
Among the most emblematic and unique mammal species in the Tropical Andes, and one of the largest predators in South America, is the spectacled or Andean bear (Tremarctos ornatus). This is the only extant bear species in South America. Spectacled bears are short-faced, with an adult weight ranging from 30 to 200 kg and body length from 0.88 to 2.22 m5,6 with adult females, being a third smaller than males7. Cuvier became the first to describe a spectacled bear specimen and suggested the scientific name of Ursus ornatus8. Cuvier had obtained the specimen at a Chilean port. As this species does not inhabit Chile, Cabrera clarified the origin of this specimen, as coming from the Libertad Department, near the city of Trujillo, in Perú9. Gervais created the genus Tremarctos, where the spectacled bear is currently included10. Gill was the first to use the current name11. Later, Kurtén and Thenius placed T. ornatus within the subfamily Tremarctinae together with other short-faced fossil bear species12,13 although this taxonomic classification is not universally accepted14,15. For example, María mentioned a purchased specimen that came from the Simitarra River (Upper Magdalena, Colombia)16 and that was previously classified as Ursus nasutus17. Four other scientific names were offered. Two (Nearctos ornatus18 and T. ornatus majori19 were based on a specimen with a type locality in southern Ecuador, probably in the province of Azuay. Another one (Ursus ornatus thomasi20 listed the Andes in southern Colombia as the type locality. The fourth’s (T. lasallei)17 type locality was Arauca, Colombia. Some of these scientific names were based on claw length, facial markings, and body proportions and were used as possible subspecies within the spectacled bear. Cabrera concluded that different geographical forms based on minimal physical differences (i.e. claw length, facial markings, and body proportions) to be inadmissible and included all of them in T. ornatus9. Therefore, T. ornatus is considered monotypic, although considerable phenotypic variation is present within the species21,22.
The geographical distribution of T. ornatus involves the countries of Venezuela, Colombia, Ecuador, Perú, and Bolivia15,23,24. Based on 18 unconfirmed sightings in northwestern Argentina, del Moral et al. suggested the presence of T. ornatus in Argentina25,26,27. After several years of debate, its presence was confirmed via genetic analysis of hair and feces collected in the Jujuy and the Salta provinces of northern Argentina28. This distribution is long (circa 4,700 km) and narrow (around 200–650 km) along the Andean mountains29. Additionally, T. ornatus inhabits a wide variety of Andean ecosystems between 200 and 4,750 masl. These ecosystems include dry forests, moist lowlands, montane forests, dry and moist shrublands, and high altitude shrublands, grasslands, and paramos7. The lower limit is close to the northwestern Peruvian desert coast, whereas the upper limit is within Carrasco National Park in Bolivia30,31,32. Projections of the effects of climate change show a general pattern toward upslope displacement of the mountain biome, suggesting that the Tropical Andes is among the most vulnerable regions to climate change33,34. The average global temperature has increased by 0.74 C° in the last century, and with a projected increase of around 3.6–5 °C by 210035, extensive changes in Andean habitat areas are expected. Therefore, all ecosystems associated with T. ornatus will probably be reduced soon.
This subfamily diverged from other branches of living bears of the genus Ursus around 10–11 million years ago (MYA) and from a chromosomal perspective is unique36,37,38. For instance, all Ursine bears have 74 chromosomes whilst Ailuropodinae (giant panda) has 42 chromosomes. However, the karyotype of the spectacled bear is completely different39,40,41,42. This species has a diploid number (2n) of 52 largely biarmed chromosomes, with 16 pairs of metacentric autosomes and nine pairs of acrocentric autosomes39,43,44. The X chromosome is relatively large and metacentric, and the Y chromosome is smaller and acrocentric44. The spectacled bear diverged from an ancestor with the ursine karyotype (2n = 74) about 10.91 MYA (9.93–11.89 MYA). It differs from that of its presumed ancestor by two fusions, two fissions, 11 centric fusions, and one inversion36,39,45,46.
For these multiple taxonomic, geographical, ecological and genetic reasons, this species is considered an umbrella and emblematic species for all the Andean countries where it inhabits. These important species need to be intensively studied from a molecular population genetic point of view because they are essential to biological conservation programs. Studies of this nature have been carried out in some of the countries where the spectacled bear lives.
Ruiz-García47 was the first to analyze 32 Colombian, 42 Ecuadorian, and 8 Venezuelan specimens for five nuclear DNA microsatellite loci (G1A, G1D, G10B, G10C and G10X). The mean level of He (expected heterozygosity = genetic diversity) for all bears was 0.38, while the level of He in the Ecuadorian population was only 0.25. These values of genetic diversity were considered very low or even depauperated. Nevertheless, these results were considered preliminary because only five markers were used with two of them being monomorphic. When Ruiz-García et al.48 enlarged the sample size to 155 specimens (including Perú and Bolivia, and more areas of Colombia and Ecuador) and the number of microsatellites was increased to nine (although two were monomorphic), the values of genetic diversity were higher (total spectacled bear sample: He = 0.56; Ecuador: He = 0.40). Later estimates of genetic diversity for Colombia49,50including more geographical regions and specimens, offered a value of He = 0.645 ± 0.211. The same occurred with the Ecuadorian spectacled bear population, when more specimens and more geographical locations were added, showing a value of He = 0.584 ± 0.18851. Nonetheless, these microsatellite genetic diversity values were still lower than that estimated in other bear species. Waits and collaborators showed that the genetic diversity values reported in studies across North America and Scandinavia that assessed genetic diversity in the brown bear with the same markers had an average of He = 0.7452. Paetkau and Strobeck determined the microsatellite genetic diversity for two continental Canadian populations of the American black bear to be around 0.853.
Additionally, an extensive analysis with microsatellites and mtDNA for the entire geographical distribution of the spectacled bear in South America was carried out by Ruiz-García and collaborators54. This work determined the existence of two possible Evolutionary Significant Units (ESUs)55 within T. ornatus using several mt genes. One ESU consisted of Venezuela, Colombia, Ecuador, and northern Peru (Northern Andean Clade; NAC) whereas the second ESU contained southern Peru and Bolivia (Southern Andean Clade; SAC). Temporal divergence times between the ESUs were estimated using a haplotype network, and the time split between them was estimated to have occurred 541,931 ± 54,193 YA. Furthermore, SAC was older than NAC, and NAC derived from SAC. Additionally, SAC showed a slightly higher level of genetic diversity than NAC. This could be explained by the central-marginal hypothesis56,57: The central distribution range of a species (oldest populations) tends to exhibit higher levels of genetic diversity than the populations located at the periphery of the distribution range (derived populations). The most relevant conclusion of this work was that this species likely originated “in situ” in South America and that the colonizing dispersion was from south to north.
Most molecular studies of the spectacled bear have occurred in Colombia47,48,49,50,54,58,59 and Ecuador47,48,49,50,51,54,58,59,60,61,62. The work of Viteri and Waits60 basically ratified the microsatellite results previously obtained by Ruiz-García’s team. However, the works of Cueva and collaborators61,62 misinterpret some of the population genetic results reported by Ruiz-García’s team. The first work61 analyzed the mt control region in 38 samples of hairs taken in the field in a very restricted area in the Metropolitan District of Quito (northern area of the country). The authors claimed to have found low levels of genetic diversity, based on Hd = 0.705 ± 0.037 and π = 0.002 ± 0.0015 as well as the existence of two differentiated spectacled bear gene pools in the area separated by the Guayllabamba River. They concluded that there was a need to establish, or revise, the current conservation strategies in the region. However, these authors61 analyzed a very small population in a very restricted area of Ecuador, and therefore did not have a sufficient broad view of the population genetic characteristics of the spectacled bear across all Ecuador49,51,54. This species does not have low mitochondrial genetic diversity levels in this country as the authors have claimed. However, the degree of differentiation among the haplotypes found was low, a result supported by both Ruiz-García et al.51,54 and Cuevas et al.61 All the spectacled bears sampled in Ecuador came from a unique genetic source and the colonization process of this species in Ecuador was very recent. Therefore, the scarce nucleotide differentiation among the haplotypes found in Ecuador was not caused by recent genetic drift originated by human habitat fragmentation or other negative human influences, rather it was due to how and when the spectacled bear colonized that area of South America. Cueva et al.62 recently reported another molecular study of the spectacled bear population in Ecuador. In this study, they analyzed DNA in hair and excrement from a small number of field samples collected from two areas of southern Ecuador (Loja and Zamora Chinchipe provinces) along with samples obtained in the Metropolitan District of Quito (Pichincha province) reported in the previous study61. They created specific mitochondrial and microsatellite primers for the spectacled bears they analyzed in three Ecuadorian provinces and used this information to show that the genetic diversity levels for both types of markers were considerably higher than that obtained by Ruiz-García et al.51 for 13 Ecuadorian provinces. Additionally, they found a very marked spatial genetic structure for the spectacled bear in Ecuador, whereas Ruiz-García et al.51 detected minimal spatial structure for this species in the same country. The latter authors concluded that, in view of the results obtained, a single genetic management plan should be created for the Ecuadorian bears. In contrast, Cueva et al.62 concluded that each of the populations they studied should be managed as different populations.
Because successful conservation programs are based on accurate population data, it is extremely important to generate high-quality and robust molecular population genetic results. Molecular population genetic results which are misunderstood and/or misinterpreted can lead to the development of inadequate conservation programs (or an unnecessary expenditure of money and time that could be invested to protect other species). Therefore, it is essential to determine which of the two studies51,62 best describes the genetic reality of the spectacled bears in Ecuador. To do this, we offer data from a new molecular analysis, which includes three data sets: sequences of six mt markers of 127 spectacled bears (108 bears studied in51, plus 19 new additional specimens) representing 13 Ecuadorian provinces, fully sequenced mitogenomes of 43 spectacled bears (obtained from the 127 specimens commented) representing 10 Ecuadorian provinces, and seven genotypified nuclear microsatellites of 88 spectacled bears of 11 Ecuadorian provinces.
The main aims of the current work are as follows. First, to determine if the new results on the genetic diversity levels found in the spectacled bears from Ecuador for mtDNA and microsatellites (with heterologous primers for both kinds of markers) are different to those previously obtained in other works51,62. Second, to estimate the degree of genetic heterogeneity and spatial genetic structure between spectacled bears from different Ecuadorian provinces to be compared with previous studies51,62. Third, to determine the effect of studying only spectacled bear populations in the most distant areas of Ecuador (Pichincha and Imbabura to the north; Loja and Zamora Chinchipe to the south) when the genetic data of spectacled bears are extracted from the intermediate zones to see if these results show a distorted view of reality. Lastly, fourth, to provide new genetics data of the Ecuadorian spectacled bear that could support the main conclusions of Ruiz-García et al.51 which suggest that the Andean cordilleras were of a residual importance for the colonization of the spectacled bear of that area of South America, and, therefore, only a single management plan should be considered for this species in Ecuador.
Results
MtDNA genes
Genetic diversity and genetic heterogeneity between northern Ecuador and southern Ecuador
A total of 127 spectacled bears were sequenced for six mtDNA genes representing 13 Ecuadorian provinces (Fig. 1). The mitochondrial genetic diversity statistics found for this data set (Hd = 0.856 ± 0.028, π = 0.0108 ± 0.0011) were undifferentiable from the values previously reported in Ruiz-García et al.51 (Hd = 0.845 ± 0.033, and π = 0.0104 ± 0.0013; t = 0.03, 1 df, p < 0.99 and t = 0.004, 1 df, p < 0.99, respectively) and in Cueva et al.62 (Hd = 0.87 ± 0.04, π = 0.0052 ± 0.0028; t = 0.146, 1 df, p < 0.91 and t = 1.011, 1 df, p < 0.48, respectively) (Table 1). On the other hand, some of the genetic heterogeneity statistics representing Pichincha (northern Ecuador) versus Loja and Zamora Chinchipe (southern Ecuador) were not significant (Table 2a), whereas others were (p < 0.05) but with a relative low magnitude (GST = 0.035; γST = 0.076; NST = 0.046; FST = 0.046). Their respective gene flow estimates were all high (for instance, Nm from GST = 13.61, and Nm from FST = 10.34).
Map of Ecuador showing the geographical points (provinces) where different spectacled bears (Tremarctos ornatus) were sampled to sequence six mitochondrial genes, complete mitogenomes and genotypified for nuclear microsatellites (n = samples sizes for each genetic dataset). The map was created by J.A-V using the QGIS v3.4 software (https://qgis.org/download/).
Spatial analyses
The Mantel’s test for all the 127 samples analyzed showed that the geographic distances did not significantly explain the genetic distances among these specimens (r = 0.0132, p = 0.238). The spatial autocorrelation analysis yielded a non-significant correlogram (V = 0.0307, p = 0.163), with only two DCs being significant (DC 6, 86–102 km, with a significant positive autocorrelation, p = 0.0229; DC 7, 102–124 km, with a significant negative autocorrelation, p = 0.0256) (Fig. 2a). In general, the correlogram’s shape showed an initial positive genetic similarity for the spectacled bears living between 0 and 20 km with a decrease genetic similarity between 20 and 37 km. Thus, this correlogram detected genetic patches of around 20–30 km in diameter. Note, the genetic similarity increased from 37 to 102 km, with a sudden decrease between 102 and 124 km (the significant negative DC 7) and slightly increased for the most distant specimens (124–240 km). However, as we previously commented, the overall correlogram was not significant. The MMA showed four weak geographical differentiation barriers which defined four areas. One area contained the southern extreme of the Napo province, and the Morona Santiago province up until the Azuay province. A second area contained a fraction of the Imbabura province and laterally extended to the Santo Domingo province. A third area covered a part of the Imbabura province and reached as far as the Napo province, including parts of the provinces of Pichincha, Cotopaxi, and Tungurahua. The fourth area covered the Cañar and Bolivar provinces, some parts of the provinces of Santo Domingo, Cotopaxi and, laterally, Loja (Fig. 2b). The BAPS analyses were carried out for individuals and for provinces (Fig. 2c). For individuals, there were two groups with an optimal partition [Log (marginal likelihood) of optimal partition = -712.94]. The specimen from the Cañar province was genetically distinct from all the remaining specimens analyzed. For provinces, the situation was the same with the number of optimal groups [Log (marginal likelihood) of optimal partition = -691.86]. The Cañar province was genetically distinct from all the other provinces which were genetically undifferentiable. Definitively, based on analysis of six mt genes, this data set showed that one specimen from the Cañar province was highly differentiated from the other 126 spectacled bears.
Analyses of the spatial structure of 127 specimens of spectacled bears sequenced at six mitochondrial (mt) genes throughout Ecuador: (a) Spatial autocorrelation analysis with eight distance classes (DCs); (b) Monmonier’s algorithm analysis (MAA) with four detected geographical barriers; the map was created by J.A-V using the ArcGIS Pro 3.2.2 software (https://pro.arcgis.com/es/pro-app/latest/get-started/download-arcgis-pro.htm); (c) Bayesian analysis of population structure (BAPS) by specimens and by provinces; the map was created by J.A-V using the ArcGIS Pro 3.2.2 software (https://pro.arcgis.com/es/pro-app/latest/get-started/download-arcgis-pro.htm).
But what is the effect of the same spatial analyses on the six mt genes of the 127 spectacled bears if only samples from the northern Pinchincha province and the southern Loja and Zamora Chinchipe provinces (total of 20 specimens) were used as Cueva et al.62 had done? The Mantel’s test did not detect any significant relationships between the geographical distances and the genetic distances (r = 0.0135, p = 0.332). The spatial autocorrelation correlogram did not show evidence of significant spatial structure globally (V = 0.0734, p = 0.626) nor for individual DCs (Fig. 3a). Nevertheless, the MAA was clearly affected by taking only specimens of the extreme geographical areas from Ecuador. The first four weak geographical barriers we detected were as follows. The first basically covered the area of Zamora Chinchipe province bordering the Loja province. The second area was in the Pichincha province extending from Antisana to San José de Minas, passing through Nanegalito and Chalpar. The third area was mostly in Loja but extended slightly into a small part of Zamora Chinchipe. The fourth area spanned from Nanegalito towards Tandapi in the Pichincha province (Fig. 3b). The BAPS analyses (Fig. 3c) for specimens and for provinces provided the same results. Based on optimal partition there was only one genetic group [Log (marginal likelihood) of optimal partition = -135.73 and Log (marginal likelihood) of optimal partition = -135.66, respectively]. Henceforth, when we eliminated all the bears from intermediate geographical areas and only left those belonging to northern (Pichincha) and southern (Loja and Zamora Chinchipe) Ecuador, as Cueva et al.62 did, three analyses (Mantel’s test, spatial autocorrelation, and BAPS) were not affected. They showed similar results to those obtained for the complete analysis with 127 spectacled bears. Nonetheless, note, a fourth analysis was completely affected indicating a supposed strong differentiation between northern and southern Ecuadorian areas (MAA).
Analyses of the spatial structure of 20 specimens of spectacled bears sequenced at six mitochondrial (mt) genes sampled in northern Ecuador (Pichincha province) and in southern Ecuador (Loja and Zamora Chinchipe provinces): (a) Spatial autocorrelation analysis with eight distance classes (DCs); (b) Monmonier’s algorithm analysis (MAA) with four detected geographical barriers; the map was created by J.A-V using the ArcGIS Pro 3.2.2 software (https://pro.arcgis.com/es/pro-app/latest/get-started/download-arcgis-pro.htm); (c) Bayesian analysis of population structure (BAPS) by specimens and by provinces; the map was created by J.A-V using the ArcGIS Pro 3.2.2 software (https://pro.arcgis.com/es/pro-app/latest/get-started/download-arcgis-pro.htm).
Mitogenomes
Genetic diversity and genetic heterogeneity
We analyzed the complete mitogenomes of 43 Ecuadorian spectacled bears representing 10 Ecuadorian provinces. This was a subset of the 127 spectacled bears analyzed for six mt genes. Of all the samples, these 43 had the greatest quantity and quality of DNA (Fig. 1). For these complete mitogenomes, the genetic diversity statistics (Table 1) yielded higher haplotype diversities than those obtained in the previous studies51,62but similar nucleotide diversities. Thus, the analysis of complete mitogenomes detected more diversity of haplotypes than did the analysis of several mt genes. Additionally, some genetic heterogeneity statistics of all 10 Ecuadorian provinces collectively did not detect significant differences globally (χ2 = 98.13, p = 0.195, HST, and GST = 0.018 with its respective Nm = 27.94). Other statistics, such as KST, KST*, Z, Z*, Snn, γST (= 0.219, and its respective Nm = 1.78), NST (= 0.244; and its respective Nm = 1.55), and FST (= 0.242, and its respective Nm = 1.56) were significant (p < 0.05). The heterogeneities were greater in the analysis of the complete mitogenome relative to the analysis of the six mt genes. However, these heterogeneities were not extremely large. All the gene flow statistics were higher than one, which is related to relatively high values of historical gene flow (Table 2b). Between province pairs, no genetic heterogeneity value was significant for the GST statistic. For the FST statistics, the most elevated heterogeneities were between Imbabura and Loja-Zamora Chinchipe (FST = 0.364, p < 0.01), and between the Napo province and Loja-Zamora Chinchipe provinces (FST = 0.442, p < 0.005). However, some other FST values were not significant (for instance, Imbabura vs. Napo, FST = 0.0009, p < 0.99 ; Azuay vs. Loja-Zamora Chinchipe, FST = 0.0137, p < 0.86) Thus, some statistics detected significant heterogeneity between some provinces and those of Loja and Zamora Chinchipe as it was noted in the previous data set, and in the two previous works51,62 but within a context of a genetic similarity.
Spatial analyses
The Mantel’s test for this data set (as well as in the previous one) did not globally reveal a significant relationship between the geographic and genetic distances (r = -0.0179, p = 0.638). The overall correlogram from the spatial autocorrelation analysis was also not significant (V = 0.0170, p = 0.995). No individual DC values were significant. Thus, even less genetic structure was detected at the mitogenomes (with a lower sample size) than with the six mt genes previously explored (with a larger sample size) (Fig. 4a). The MAA detected four possible areas restricted by some geographical barrier especially in the northern area of Ecuador (Fig. 4b). The first is an area that extends from a part of Imbabura, through Pichincha (Tandapi), Santo Domingo, Tungurahua, Napo (especially the zone of Cosanga), to part of the provinces of Azuay and Morona Santiago. The second area extends from part of Imbabura, through another area of Pichincha, as well as Tungurahua, Bolivar to Zamora Chinchipe, collaterally brushing the area of Cosanga (Napo Province). The third area comprises Intag, Cotacachi, Zuleta, and Alto Chocó and a fourth area spans from Tungurahua to Napo, including a major part of Oyacachi and Cosanga. The BAPS analysis for individuals detected two populations as the number for an optimal partition (Log [marginal likelihood] of optimal partition = -894.39). One population was basically found in the southern area of Ecuador but included some specimens that had characteristics typically found in the northern area. The second population was distributed in northern Ecuador and had diverse specimens (in three locations) with genetic characteristics typically from southern Ecuador. These specimens were not geographically isolated from each other. Nevertheless, when the provinces were used in the analysis, only one population was detected as optimal partition (Log [marginal likelihood] of optimal partition = -949.26). We did not detect any differentiated “genetic pools” in Ecuador with this procedure (Fig. 4c), which supports the findings of the first data set.
Analyses of the spatial structure of 43 specimens of spectacled bears sequenced for complete mitogenomes throughout Ecuador: (a) Spatial autocorrelation analysis with eight distance classes (DCs); (b) Monmonier’s algorithm analysis (MAA) with four detected geographical barriers; the map was created by J.A-V using the ArcGIS Pro 3.2.2 software (https://pro.arcgis.com/es/pro-app/latest/get-started/download-arcgis-pro.htm); (d) Bayesian analysis of population structure (BAPS) by provinces; the map was created by J.A-V using the ArcGIS Pro 3.2.2 software (https://pro.arcgis.com/es/pro-app/latest/get-started/download-arcgis-pro.htm).
Just as we did for the first data set, we pursued an answer to the question “What would be the result if we only analyzed the mitogenomes of 24 spectacled bears sampled in the most northern (Imbabura-Pichincha) and southern (Loja-Zamora Chinchipe) areas of Ecuador?” The Mantel’s test, in this case, offered a significant negative relationship between geographic and genetic distances (r = -0.124, p = 0.007). That is, the specimens that were geographically closer were more genetically differentiated from each other. In agreement with this result, the overall correlogram from the spatial autocorrelation analysis was not significant (V = 0.0734, p = 0.534). However, the first DC was significantly negative (1 DC: 0–4 km, p = 0.019), supporting strong genetic differentiation among the closest specimens (Supplementary Fig. S1a). The MAA agrees quite well with the previous analysis. There were four geographical areas differentiated by weak barriers: (1) An area which covered the most northern region of Imbabura up until Tandapi (Pinchicha); (2) The most southern area of Imbabura which extends to Tandapi; (3) A cut-off area between Pichincha and Loja; (4) An area which contained Loja and Zamora Chinchipe (Supplementary Fig. S1b). The BAPS analysis of complete mitogenomes from 24 specimens (Imbabura-Pichincha vs. Loja-Zamora Chinchipe) showed that the most optimal partition was two genetic populations (Log [marginal likelihood] of optimal partition = -506.46). The southern and northern specimens formed different populations. Nonetheless, one of the most northern specimens sampled (Barcelona in the Imbabura province) belonged to the southern “gene pool.” However, when we applied the BAPS analysis for provinces, the optimal partition was one (Log [marginal likelihood] of optimal partition = -516.18) (Supplementary Fig. S1c).
Demographic changes
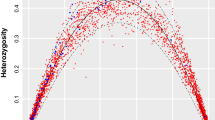
Taking into consideration the mitogenome information and using different statistics and the mismatch distributions, we studied the possible historical demographic change of the Ecuadorian spectacled bear population (Table 3; Fig. 5). For the overall Ecuadorian sample, the five statistics used as well as the mismatch distribution supported significant evidence of female population expansions. With the assumptions explained in the material and methods, t = 2µt = 5.052 which is equivalent to an initial population expansion around 27,300 years ago (YA). For the northern Ecuadorian spectacled bear population (Imbabura and Pichincha provinces), all the statistics and the mismatch distribution were significant indicating female population expansion. With t = 2µt = 4.055, this population expansion began around 22,000 YA. For the southern Ecuadorian spectacled bear population (Loja and Zamora Chinchipe provinces) only three statistics as well as the mismatch distribution presented significant values agreeing with a female population expansion (in this case, the lower number of significant statistics is related to the smaller sample size for this geographical area). With t = 2µt = 8.333, and with the same conditions as in the previous analyses, there was an initial population expansion around 45,100 YA. Thus, the oldest population expansion seems to have started in the south of present-day Ecuador.
Microsatellites
Genetic diversity and Hardy-Weinberg equilibrium
A total of 88 Ecuadorian spectacled bears representing 11 Ecuadorian provinces were completely genotypified for seven nuclear DNA microsatellites (Fig. 1). The He obtained in the current work was 0.594 ± 0.120 and the values from Ruiz-García et al.51 and Cueva et al.62 were He = 0.584 ± 0.188 (t = 0.22, 1df, p < 0.85) and He = 0.60, respectively (they did not show the standard error of this statistic, but their value is inside the range between the current one and that by Ruiz-García et al.51 (Table 1). These values were statistically undifferentiable. We reported here a value of MNA = 6.714 ± 3.251. Ruiz-García et al.51 estimated MNA = 5.857 ± 3.076. These values were statistically undifferentiable (t = 0.728, 1df, p < 0.60). Cueva et al.62 did report an MNA of 5.0, which is lower than what we report here. However, Cueva et al.62 did not provide a standard error and therefore we cannot test for a significant difference, but this value is within the range of values reported by Ruiz-García et al.51 and the current work.
The HWE analysis for the overall Ecuadorian sample showed that five out of seven microsatellites studied had a significant excess of homozygotes (Table 4). If the Bonferroni’s correction is applied, four microsatellites showed excess of homozygotes (G10B, G10L, G10M, and G10P). The seven microsatellites taken together also showed a significant homozygote excess (χ2 = 91.28, df = 14, p < 0.0000001) with an average homozygote excess for the W-C statistic of 26.8% (FIS = 0.268) and of 17% for the R-H statistic (FIS = 0.170). By province, one of them showed a significant excess of homozygotes (Napo Province: p = 0.0000429). The remaining 10 provinces studied did not show a significant excess of homozygotes. By marker, only two out of seven microsatellites showed a significant homozygote excess (G10B: Fisher’s method, p = 0.0023; and G10L: Fisher’s method, p = 0.0097).
Genetic heterogeneity
With the Fisher’s method and exact probabilities, three out of seven microsatellites showed significant heterogeneity (G10M, G10P, and G10X) for the spectacled bears from Ecuador. Globally, the seven microsatellites taken together also yielded significant heterogeneity (χ2 = 40.07, df = 14, p = 0.00025) (Table 5). With the log-likelihood G statistic and 10,000 randomizations only G10P showed significant genetic heterogeneity across Ecuador. The values of the RST statistic ranged, depending on the procedure used, around 6–9% (weighted RST = 0.063; unweighted RST = 0.087; Goodman’s RST = 0.088). These were relatively small values of genetic heterogeneity. Based on the private allele method, the estimate of gene flow (Nm) involving microsatellites for all the Ecuadorian spectacled bear populations was 2.4. This value could be considered relatively high. The genetic heterogeneity by population pairs with the Fisher’s method only showed five out of 55 comparison pairs (9.09%) as significant at the level p < 0.05. These were the cases of Pichincha vs. Napo (χ2 = 21.92, df = 12, p = 0.038), Napo vs. Cañar (χ2 = 14.55, df = 6, p = 0.024), Napo vs. Azuay (χ2 = 22.58, df = 12, p = 0.032), Napo vs. Loja (χ2 = 35.31, df = 14, p = 0.0013), and Azuay vs. Loja (χ2 = 24.32, df = 14, p = 0.042). Napo province had one of the greater sampling sizes and presented more cases of genetic heterogeneity relative to the other provinces. Nonetheless, when the Bonferroni’s correction was applied, no comparison pairs were significant (p < 0.0009). A population assignment analysis was completed for the five most polymorphic microsatellites used. All the procedures indicated few to very few specimens that were well assigned to their respective geographical populations. Furthermore, the percentages of first-generation migrants were very high. The procedures identifying the highest number of correctly assigned individuals were developed by Paetkau et al.63 and Rannala and Mountain64. In those cases, twelve specimens were well assigned (14.6% of specimens). The Baudouin and Lebrun method65 detected 76 specimens as first-generation migrants (86.36%). Slightly lower, the Rannala and Mountain and Nei’s DA distance’ methods64,66 detected 70 specimens as first-generation migrants (79.54%). In summary, all the microsatellite results indicated a limited population structure for the spectacled bear in Ecuador.
Spatial genetic structure
The results of Mantel’s test of microsatellites and the results of the same test on the mt genes and mitogenomes were slightly different. The overall relationship between the geographic and the genetic distances was significantly positive (r = 0.105, p = 0.008). Although this value was significant, the geographical distances only explained 1.1% of the genetic distances (Fig. 6a). The spatial autocorrelation analysis showed an overall significant correlogram (V = 0.042, p = 0.0034). This contrasted with that observed for mt genes and mitogenomes. By individual DC, the fourth DC showed significant positive autocorrelation (4 DC: 58–81 km, p = 0.0001), and the sixth and eighth DCs presented significant negative autocorrelation (6 DC: 95–107 km, p = 0.0044; 8 DC: 140–218 km, p = 0.0267). Thus, from 0 to 81 km, the spatial trend showed a close genetic relationship among the specimens. There was a strong and significant genetic resemblance between 58 and 81 km, whereas between 81 and 218 km, there was significant genetic dissimilarity. This dissimilarity was especially marked from 95 to 107 km and from 140 to 218 km. Therefore, this new autocorrelation spatial analysis with microsatellites showed more spatial structure than that found for individual mt genes and mitogenomes as well as to that found with microsatellites reported by Ruiz-García et al.51 (Fig. 6b). The MMA showed the existence of areas differentiated by possible geographical barriers. The first area was in the Napo province (especially the zone of Cosanga) bordering with the Tungurahua province. The second area covered the provinces of Cañar, Azuay, Morona Santiago, Loja, and Zamora Chinchipe. The third area comprised basically the northern and central Azuay, whilst the fourth area comprised the provinces of Imbabura and Pichincha until Napo (especially Cayambe-Coca National Park) and Santo Domingo (Fig. 6c). These results were in clear agreement with the spatial autocorrelation analysis indicating certain spatial structure for microsatellites, which was not found for individual mt genes and mitogenomes. Based on the BAPS analysis for specimens, seven was the optimal number of groups (Log [marginal likelihood] of optimal partition = -692.73, p = 0.951) (Fig. 6d). Five of these groups were in southern Ecuador. Five groups were also found in northern Ecuador. Two groups were exclusive to southern Ecuador and another two small groups were exclusive to northern Ecuador. Nevertheless, when we carried out the BAPS analysis by provinces, the optimal partition number of groups was one (Log [marginal likelihood] of optimal partition = -702.90, p = 0.999) (Fig. 6e). Although significant spatial structure was determined for the microsatellites, the magnitude of the genetic heterogeneity found for the Ecuadorian spectacled bear at these molecular markers was not extremely high. Therefore, beyond local genetic fluctuations caused by gene drift, the Ecuadorian spectacled bear population represents a unique gene pool.
Analyses of the spatial structure of 88 specimens of spectacled bears analyzed at seven nuclear microsatellites throughout Ecuador: (a) Mantel’s test (b) Spatial autocorrelation analysis with eight distance classes (DCs); (c) Monmonier’s algorithm analysis (MAA) with four detected geographical barriers; the map was created by J.A-V using the ArcGIS Pro 3.2.2 software (https://pro.arcgis.com/es/pro-app/latest/get-started/download-arcgis-pro.htm) (d) Bayesian analysis of population structure (BAPS) by specimens; the map was created by J.A-V using the ArcGIS Pro 3.2.2 software (https://pro.arcgis.com/es/pro-app/latest/get-started/download-arcgis-pro.htm) (e) Bayesian analysis of population structure (BAPS) by provinces; the map was created by J.A-V using the ArcGIS Pro 3.2.2 software (https://pro.arcgis.com/es/pro-app/latest/get-started/download-arcgis-pro.htm).
We also determined what would occur when microsatellites of just the northern (Imbabura and Pichincha) and southern (Loja and Zamora Chinchipe) Ecuadorian spectacled bear populations were analyzed. When only Imbabura-Pichincha and Loja-Zamora Chinchipe were considered, the Mantel’s test, and the spatial autocorrelation analysis (V = 0.0334, p = 0.413) did not detect significant spatial structure as they did with the complete microsatellite data set. The MAA detected geographical barriers in the following zones: (1) An area covering a major part of Imbabura and Pichincha; (2) An area from eastern Pichincha until Zamora Chinchipe, including Loja; (3) An area that differentiated one part of Zamora Chinchipe with a fraction of Loja; (4) A little area in western Pichincha (Supplementary Fig. S2a). Finally, the BAPS analysis with specimens detected six possible groups as the optimal partition (Log [marginal likelihood] of optimal partition = -308.30, p = 0.611). For Loja-Zamora Chinchipe five different groups (one exclusive of this area) were found. Similarly, for Imbabura-Pichincha there were five groups with one exclusive to this area (Supplementary Fig. S2b). When the BAPS analysis considered provinces, however, the optimal partition was one group (Log [marginal likelihood] of optimal partition = -317.94, p = 0.999) (Supplementary Fig. S1c).
Discussion
New results on the molecular population genetics of the spectacled bear in Ecuador
This is the first study to use complete mitogenomes of the spectacled bear to make population genetic inferences. The information from this study is relevant to the conservation of the spectacled bear in Ecuador, especially, the one related to the following topics:
The mt genetic diversity levels obtained for the six mt genes in this work were high and identical to those estimated in Ruiz-García et al.51. However, the mitogenome result for the haplotypic diversity was substantially higher than for the six mt gene data set, although the nucleotide diversity was similar in both mt data sets. This highlights the fact that when studying a larger number of genes, a greater genetic diversity (haplotypic diversity) can be detected. Note, however, relative genetic diversity (nucleotide diversity) is independent of sample size and the length of sequence analyzed. The genetic diversity of mtDNA for the Ecuadorian spectacled bear population is considerably high. In contrast, the microsatellite genetic diversity level for the Ecuadorian spectacled bears is only medium (as it was also shown by Ruiz-García et al.51. Although the effects of ascertainment bias are not discarded as we will discuss later, it seems clear that the mt results reflect the original colonization events of the different maternal lineages more than the action of recent anthropogenic perturbations, whereas microsatellite genetic diversity may be more influenced by more recent demographic events and fragmentation caused by human actions and climatological changes.
The new mt genetic results with both six mt genes and mitogenomes revealed certain genetic heterogeneity among the spectacled bear samples of several different Ecuadorian provinces, including significant heterogeneity between the most northern and most southern Ecuadorian areas, such as it was reported by Ruiz-García et al.51. However, this heterogeneity, although significant, is relatively small and it didn’t induce any kind of significant spatial structure. In fact, the spatial genetic structure analyses with both six mt genes and mitogenomes revealed scarce spatial structure for this species in Ecuador. Our analysis with individuals of the mitogenome data detected two genetic clusters in Ecuador that were geographically mixed. Nonetheless, when the specimens were pooled by provinces, only one genetic cluster was detected for all Ecuador for both the six mt gene and the mitogenome data sets. Of note, there was genetic differentiation of the Cañar specimen bordering the Guayas province (this specimen should belong to another differentiated gene pool).
The new microsatellite data set represented 11 Ecuadorian provinces and detected more significant spatial structure than that reported by Ruiz-García et al.51who only analyzed seven provinces. Up to seven different genetic clusters were detected in this new analysis but mixed in different areas of Ecuador. However, when they were analyzed by provinces, a unique genetic cluster appeared. In line with this, the geographical assignation analysis revealed low levels of correct assignations among the spectacled bears of diverse Ecuadorian provinces.
A fundamental new result of the current work was the comparison between the population genetic results for the global sample of Ecuador with respect to the samples obtained only in the northernmost (Imbabura and Pichincha provinces) and southernmost (Loja and Zamora Chinchipe provinces) distribution of that country. When only the specimens from the most extreme geographical areas of Ecuador were analyzed, some procedures tended to artificially exaggerate the genetic differences between these areas. This occurs because the information of the intermediate geographical area is lost. This is verifiable by comparing the BAPS analyses of the complete mitogenome data set of 43 specimens with the data set of 24 specimens (only the northernmost and southernmost Ecuadorian specimens). In the first analysis, we found four specimens misclassified. The northern Ecuadorian area had two specimens with mitogenome characteristics typical of the southern stock (one from Barcelona, Imbabura province, and the other from Tungurahua province). In contrast, we found two specimens with mitogenomes typical of the northern stock in the southern part of the Azuay province, on the border with the Loja and Zamora Chinchipe provinces. Thus, we detected four misclassified specimens out of the 43 specimens analyzed. In the case of the second analysis, we only found one misclassified specimen in the northern area (the specimen from Barcelona, Imbabura province). Henceforth, when we reduced the sample approximately to 50% (24 vs. 43), we decreased the probability to detect specimens “misclassified or mixed” by 4. Similarly, the five spatial analyses undertaken with only the most extreme geographically sampled specimens (North-South) for mt genes and mitogenomes clearly showed biases and distortions of the spatial structure with respect to the complete data set, including many spectacled bear specimens between these geographically extreme areas. This distortion and bias were more exaggerated for the mitogenome data (less specimens analyzed) than for the six mt genes (more specimens analyzed). When only microsatellites from specimens collected in Imbabura-Pichincha (northern Ecuador) and Loja-Zamora Chinchipe (southern Ecuador) were analyzed, the results were again biased and distorted. Any summaries based on these results would be inadequate as foundational information to develop a conservation program for this emblematic species in Ecuador. When we analyzed all spectacled bears, this southern and northern pattern disappeared. This shows how important it is to sample across all of Ecuador rather than just the most extreme areas of this country.
The comparison between the current work and Ruiz-García et al.51 and that from Cueva et al.62 in reference to the possible genetic structure of the spectacled bear in Ecuador
Until the current paper, there were only two molecular genetics studies of the spectacled bear in Ecuador, and supposedly, they had dissimilar findings. The first one authored by Ruiz-García et al.51 analyzed six mt genes of 108 specimens sampled in 13 Ecuadorian provinces. The fragments of these six mt genes totaled 2,901 base pairs (bp). Additionally, seven nuclear DNA microsatellites of 92 specimens from seven Ecuadorian provinces were analyzed. Based on the analysis of mt genes, the overall genetic diversity found for Ecuador in that study was H = 0.845 ± 0.033, and π = 0.0104 ± 0.0013. For northern Ecuador, the genetic diversity was H = 0.818 ± 0.041, and π = 0.0082 ± 0.0021, whereas for southern Ecuador, these statistics were H = 0.910 ± 0.045, and π = 0.0154 ± 0.0041. These genetic diversities were relatively high, such as it was detected in the current work. A maximum likelihood tree, with these mt sequences, indicated that there were only a few small geographical clusters with most specimens intermixed independently of the area where they were sampled (in general, all the bootstraps were low). Three sampling schemes were used to measure the genetic heterogeneity of the spectacled bear in Ecuador (by province, by Andean cordillera, and between northern and southern Ecuador). A relatively low genetic heterogeneity was found among the provinces (with the Benjamini-Hochberg correction, only 6.1% of the province pair comparisons were statistically significant). By cordillera, there was no significant differentiation. In contrast, a significant genetic heterogeneity was found between northern and southern Ecuador (FST = 0.059, p = 0.000001). However, the genetic heterogeneity was relatively small (around 6%). The analysis of the nuclear DNA microsatellites indicated a medium genetic diversity (H = 0.584 ± 0.188; pages 203–204). Three out of the seven microsatellites (and the seven microsatellites taken together) showed significant genetic heterogeneity by province. With the Benjamini-Hochberg correction, only two out of the 21 province pair comparisons were significant (the southern Ecuadorian province, Loja, was differentiated from the other two more northern Ecuadorian provinces, Azuay and Napo). By cordillera, two microsatellites (and the seven taken together) had significant genetic heterogeneity but with the correction, they were not significant. In sum, there was significant heterogeneity at the mt and nuclear markers between southern and northern Ecuadorian spectacled bears, but the amount was relatively small. Additionally, several spatial structure analyses were undertaken (Mantel’s test, and spatial autocorrelation) but no significant structure was detected across all of Ecuador.
Ruiz-García et al.51 also commented on ascertainment bias, human perturbations, historical gene flow, climatological changes, and size of management unit. First, ascertainment bias can be an issue when using heterologous microsatellites to determine genetic diversity. On page 204 the authors state “A possible explanation to this paradox (high mitochondrial genetic diversity, low or moderate microsatellite genetic diversity) is the effect of ‘ascertainment bias’ (Ellegren et al. 1995, 1997; Amos et al. 1996).” Second, the authors commented that the spatial patterns they detected, especially for the mtDNA, were not the product of recent human perturbations, but they were the product of how and when the different spectacled bear maternal lineages colonized this small portion of South America. Third, the possible existence of historical gene flow from South to North in the Ecuadorian territory (as well as in all South America54 could be important to our understanding of this species’ colonization in South America. Fourth, climatological changes (glaciations) at the end of the Pleistocene affected the demographic history of the spectacled bear in Ecuador. Fifth, all the spectacled bears in Ecuador should be treated as a unique Management Unit (MU) for conservation purposes.
Four years later Scientific Reports published the second one authored by Cueva et al.62 which concluded that the Ecuadorian spectacled bear population has a strong population genetic structure. Furthermore, the article claimed that the results presented by Ruiz-García et al.51 were wrong because the molecular markers used were inadequate. Cueva et al.62 analyzed the mt control region (HVR1) from a sample of 74 specimens (three Ecuadorian provinces, two in southern Ecuador [Loja and Zamora Chinchipe] and one in northern Ecuador [Quito]). Thus, the number of samples (74 vs. 108 specimens) and the number of Ecuadorian provinces sampled (three versus 13) were substantially lower than in the previous work. Additionally, the total length of the sequences was smaller in the second work (462 bp) compared to the first work (2,901 bp). The primers used in the second work were specifically created for the spectacled bear, however, the primers of the first work, and the current ones, were for all species of bears. The samples collected by Cueva et al.51 consisted of 26 fecal ones from Loja (but in no case did the authors explain how they determined the number of specimens which corresponded to these 26 fecal samples), 12 hair ones obtained through field work conducted in Zamora Chinchipe, and 36 hair ones obtained through field work conducted in Quito. Information about the last set had been previously published61. Given the descriptions of these samples, the quantity and quality of the DNA used in this second work were considerably lower than the DNA used in the first work and the current one. Remember, in the study by Ruiz-García et al.51 around 75% of the samples consisted of blood and hairs obtained from live specimens, and by pieces of skin (especially) and teeth of hunted specimens. Notably, the quantity and quality of DNA used by Ruiz-García et al.51and in the current analysis, was considerably better than that used by Cueva et al.62. This is a very relevant point because Cueva et al.62 had such a small amount and poor quality of DNA that they could not amplify their samples with standard mt primers for bears or with heterologous microsatellite markers designed for U. americanus as Ruiz-García et al.51 had done. Ruiz-García et al.51 had sufficiently high-quality DNA to be able to effectively use standard mt and heterologous microsatellite primers on the samples they used. In contrast, Cueva et al.62 created specific mt and microsatellite markers for the spectacled bear to try to amplify the DNA obtained from dried hairs and feces obtained in the field, which are of lower quality relative to a large fraction of the samples used by Ruiz-García et al.51 and in the current work.
The genetic diversities obtained for the control region for the overall Ecuadorian sample were H = 0.82 ± 0.02 and π = 0.0077 ± 0.0044 in that study62. These values were statistically undifferentiable from those obtained in the first work and in the present work, such as we previously showed, independently of the use of mt specific or non-specific primers for the spectacled bear. Additionally, Cueva et al.62 showed a maximum likelihood tree (without bootstraps in the nodes) and a haplotype network (Fig. 1b, and d in their work) that clearly showed nine specimens from Loja (southern Ecuador) that were more related to the bears from Quito (northern Ecuador) than to other bears from southern Ecuador. This result agrees quite well with the phylogenetic tree of the first work51 where some specimens from northern and southern Ecuador were intermixed. Therefore, the first data set used by Cueva et al.62 ratified the results of Ruiz-García et al.51which had already detected a difference between northern and southern Ecuadorian spectacled bears but in a context of mixing throughout Ecuador. However, Cueva et al.62 provided a second data set with a greater differentiation between the northern and southern Ecuadorian spectacled bears. This second data set only had 19 specimens (nine from Quito, five from Loja and seven from Zamora Chinchipe). Note that Quito (northern Ecuador) and Loja (southern Ecuador), which were the provinces with the two original greatest sample sizes (36 and 26 samples, respectively) and which showed intermixed bears, were extremely reduced (nine and five, respectively) in the second data set, eliminating the specimens shared between northern Ecuador (Quito, Pichincha) and southern Ecuador (Loja). The authors added sequences of a second mt gene to the second data set (COXII), which in some mammalian taxa, such as in Primates67,68,69,70,71,72,73 can show high levels of genetic diversity, but in bears has shown a very limited genetic diversity. Therefore, the authors obtained a strong differentiation between northern and southern Ecuadorian spectacled bears, by extremely reducing the sample size (especially in the northern and southern areas which showed intermixing) and by introducing sequences of a mt gene with a very limited genetic diversity in bears. Nevertheless, despite of this, the maximum likelihood tree for control region + COXII continued to show one specimen from Loja (southern Ecuador) more related to the northern spectacled bears than to the southern ones (their Fig. 1c, d). Similarly, the genetic diversity estimates for these 19 spectacled bears based on only the control region (H = 0.85 ± 0.04 and π = 0.0083 ± 0.0048) or based on both mt markers (control region + COXII) (H = 0.87 ± 0.04 and π = 0.0052 ± 0.0044) were not statistically different from the values showed by Ruiz-García et al.51 and those presented here. In summary, the use of mt primers specifically created for the spectacled bear (which is useful when the quality of DNA is poor as it was in Cueva et al.62 along with a small second data set were insufficient to refute the results presented by Ruiz-García et al.51 in reference to the mt genetic diversity levels. These results only served to support the findings of Ruiz-García et al.51. Therefore, discussing the use of specific mt primers for the spectacled bear to improve the estimates of genetic diversity is not helpful.
Additionally, Cueva et al.62 used 10 new DNA microsatellites specifically designed for the spectacled bear. The authors remarked, at least, seven times throughout their work, that the seven heterologous microsatellites used by Ruiz-García et al.51 underestimated the real levels of genetic diversity of microsatellites for the spectacled bears in Ecuador. Cueva et al.62 analyzed 10 homologous microsatellites in 36 specimens (22 from Quito, seven from Loja, and seven for Zamora Chinchipe) from three Ecuadorian provinces. In contrast, Ruiz-García et al.51 analyzed seven heterologous microsatellites of 92 specimens collected from seven different Ecuadorian provinces. The first authors determined an expected heterozygosity (He) of 0.60 for their overall sample, with He values for the three provinces ranging from 0.53 to 0.64, and with a MNA of 5. However, these microsatellite results were not significantly different from results reported by Ruiz-García et al.51or even by Ruiz-García49and the results herein. Ruiz-García et al.51 reported an He of 0.584 ± 0.188 (pages 203–204) for the overall sample of Ecuadorian spectacled bears. This value is practically identical to that found by Cueva et al.62. Furthermore, the new results reported in the current work and based on an analysis of seven heterologous microsatellites in 88 specimens collected from 11 different Ecuadorian provinces showed an He of 0.594 ± 0.162 and a MNA of 6.714 ± 3.251. These values were not statistically different (as we previously showed) from those based on homologous microsatellites and presented by Cueva et al.62. Even our new MNA values are slightly higher than that reported by the previous authors. Therefore, claims by Cueva et al.62that their homologous microsatellite results showed higher levels of genetic diversity (and more real ones) relative to those previously published by Ruiz-García’s team49,51are not supported with evidence. The previous and current results obtained with heterologous microsatellites adequately describe the possible genetic structure of the spectacled bear in Ecuador and were not especially influenced by ascertainment bias. In fact, Ruiz-García et al.51,54 were the first to draw attention to this possible phenomenon affecting microsatellites results in the spectacled bear.
Cueva et al.62 commented that the STRUCTURE analyses of their microsatellite data showed the best result with k = 2 (two populations; their Fig. 1e). However, five specimens from northern Ecuador had some degree of genetic mixing from the southern population. Moreover, because they showed an analysis with k = 3 (three populations; Fig. 1e) the genetic pool of the most southern Ecuadorian population (Zamora Chinchipe) clearly was mixed with the genetic profiles of the spectacled bears from Loja and Quito. In fact, when they used k = 3 (to differentiate the two southern populations), the characteristic genetic component from the Zamora Chinchipe population (the most southern Ecuadorian population) was present in all the specimens studied at Loja, and that this southern genetic component was very important in six specimens from Quito (northern population). This supports the affirmation of Ruiz-García et al.50,51,54 that the pattern of gene flow for the spectacled bear in South America is from South to North and not North to South as it has been historically considered. This information could be extremely important in our understanding the origin and colonization routes of this bear species in South America. Nevertheless, Cueva et al.62 overlooked this extremely important result.
Two other microsatellite results in the paper by Cueva et al.62 deserve some additional comments. First, the authors showed in their Table 3, the assumed results of FST for each one of the three populations they studied. However, it is conceptually impossible to estimate values of FST for individual populations, because this statistic is used to measure genetic heterogeneity among populations and not within populations. The FST value for the overall sample (FST = 0.14) could be correct, but the FST statistics for Quito, Loja, Zamora Chinchipe, individually, are impossible to calculate. Thus, some mistake was carried out in the calculation of this statistic. Second, they did obtain the overall and the specific population values of the Wright FIS, which in this case are correct (see their Table 3). This statistic measures the excess or defect of homozygotes and/or heterozygotes in a sample. They obtained an overall value of FIS = 0.16 (16% of homozygote excess) and, for Quito, the value was FIS = 0.26 (26% of homozygote excess), although they did not indicate the statistical significance of these values. In the current work, we obtained an overall FIS = 0.20, which is a statistically significant value (Fisher’s method: p = 0.000967; 1,000 randomizations: p = 0.001). These two values were similar. This means that our homozygote excess is not the primary product of ascertainment bias such as Cueva et al.62 have reiteratively claimed. Using specific homologous microsatellites, Cueva et al.62 obtained a very similar excess of homozygotes in their data. Even if our microsatellite data were influenced by ascertainment bias, the results of Cueva et al.62 were also influenced by a preferential allele amplification or similar phenomenon. Homozygote excess in some Ecuadorian areas, or in some microsatellites, could be the product of certain levels of endogamy (less probable because all loci were not affected by the same homozygote excess). It could also be the product of a Wahlund effect due to some subdivision or fragmentation in some areas of Ecuador within the general context of one basic genetic pool for the Ecuadorian spectacled bears. This would be logical for species such as the spectacled bear that have a small population size.
Because Cueva et al.62 did not analyze spectacled bears between these two extreme areas, an inadequate bias was likely generated similar to that we showed in this work when extracting the intermediate spectacled bears. Consider the following example. Imagine that a human family is randomly chosen from a certain neighborhood in a large city and genetically compared with another family, also chosen at random, from another neighborhood in the same city. It is possible, due to “genetic drift” when sampling that the families are genetically different. However, if we genetically analyze many families in each of the two neighborhoods, the genetic differences between the two neighborhoods are minimized. The first part of the example represents what Cueva et al.62 did whereas the second part of the example story represents the current work and Ruiz-García et al.51. Cueva et al.62 originally used a sample size of 74 specimens and detected nine specimens from southern Ecuador (9/74 = 12.16%) within the group of the northern Ecuadorian spectacled bears. Nonetheless, when they reduced their sample to 19 specimens (although they added one more mt gene [one that was scarcely polymorphic]), they only found one southern specimen in the northern stock (1/19 = 5.3%). The result was a strong bias (a “strong genetic drift” in the small data set), which induced an extreme genetic differentiation between the southern and northern Ecuadorian spectacled bears.
In fact, this strong bias is observed when we compare the works of Cueva et al.61 and Cueva et al.62. Cueva et al.61 reached two main conclusions from analyzing the mt control region of 38 spectacled bears from Quito. One, this spectacled bear population had a very low level of mt genetic diversity (Hd = 0.705 ± 0.037 and π = 0.00197 ± 0.00156). Second, there were possibly two different populations separated by the Guayllabamba River, although the genetic distances between the haplotypes of these populations were very low. However, their newer findings62 for a wider geographical sample in Ecuador and with the same mt control region were Hd = 0.85 and π = 0.8%. In other words, Hd and π increased by 18% and 400% respectively. These values of mt genetic diversity are high and identical to those reported by Ruiz-García et al.51 and herein. Therefore, the researchers’ two papers are in conflict due to the “strong genetic drift” in the small data set. Furthermore, criticism by Ruiz-García et al.51of the small area sampled (just near Quito) and the data not representing all of Ecuador, is justified. There was also disagreement between Cueva et al.61 and Cueva et al.62 about the existence of two different groups of spectacled bears in the area of Quito due to this “strong genetic drift” in the small data sets. Two groups were observable with the analysis of the mt control region of 74 specimens but disappeared when the researchers markedly reduced the sample size for the mt control region and COXII. The reduction enhanced the differentiation of the 19 northern and southern Ecuadorian spectacled bears. Therefore, based on their decisive analysis in 202462, there is a single unique gene pool for the spectacled bears from Quito and not two slightly differentiated genetic groups as claimed by Cueva et al.61.As we showed in the current work, each time a smaller number of specimens is analyzed, more “artificial genetic drift” is introduced into the data. We buffered this “artificial gene drift” by increasing the number of specimens analyzed and expanding the geographical areas analyzed as much as possible. In short, many of the results by Cueva et al.62 were statistically undifferentiable from the new results herein, and those from Ruiz-García et al.51.
Based on findings of the spectacled bear to date, it is more logical to have a single management plan for the spectacled bear in Ecuador than to have separate management plans for four different populations as suggested by Cuevas et al.61,62. There is little difference in genetic structure of the spectacled bear population across the relatively small South American country of Ecuador. Therefore, we suggest a unified national approach for conservation of the spectacled bear in Ecuador unlike what was found in Colombia, where at least three different management units (MUs) were found50. We suggest that future studies conduct nuclear marker studies to make accurate estimates of current and past gene flow between spectacled populations in different areas of Ecuador. Additionally, it would be useful to analyze MHC genes to understand the evolutionary potential of the spectacled bears facing possible new contagious diseases.
Methods
Samples
Three molecular data sets were analyzed in this study. The first enclosed a sample of 127 spectacled bears (108 bears studied by Ruiz-García et al.51plus 19 new additional specimens) sequenced at six mt markers (ND5, 16 S rRNA, 12 S rRNA, COXI, COXII and control region) representing 13 Ecuadorian provinces (from north to south, Imbabura, Pichincha, Sucumbíos, Santo Domingo de Tsáchilas, Napo, Cotopaxi, Tungurahua, Bolivar, Cañar, Morona Santiago, Azuay, Loja, and Zamora Chinchipe). The 19 new specimens were from the following localities: four samples from Pichincha province, eight samples from Papallacta (Napo province), two samples from Cotopaxi province, and five samples from Valle de Leito, Llanganates NP (Tungurahua province).
A second data base consisted of completely sequenced mitogenomes of a sample of 43 spectacled bears (obtained from the 127 specimens previously mentioned and representing 10 Ecuadorian provinces). The specimens from this data set were from Imbabura, Pichincha, Sucumbíos, Santo Domingo de Tsáchilas, Napo, Tungurahua, Morona Santiago, Azuay, Loja, and Zamora Chinchipe.
The third molecular database was made up of seven genotypified nuclear microsatellites (G1D, G10B, G10C, G10L, G10M, G10P, and G10X designed for Ursus americanus) of 88 specimens. Eleven microsatellites were used at first, but four of them showed irregular patterns or were completely monomorphic and were deleted. The specimens came from 11 Ecuadorian provinces (Imbabura, Pichincha, Sucumbíos, Santo Domingo de Tsáchilas, Napo, Tungurahua, Cañar, Morona Santiago, Azuay, Loja, and Zamora Chinchipe). Some of the specimens analyzed by Ruiz-García et al.51 for microsatellites were deleted because they had incomplete genotypes for all seven microsatellites.
Approximately 75% of the samples were from blood and fresh hairs from specimens captured in the field (studies of radiotelemetry by A. Castellanos). This percentage also included samples of skin, teeth and bone collected in Andean communities, where these specimens were hunted by local people. The remaining 25% were from dried hairs found in the field. The ethical approval for animal studies was obtained from Ministerio del Ambiente, Agua y Transición Ecológica from Ecuador (065-2024-EXP-CM-DBI/MAATE, No.048.October-2018-MEPN, MAE DNB-CM-2019-0126, and MAATE-ARSFC-2022-2583). All the animals studied carried out in accordance with ARRIVE guidelines. All the captures of spectacled bears were carried out in accordance with relevant guidelines and regulations from the Ecuadorian MAATE.
Molecular procedures
DNA from blood and skin was extracted using the phenol-chloroform procedure74. Skin samples were crushed and macerated with a pestle and mortar and then added to a 300 µl of lysis buffer. Two microliters of proteinase-k and 6 µl of DTT were added and the solution digested at 56 °C for two days. A solution of phenol-chloroform-isoamyl alcohol (25:24:1) was added to the digested sample to obtain DNA. To extract DNA from hair we added 200 µl of 10% Chelex resin, 7 µl of DDT, and 4 µl of proteinase-k and digested at 56 °C overnight75. To prep osseous tissues for DNA extraction, bone samples were ground to a powder. DNA was extracted from one to five grams of bone powder with the Blood and Tissue Kit (Qiagen, Inc.) following the protocol provided by the manufacturer.
For the first molecular data set, we used PCR amplifications and primers at the six mt genes (ND5, 16 S rRNA, 12 S rRNA, COXI, COXII, and control region)76,77,78,79. We analyzed up to 2,901 base pairs (bp) of these six mitochondrial genes. All amplifications, including positive and negative controls, were checked in 2% agarose gels. The gels were visualized in a Hoefer UV Transilluminator. Both mt DNA strands were sequenced directly using BigDye Terminator v3.1 (Applied Biosystems, Inc.). We used a 377 A (ABI) automated DNA sequencer and sequenced in both directions to ensure sequence accuracy.
For the second molecular data set, only the mt DNA from 43 specimens was of sufficient quality to be used in Illumina procedures. Standard Illumina fragment libraries (∼300-bp average insert size) were prepared for these 43 mt DNAs of high quality and were sequenced to ∼0.3× genome-wide depth of coverage on the Illumina HiSeq 2000 platform. We generated the 43 mitochondrial genome assemblies with SOAPdenovo2 v2.4.0 software (https://github.com/aquaskyline/SOAPdenovo2)80 by evaluating a series of k-mer sizes. Following assembly, all raw Illumina reads were mapped to assembled contigs using BWA-MEM v2.0 (https://github.com/bwa-mem2/bwa-mem2)81 to assess coverage depth. Mapping results were analyzed using SAMtools v1.22 software (https://github.com/samtools/)82including removal of PCR duplicates. Mitochondrial contigs were identified with ElasticBLAST v.1.4.0 software (https://blast.ncbi.nlm.nih.gov/Blast.cgi)83 and compared to other Ursidae mitochondrial genomes and confirmed by read-depth statistics.
The third molecular data set consisted of seven microsatellites (G1D, G10B, G10C, G10L, G10M, G10P and G10X, originally developed for the black bear [U. americanus])53,84. The final PCR volume reaction for nuclear microsatellites (DNA obtained by the phenol–chloroform procedure) was 25 µl with 2.5 µl of 3 mM MgCl2, 2.5 µl of 10x buffer, 2 µl of 1 mM dNTPs, 10 pmol of each primer (forward and reverse), 13 µl of H2O, 2 µl of DNA, and one unit of Taq Polymerase. For the PCR with template DNA extracted from hairs, using 10% Chelex resin, the overall volume was 50 µl, with 20 µl of DNA and twofold amounts of all other reactants. The PCR reactions were carried out in a BioRad thermocycler. The temperatures were 95 °C for 5 min, 35 cycles of 1 min at 95 °C, 1 min at the most accurate annealing temperature (52, 55–60 °C depending on the microsatellite) and 1 min at 72 °C. Following the cycles, the sample was kept 5 min at 72 °C. The PCR amplification products were run in denaturant 6% polyacrylamide gels and visualized with a Hoefer SQ3 sequencer vertical camera. They were stained with silver nitrate. The PCR reactions were repeated three times for DNA extracted from hairs, teeth and bones to confirm the genotypes obtained from these tissues. Therefore, genotyping errors due to allelic dropout were minimized.
Statistical population analysis for individual Mt genes and mitogenomes
The following genetic diversity statistics were used for the 127 spectacled bears analyzed at the six mt genes as well as for the 43 specimens analyzed at their mitogenomes: haplotype diversity (Hd), nucleotide diversity (π), and θ statistic by sequence. These genetic diversity statistics were calculated using the DNAsp v5.1 software (http://www.ub.edu/dnasp/)85.
Both, for individual mt genes and for mitogenomes, we calculated overall statistical heterogeneity indices (table of contingency, HST, KST, KST*, γST, NST, and FST86. Indirect gene flow estimates were obtained assuming an infinite island model87. Significance was estimated with permutation tests using 10,000 replicates. We also estimated genetic heterogeneity between northern (Pichincha) and southern (Loja and Zamora Chinchipe) Ecuadorian province pairs with the data set of six mt genes as well as for the 10 Ecuadorian provinces with the complete mitogenome data set. For this task, we used exact probability tests with Markov chains, using 10,000 dememorization parameters, 20 batches, and 5,000 iterations per batch. All the heterogeneity statistics were calculated with the DNAsp v5.1 (http://www.ub.edu/dnasp/) and Arlequin v3.5.1.2 programs (https://cmpg.unibe.ch/software/arlequin35/)85,88.
Different spatial analyses were applied to the six mt DNA genes as well as to the mitogenomes. The first spatial analysis was a Mantel’s test89. It was used to detect possible overall relationships between a genetic matrix among the individuals of Andean bears sampled in Ecuador (Kimura 2P genetic distance90 and the geographic distance matrix among the individuals. In this study, Mantel’s test was normalized according to Smouse and collaborators91. This procedure transforms the statistic into a correlation coefficient. Both genetic and geographic distances were logarithmized. The probability was estimated by means of 10,000 replicates. A spatial autocorrelation analysis was carried out using the Ay statistic with eight distance classes (DCs). Eight DCs were chosen because the number of comparison pairs within each DC were very similar and high enough to reach statistical significance. Ay can be interpreted as the average genetic distance between pairs of individuals that fall within a specified DC. Ay takes on a value of 0 when all individuals within a DC are genetically identical and a value of 1 when all individuals within a DC are completely dissimilar. The probability for each DC was obtained using 10,000 randomizations. The eight DCs were as follows: 0–20, 20–37, 37–53, 53–68, 68–86, 86–102, 102–124, and 124–240 km for the six mt genes data set and 0–17, 17–40, 40–53, 53–69, 69–87, 87–99, 99–111, and 111–199 km for the mitogenome data set. The real distances among the specimens studied are larger than those used in this analysis because we used geographic coordinates that do not consider the geomorphology of the area studied. These analyses were carried out with AIS v1.0 software (http://www.marksgeneticsoftware.net)92. Another spatial analysis was carried out with the Monmonier’s Algorithm Analysis (MAA)93 with the same software92. This geographical regionalization procedure is used to detect the locations of putative barriers to gene flow by iteratively identifying sets of contiguous, large genetic distances along connectivity networks94,95,96. A Delaunay triangulation was used to generate the connectivity network among sampling points97,98. A graphical representation of putative ‘‘barriers’’ inferred by the algorithm is superimposed over the connectivity network to detect rapid identification of important geographical features reflected by the genetic dataset. In this case, we used this procedure to detect the four most important geographical barriers to the six mt genes and the mitogenomes of the spectacled bears in Ecuador. Finally, we conducted a Bayesian Analysis of Population Structure with the software BAPS v6.0 (https://github.com/ocbe-uio/BAPS)99using mixed and unmixed models to group genetically similar individuals into panmictic genetic clusters. Calculations were performed, with the number of k clusters varying from 1 to 20. Five replicates were carried out for each k value to determine convergence among the results. All these spatial analyses were repeated for the six mt gene and mitogenome data sets but intermediate specimens and provinces between the most extreme areas of Ecuador (Imbabura and Pichincha in the north and Loja and Zamora Chinchipe in the south) were not included. This was carried out to simulate the same geographical conditions used by Cueva and collaborators62 and to determine whether sampling only from the most extreme areas of Ecuador could introduce some kind of bias and distortion in the results.
For the mitogenome data set, we relied on two procedures to determine a possible historical female population change in the overall Ecuadorian Andean bear sample as well as for the northern (Imbabura and Pichincha) and the southern (Loja and Zamora Chinchipe) areas. First, we used the Fu and Li D* and F* tests100, the Fu FS statistic101the Tajima D test102 and the R2 statistic103. A 95% confidence interval and probabilities were obtained with 10,000 coalescence permutations. Second, the mismatch distribution (pairwise sequence differences) was obtained following the method of Rogers and Harpending, and Rogers et al.104,105. We used the raggedness rg statistic to determine the similarity between the observed and the theoretical curves generated in this procedure106,107. The DNAsp v5.1 (http://www.ub.edu/dnasp/) software was used for this task. The female sexual maturity ranged from four to seven years108. Based on this, we used six years for a generation time and determined the time when possible population expansions began.
Statistical population analysis for nuclear microsatellites
For microsatellites, the mean number of alleles per locus (MNA), and the expected heterozygosity (He)109 were calculated for the overall Ecuadorian spectacled bear sample studied. He values were arcsine transformed prior to statistical analysis110. Kruskal–Wallis (KW) tests were performed to determine significant differences in genetic diversity among diverse populations111. If significant differences were found, a Dunn’s test was performed on pairwise comparisons to determine which pair of groupings was driving the significant difference (https://cran.r-project.org/web/packages/dunn.test)111,112.
The Hardy–Weinberg equilibrium (HWE) for the overall Ecuadorian spectacled bear population was estimated using several different analysis strategies of the microsatellites. The Robertson and Hill f statistic (R-H) and the Weir and Cockerham FIS statistic (W-C) were used to calculate the degree of excess or deficit of homozygosity and heterozygosity (complete enumeration) of the overall sample113,114. To measure the exact probabilities of the G test, the Markov chain method, with a 10,000-dememorization number, 20 batches and 5,000 iterations per batch, was used, following the Genepop v4.2.1 Program (https://kimura.univ-montp2.fr/~rousset/Genepop4.2)115. The HWE was simultaneously analyzed by locus and by population using Fisher’s method115.
To determine the genetic heterogeneity among spectacled bear populations in Ecuador, microsatellites were analyzed separately as well as collectively taken all markers. One strategy used exact tests with Markov chains, 10,000 dememorizations parameters, 20 batches, 5,000 iterations per batch, for both genotypic and genic frequencies with the Genepop v4.2.1 Program (https://kimura.univ-montp2.fr/~rousset/Genepop4.2)115. A second strategy used the Wright F-statistics116 with the Michalakis and Excoffier procedure117. To measure the significance of FST, we used 10,000 randomizations of genotypes among populations and did not assume random mating within populations by means of the log-likelihood G test118. Also, we used the RST statistic (more indicated for microsatellites than FST) to measure genetic heterogeneity among the Ecuadorian spectacled bear samples119. Possible theoretical gene-flow estimates among spectacled bear populations were measured using the private allele model120 and from FST statistics. These statistics were obtained with the FSTAT v2.9.4 software (https://www2.unil.ch/popgen/softwares/fstat.htm)119.
We developed diverse assignment analyses for microsatellites by using the GENECLASS 2 software (https://www1.montpellier.inrae.fr/CBGP/software/GeneClass/GeneClass2/)121 with Bayesian, frequency, and genetic-distance procedures. The assignation analyses were carried out without simulations and served to estimate the probabilities of individuals belonging to, or excluding from, the original populations where they were a priori assigned (p < 0.05). Some assignation analyses were also completed with 10,000 resampling simulations by means of the Monte Carlo technique and with the procedure used by Paetkau et al.63. Additionally, we estimated the possible existence of first-generation migrants in the different Ecuadorian spectacled bear populations found in the diverse provinces by using the Bayesian, frequency, and genetic distance procedures. To assist in the determination of this estimate, we considered the relationship L = Lhome/Lmax. This is the ratio of the likelihood computed from the population where the individual was sampled (Lhome) over the highest likelihood value among all population samples including the population where the individual was sampled (Lmax).
Finally, the five spatial structure procedures, used for the six mt genes and the mitogenome data sets, were also applied to the microsatellite data set (Mantel’s test, spatial autocorrelation, MAA, and BAPS). For the spatial autocorrelation analysis, the following DCs were used: 0–22, 22–46, 46–58, 58–81, 81–95, 95–107, 107–140, and 140–218 km. In this spatial autocorrelation analysis as well as in the two analyses carried out with the mitochondrial data sets, the DCs were constructed using unequal distances with equal sample sizes within the DCs.
Data availability
Some GenBank accession numbers of the spectacled bears analyzed are PP267344.1-PP267398.1, MN163059.1-MN163108.1, PV762057-PV762161 and [SUB15373612](https:/submit.ncbi.nlm.nih.gov/subs/genbank/SUB15373612) . The data sets generated and analyzed during the current study are available from the corresponding author on reasonable request at the e-mails: [mruizgar@yahoo.es](mailto: mruizgar@yahoo.es) , and [mruiz@javeriana.edu.co](mailto: mruiz@javeriana.edu.co) .
References
Myers, N., Mittermeier, R. A., Mittermeier, C. G., Da Fonseca, G. A. & Kent, J. Biodiversity hotspots for conservation priorities. Nature 403, 853 (2000).
Killeen, T. J., Douglas, M., Consiglio, T., Jørgensen, P. M. & Mejia, J. Dry spots and wet spots in the Andean hotspot. J. Biogeog. 34, 1357–1373 (2007).
Lutz, D. A., Powell, R. L. & Silman, M. R. Four decades of Andean timberline migration and implications for biodiversity loss with climate change. PLoS ONE. 8, e74496. https://doi.org/10.1371/journal.pone.0074496 (2013).
Patterson, B. D., Solari, S. & Velazco, P. M. The role of the Andes in the diversification and biogeography of Neotropical mammals. In Bones, Clones and Biomes. The History and Geography of Recent Neotropical Mammals (eds. Patterson, B. D. & Costa, L. P.) 51–378 (The University of Chicago Press, 2012).
Macdonald, D. The New Encyclopedia of Mammals (Oxford University Press, 2001).
Paisley, S. Andean bear and people in Apolobamba, Bolivia: culture, conflict and conservation. PhD thesis, University of Kent, Kent, UK (2001).
García-Rangel, S. On Andean bear (Tremarctos ornatus) natural history and conservation. Mammal Rev. 42, 85–119 (2012).
Cuvier, F. Des Dents Des Mammiferes Considerees Comme Caracteres Zoologiques. Monographie (Le Normant, 1825).
Cabrera, A. Catálogo De Los Mamíferos De America Del Sur 1 (Metatheria, Unguiculata, Carnivora) (Imprenta y Casa Editorial Conil, 1957).
Gervais, P. Tremarctos. Hist. Nat. Mamm. 2, 20 (1855).
Gill, T. On the genera Tremarctos, gervais (Nearctos, Gray), and Aelurina, gervais (Ailurogale, Fitz). Annals Magaz Nat. Hist. Including Zool. Bot. Geol. 13, 15–16 (1874).
Kurtén, B. Pleistocene bears of North america, II: genus Arctodus, short-faced bears. Acta Zool. Fenn. 117, 1–160 (1967).
Thenius, E. Zur stammesgeschichtlichen herkunft von tremarctos (Ursidae, Mammalia). Z. Säugetierk. 41, 109–114 (1976).
Wilson, D. E. & Reeder, D. M. Mammal Species of the World. A Taxonomic and Geographic Reference 3rd edn (Johns Hopkins University, 2005).
Garshelis, D. L. Family ursidae. In Handbook of the Mammals of the World: Volume 1. Carnivores (eds. Wilson, D. E. & Mittermeier, R. A.) 448–497 ( Lynx Edicions, 2009).
María, A. Tremarctos lasallei. Bol. Soc. Colom Cien Nat. 13, 113–118 (1924).
Sclater, P. L. Remarks upon a bear recently added to the Society’s Menagerie (Plate VIII). Proc. Zool. Soc. London 1868, 71–73 (1868).
Gray, J. E. On the skull of the spectacled bear of Peru and of the Helarctos from Malacca and Java. Annals Magaz Nat. Hist. Including Zool. Bot. Geol. 12, 182–183 (1873).
Thomas, O. On the bear of Ecuador. Annals Magaz Nat. Hist. Including Zool. Bot. Geol. 9, 215–217 (1902).
Hornaday, W. The spectacled bear. Zool. Soc. Bull. 45, 747–748 (1911).
Krause, J. et al. Mitochondrial genomes reveal an explosive radiation of extinct and extant bears near the Miocene-Pliocene boundary. BMC Evol. Biol. 8, 1–12 (2008).
Kitchener, A. C. Taxonomic issues in bears: impacts on conservation in zoos and the wild, and gaps in current knowledge. Inter Zoo Yearbook. 44, 33–46 (2010).
Mondolfi, E. Notes on the distribution, habitat, food habits, status and conservation of the spectacled bear (Tremarctos ornatus) in Venezuela. Mammalia 53, 525–544 (1989).
Peyton, B. et al. Status of wild Andean bears and policies for their management. Ursus 10, 87–100 (1988).
Del Moral, J. F. & Bracho, A. E. Indicios indirectos de La presencia Del Oso Andino (Tremarctos ornatus cuvier, 1825) En El Noroeste de Argentina. Rev. Mus. Argen Cienc. Nat. 11, 69–76 (2009).
Del Moral, J. F. & Lameda, F. I. Registros de ocurrencia Del Oso Andino (Tremarctos ornatus cuvier, 1825) En Sus límites de Distribución nororiental y Austral. Rev. Mus. Argen Cienc. Nat. 13, 7–19 (2011).
Rumiz, D. I. et al. El Ucumar (Tremarctos ornatus), Mito y Realidad de Su presencia En La Argentina. Mastozool Neotrop. 19, 359–366 (2012).
Cosse, M., Del Moral Sachetti, J. F., Mannise, N. & Acosta, M. Genetic evidence confirms presence of Andean bears in Argentina. Ursus 25, 163–171 (2014).
Rodríguez, E. D. et al. Ecoregional strategy for the conservation of the spectacled bear (Tremarctos ornatus) in the Northern Andes. WWF Colombia, Fundación Wii, EcoCiencia, Wildlife Conservation Society, and Red Tremarctos. Bogotá, Colombia (2003).
Peyton, B. Ecology, distribution, and food habits of spectacled bears, Tremarctos ornatus, in Peru. J. Mammal. 61, 639–652 (1980).
Sánchez-Mercado, A., Ferrer-Paris, J. R., Yerena, E. & García Rangel, S. Rodríguez-Clark, K. M. Factors affecting poaching risk to vulnerable Andean bears Tremarctos ornatus in the cordillera de mérida, venezuela: space, parks and people. Oryx 42, 437–447 (2008).
Figueroa, J. & Stucchi, M. El Oso Andino: Alcances Sobre Su Historia Natural (Lima, 2009).
B. L.J. et al. Impacts of climate change on the world’s most exceptional ecoregions. Proc. Natl. Acad. Sci. USA. 108, 2306–2311 (2011).
Tovar, C., Arnillas, C. A., Cuesta, F. & Buytaert, W. Diverging responses of tropical Andean biomes under future climate conditions. PloS ONE. 8, e63634 (2013).
IPCC. Summary for Policymakers. Climate Change 2013: the Physical Science Basis. Contribution of Working Group I To the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, Intergovernmental Panel on Climate Change (Cambridge University Press, 2013).
Yu, L., Li, Q. W., Ryder, O. A. & Zhang, Y. P. Phylogeny of the bears (Ursidae) based on nuclear and mitochondrial genes. Mol. Phylogenet Evol. 32, 480–494 (2004).
Kutschera, V. E. et al. Bears in a forest of gene trees: phylogenetic inference is complicated by incomplete lineage sorting and gene flow. Mol. Biol. Evol. 31, 2004–2017 (2014).
Kumar, V. et al. The evolutionary history of bears is characterized by gene flow across species. Sci. Rep. 7, 46487 (2017).
Nash, W. G. & O’Brien, S. J. A comparative chromosome banding analysis of the ursidae and their relationship to other carnivores. Cytogenet. Cell. Genet. 45, 206–212 (1987).
O’Brien, S. J. & Knight, K. The future of the giant panda. Nature 325, 758–759 (1987).
Goldman, D., Giri, P. R. & O’Brien, S. J. Molecular genetic-distance estimates among the ursidae as indicated by one- and two-dimensional protein electrophoresis. Evolution 43, 282–295 (1989).
Waits, L. P., Sullivan, J., O’Brien, S. J. & Ward, R. H. Rapid radiation events in the family ursidae indicated by likelihood phylogenetic Estimation from multiple fragments of MtDNA. Mol. Phylogenet Evol. 13, 82–92 (1999).
Newnham, R. (ed Davidson, W.) Comparative study of the karyotypes of several species in carnivora including the giant panda (Ailuropoda melanoleuca). Cytogenetics 5, 152–163 (1966).
Newnham, R. & Davidson, W. Authors addendum to: comparative study of the karyotypes of several species in carnivora, including the giant panda (Ailuropoda melanoleuca). Cytogenetics 6, 156–157 (1967).
Tian, Y., Nie, W., Wang, J., Ferguson-Smith, M. A. & Yang, F. Chromosome evolution in bears: reconstructing phylogenetic relationships by cross-species chromosome painting. Chromosome Res. 12, 55–63 (2004).
Wienberg, J. The evolution of the eutherian chromosomes. Genomes Evol. 14, 657–666 (2004).
Ruiz-Garcia, M. Molecular population genetic analysis of the spectacled bear (Tremarctos ornatus) in the Northern Andean area. Hereditas 138, 81–93 (2003).
Ruiz-García, M., Orozco-terWengel, P., Castellanos, A. & Arias, L. Microsatellite analysis of the spectacled bear (Tremarctos ornatus) across its range distribution. Genes Genet. Syst. 80, 57–69 (2005).
Ruiz-García, M. The genetic demography history and phylogeography of the Andean bear (Tremarctos ornatus) by means of microsatellites and MtDNA markers. In Molecular Population Genetics, Evolutionary Biology and Conservation of Neotropical Carnivores (eds. Ruiz-García, M. & Shostell, J. M.) 129–158 (Nova Science, 2013).
Ruiz-García, M., Arias, J. Y., Restrepo, H., Cáceres-Martínez, C. H. & Shostell, J. M. The genetic structure of the Andean bear (Tremarctos ornatus; Ursidae, Carnivora) in Colombia by means of mitochondrial and micro satellite markers. J. Mammal. 101, 1072–1090 (2020).
Ruiz-García, M., Castellanos, A., Arias-Vásquez, J. Y. & Shostell, J. M. Genetics of the Andean bear (Tremarctos ornatus; Ursidae, Carnivora) in ecuador: when the Andean cordilleras are not an obstacle. Mitochondrial DNA Part. A. 31, 190–208 (2020).
Waits, L., Taberlet, P., Swenson, J. E., Sandegren, F. & Franzén, R. Nuclear DNA microsatellite analysis of genetic diversity and gene flow in the Scandinavian brown bear (Ursus arctos). Mol. Ecol. 9, 421–431 (2000).
Paetkau, D. & Strobeck, C. Microsatellite analysis of genetic variation in black bear populations. Mol. Ecol. 3, 489–495 (1994).
Ruiz-García, M., Arias-Vásquez, J. Y., Castellanos, A., Kolter, L. & Shostell, J. M. Molecular evolution (mitochondrial and nuclear microsatellites markers) in the Andean bear (Tremarctos ornatus; Ursidae, Carnivora): how many ESUs are there? In Conservation genomics of mammals (eds. Ortega J. & Maldonado, J. E.)165–193 (Springer, 2020).
Moritz, C. Defining evolutionarily significant units for conservation. Trends Ecol. Evol. 9, 373–375 (1994).
Dobzhansky, T. Evolutionary oscillations in Drosophila pseudoobscura. In Ecological Genetics and Evolution (ed. Ford, E. B.) 109–133 (Blackwell Scientific, 1971).
Eckert, C. G., Samis, K. E. & Lougheed, S. C. Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol. Ecol. 17, 1170–1188 (2008).
Ruiz-García, M. & de Genética Poblaciones: Teoría y Aplicación a La conservación de Mamíferos Neotropicales (Oso Andino y Delfín rosado). Bol. Real. Soc. Esp. Hist. Nat. 102, 99–126 (2007).
Ruiz-García, M., Orozco-terWengel, P., Payán, E. & Castellanos, A. Genética de poblaciones molecular Aplicada al estudio de Dos grandes carnívoros (Tremarctos ornatus – Oso andino, Panthera onca - jaguar): lecciones de conservación. Bol. Real. Soc. Esp. Hist. Nat. 98, 135–158 (2003).
Viteri, M. P. & Waits, L. P. Identifying polymorphic microsatellite loci for Andean bear research. Ursus 20, 102–108 (2009).
Cueva, D. F., Gutierrez, B., Bruque, G., Molina, S. & Torres, M. L. Mitochondrial DNA reveals low genetic diversity in Ecuadorian Andean bears. Ursus 29, 43–51 (2018).
Cueva, D. F. et al. Evidence of population genetic structure in Ecuadorian Andean bears. Sci. Rep. 14, 2834. https://doi.org/10.1038/s41598-024-53003-5 (2024).
Paetkau, D., Slade, R., Barden, M. & Estoup, A. Genetic assignment methods for the direct, real-time Estimation of migration rate: a simulation based exploration of accuracy and power. Mol. Ecol. 13, 55–65 (2004).
Rannala, B. & Mountain, J. L. Detecting immigration by using multilocus genotypes. Proc. Nat. Acad. Sci. USA 94, 9197–9201 (1997).
Badouin, L. & Lebrun, P. An operational Bayesian approach for the identification of sexually reproduced cross-fertilized populations using molecular markers. In Proceedings of the international symposium on molecular markers for characterizing genotypes and identifying cultivars in horticulture (eds. Dore, C. et al.) 6–9 (2000).
Nei, M., Tajima, F. & Tateno, Y. Accuracy of estimated phylogenetic trees from molecular data. J. Mol. Evol. 19, 153–170 (1983).
Andrews, T. D. & Easteal, S. Evolutionary rate acceleration of cytochrome c oxidase subunit in Simian primates. J. Mol. Evol. 50, 562–569 (2000).
Collins, A. C. & Dubach, J. M. Phylogenetic relationships of spider monkeys (Ateles) based on mitochondrial DNA variation. Int. J. Primatol. 21, 381–420 (2000).
Ascunce, M. S., Hasson, E. & Mudry, M. D. COII: A useful tool for inferring phylogenetic relationships among new world monkeys (Primates, Platyrrhini). Zool. Scrip. 32, 397–406 (2003).
Ruiz-García, M., Pinedo-Castro, M. & Shostell, J. M. How many genera and species of woolly monkeys (Atelidae, platyrrhine, Primates) are? First molecular analysis of Lagothrix flavicauda, an endemic Peruvian primate species. Mol. Phylogenet Evol. 79, 179–198 (2014).
Ruiz-García, M., Luengas, K., Leguizamón, N., Thoisy, B. & Gálvez, H. Molecular phylogenetics and phylogeography of all the Saimiri species (Cebidae, Primates) inferred from Mt COI and COII gene sequences. Primates 56, 145–161 (2015).
Ruiz-García, M. et al. Phylogeography of the mantled howler monkey (Alouatta palliata; atelidae, Primates) across its geographical range by means of mitochondrial gene analyses and some insights in the phylogeny of Alouatta. Folia Primatol. 88, 421–454 (2017).
Ruiz-García, M., Albino, A., Pinedo-Castro, M. & Shostell, J. M. First molecular phylogenetics analysis of the Lagothrix taxon living in Southern Peru and Northern bolivia: Lagothrix lagotricha tschudii (Atelidae, Primates), a new subspecies. Folia Primatol. 90, 215–239 (2019).
Sambrock, J., Fritsch, E. & Maniatis, T. Molecular Cloning: a Laboratory Manual 2nd edn (Cold Spring Harbor Laboratory Press, 1989).
Walsh, P. S., Metzger, D. A. & Higuchi, R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 10, 506–513 (1991).
Folmer, O., Black, M., Hoeh, W., Lutz, R. & Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotech. 3, 294–299 (1994).
Zhang, B. et al. Genetic viability and population history of the giant panda, putting an end to the evolutionary dead end? Mol. Biol. Evol. 24, 1801–1810 (2007).
Zhang, Y. P. & Ryder, O. A. Mitochondrial DNA sequence evolution in the arctoidea. Proc. Nat. Acad. Sci. USA. 90, 9557–9561 (1993).
Zhang, Y. P. & Ryder, O. A. Phylogenetic relationships of bears (the Ursidae) inferred from mitochondrial DNA sequences. Mol. Phylogenet Evol. 3, 351–359 (1994).
Luo, R. et al. SOAPdenovo2: an empirically improved memory-efficient short read de Novo assembler. Gigascience 1, 18 (2012).
Li, H. & Durbin, R. Fast and accurate short read alignment with burrows Wheeler transform. Bioinformatics 25, 1754–1760 (2009).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Altschul, S. F. et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 (1997).
Paetkau, D., Calvert, W., Stirling, I. & &Strobeck, C. Microsatellite analysis of population structure in Canadian Polar bears. Mol. Ecol. 4, 347–354 (1995).
Librado, P. & Rozas, J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452 (2009).
Hudson, R., Boss, D. D. & Kaplan, N. L. A statistical test for detecting population subdivision. Mol. Phylogenet Evol. 9, 138–151 (1992).
Wright, S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution 19, 395–420 (1965).
Excoffier, L. & Lischer, H. E. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol. Ecol. Res. 10, 564–567 (2010).
Mantel, N. A. The detection of disease clustering and a generalized regression approach. Cancer Res. 27, 209–220 (1967).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Smouse, P. E., Long, J. C. & Sokal, R. R. Multiple regression and correlation extension of the mantel test of matrix correspondence. Syst. Zool. 35, 627–632 (1986).
Miller, M. P. Alleles in space: computer software for the joint analysis of interindividual Spatial and genetic information. J. Hered. 96, 722–724 (2005).
Monmonier, M. S. Maximum-difference barriers: an alternative numerical regionalization method. Geogr. Anal. 5, 245–261 (2010).
Dupanloup, I., Schneider, S. & Excoffier, L. A simulated annealing approach to define the genetic structure of populations. Mol. Ecol. 11, 2571–2581 (2002).
Manel, S., Schwartz, M. L., Luikart, G. & Taberlet, P. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol. Evol. 18, 189–197 (2003).
Manni, F., Guerard, E. & Heyer, E. Geographic patterns of (genetic, morphologic, linguistic) variation: how barriers can be detected by using monmonier’s algorithm. Hum. Biol. 76, 173–190 (2004).
Watson, D. F. Contouring: a Guide To the Analysis and Display of Spatial Data (Pergamon, 1992).
Brouns, G., De Wulf, A. & Constales, D. Delaunay triangulation algorithms useful for multibeam echosounding. J. Surv. Eng. 129, 79–84 (2003).
Corander, J., Marttinen, P., Sirén, J. & Tang, J. B. A. P. S. Bayesian Analysis of Population Structure. Department of Mathematics, Åbo Akademi University: Helsinki, Finland (2013). http://www.abo.fi/mnf/mate/jc/smack_index_eng.html.
Fu, Y. & Li, W. Statistical tests of neutrality of mutations. Genetics 133, 693–709 (1993).
Fu, Y-X. Statistical tests of neutrality against population growth, hitchhiking and background selection. Genetics 147, 915–925 (1997).
Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989).
Ramos-Onsins, S. E. & Rozas, J. Statistical properties of new neutrality tests against population growth. Mol. Biol. Evol. 19, 2092–2100 (2002).
Rogers, A. R. & Harpending, H. C. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 9, 552–569 (1992).
Rogers, A. R., Fraley, A. E., Bamshad, M. J., Watkins, W. S. & Jorde, L. B. Mitochondrial mismatch analysis is insensitive to the mutational process. Mol. Biol. Evol. 13, 895–902 (1996).
Harpending, H. C. Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Hum. Biol. 66, 591–600 (1994).
Harpending, H. C., Sherry, S. T., Rogers, A. R. & Stoneking, M. Genetic structure of ancient human populations. Curr. Anthropol. 34, 483–496 (1993).
Stirling, I. Bears: Majestic Creatures of the Wild 240 (Rodale, 1993).
Nei, M. Analysis of gene diversity in subdivided populations. Proc. Nat. Acad. Sci. USA. 70, 3321–3323 (1973).
Archie, J. W. Statistical analysis of heterozygosity data: independent sample comparisons. Evolution 39, 623–637 (1985).
R Core Team. R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2019).
Dinno, A. Dunn’s Test of Multiple Comparisons Using Rank Sums. R package version, 1.4 (2017, accessed 12 Oct 2020). https://www.cran.r-project.org/web/%20packages/dunn.test/index.html.
Robertson, A. & Hill, W. G. Deviations from Hardy-Weinberg proportions, sampling variances and use in Estimation of inbreeding coefficients. Genetics 107, 703–718 (1984).
Weir, B. S. & Cockerham, C. C. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984).
Raymond, M. & Rousset, F. An exact test for population differentiation. Evolution 49, 1280–1283 (1995).
Wright, S. The genetical structure of populations. Ann. Eugenet. 15, 323–354 (1951).
Michalakis, Y. & Excoffier, L. A generic Estimation of population subdivision using distances between alleles with special reference to microsatellite loci. Genetics 142, 1061–1064 (1996).
Goudet, J., Raymond, M., Demeeus, T. & Rousset, F. Testing differentiation in diploid populations. Genetics 144, 1933–1940 (1996).
Goodman, S. J. RST calc: a collection of computer programs for calculating estimates of genetic differentiation from microsatellite data and determining their significance. Mol. Ecol. 6, 881–885 (1997).
Slatkin, M. Rare alleles as indicators of gene flow. Evolution 39, 53–65 (1985).
Piry, S. et al. GENECLASS2: a software for genetic assignment and first-generation migrant detection. J. Hered. 95, 536–539 (2004).
Acknowledgements
AcknowledgmentsThe authors wish to express their gratitude to the Academic Vicerectory of the Pontificia Universidad Javeriana at Bogotá (Colombia) and to Corponariño (Aida Delgado, Diana Ocaña Saltikova) at Pasto (Nariño, Colombia) for providing monetary resources to carry out this research. Thanks also to Termas Papallacta (Ecuador) and to the Zoo Conservation Outreach Group (Kansas City Zoo) as well as to Melchor Ascanta, David Jackson, and Rodrigo Ascanta for their help in obtaining spectacled bear samples across Ecuador. Thanks also to the Instituto Nacional de Biodiversidad from Ecuador (INABIO) for their logistic support and approval to conduct research in Ecuador.
Author information
Authors and Affiliations
Contributions
M.R-G: conceptualization, sample collection, formal analysis, supervision, funding acquisition, methodology, writing—original draft, writing—review and editing. A.C: sample collection, methodology, writing—review and editing. J.A-V: funding, formal analysis, writing—review and editing. J.M.S: formal analysis, supervision, writing—review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ruiz-García, M., Castellanos, A., Árias-Vásquez, J.Y. et al. Effects of the Ecuadorian Andes on genetic structure of the spectacled bear with new genetic datasets. Sci Rep 15, 32735 (2025). https://doi.org/10.1038/s41598-025-17839-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-17839-9