Abstract
Lung cancer ranks among the top causes of cancer-related deaths globally. Prompt diagnosis and timely intervention play a vital role in enhancing patient prognosis and increasing survival chances. Although C15ORF48 expression is generally elevated in tumor tissues, it is reduced in specific cancers like colorectal cancer, and its role across pan-cancer remains unclear. This research investigates the expression, immune infiltration, and predictive significance of C15ORF48 in pan-cancer using multiple databases, including TIMER2.0, GEPIA, and the Human Protein Atlas. Additionally, we use cBioPortal, PhosphoNET, and AlphaFold to analyze the distribution of genetic alterations, phosphorylation sites, and pathogenic hotspots in C15ORF48. C15ORF48 expression is generally elevated in tumor versus normal tissues, and its distribution in lung tissues is predominantly cytoplasmic. C15ORF48 expression is positively correlated with the level of expression shows a positive association with the degree of CD8 + T cell infiltration.,etc. Furthermore, higher C15ORF48 expression is associated with poorer survival in lung adenocarcinoma, etc. Genetic alterations in C15orf48, including S29F, T38I, and K61N, and predicted phosphorylation sites such as S28, S29, T38, and T53, are identified across various cancers. Additionally, pathogenic hotspots, including G25D, S28K, and T38W, are highlighted. The results indicate that C15ORF48 could play a significant role in pan-cancer, particularly in lung cancer, and may serve as a potential biomarker for prognosis and targeted therapies.
Similar content being viewed by others
Introduction
Cancer is the second leading cause of death in the United States and the primary cause among individuals under 85 years. Incidence rates rose gradually from 1975, peaking at approximately 500 per 100,000 around 1990, before declining and stabilizing at about 470 per 100,000 by 20201. Lung cancer ranks as the second most prevalent malignancy globally and remains a primary contributor to cancer-associated mortality2. And lung cancer as the leading cause of cancer-related deaths, with an estimated 2.5 million new cases and 1.8 million deaths in 20223. In the United States alone, an estimated 234,580 new cases of lung and bronchial cancer are projected for 2024, with approximately 125,070 deaths attributed to these diseases, accounting for about 21% of all cancer-related mortality1. Cancer treatment has advanced from early surgical and radiotherapeutic approaches to contemporary multimodal strategies incorporating chemotherapy, targeted agents, and immunotherapies4. Looking ahead, future therapies will prioritize early detection, advanced immunotherapies, nanomedicines, stem cell interventions, and emerging techniques5. For lung cancer in particular, timely diagnosis and timely therapy of lung cancer are crucial for improving patient survival rates6. However, there is still a lack of reliable biomarkers for early diagnosis. Therefore, identifying new biomarkers and targeted therapeutic pathways remains an urgent research challenge.
C15ORF48 was initially discovered in studies of esophageal squamous cell carcinoma (ESCC)7 exhibits high expression levels in esophageal tissues and also significantly expressed in stomach and intestines8. The role of C15ORF48 remains unclear, but it has been identified as being associated with inflammatory responses9. Its expression has been shown to be reduced in colorectal cancer10. Methylation of C15orf48, which is associated with gene downregulation, has been confirmed in cervical squamous cell carcinoma cells11. Future studies could employ CRISPR screening to identify C15ORF48 as a drug resistance target, similar to approaches revealing key genes for trametinib resistance12. This could elucidate C15ORF48’s role in resistance mechanisms and guide targeted therapies. Abnormal expression of this gene has also been observed in several other types of cancer13,14. Recent studies have advanced our understanding of tumor microenvironment (TME) dynamics and biomarker discovery in pan-cancer contexts, providing a robust framework for integrating multi-omics data to predict immunotherapy responses. For instance, tools like IOBR enable systematic TME deconvolution and signature scoring, facilitating the identification of immune-related patterns across malignancies15. Similarly, optimized dynamic network biomarkers have deciphered high-resolution heterogeneity in thyroid cancers, highlighting critical transitions in progression that could inform targeted interventions16. Furthermore, machine learning frameworks such as iMLGAM have demonstrated potential in predicting pan-cancer immunotherapy outcomes through genetic algorithm-driven multimatics analysis17. These approaches underscore the need for comprehensive analyses like ours, which integrate expression, mutations, and modifications for C15ORF48.
At present, research on this gene has mainly focused on adenosquamous carcinoma and colorectal cancer, with relatively little exploration in other types of cancer. In the future, additional investigations are required to broaden the research focus and clarify the involvement of C15ORF48 in pan-cancer. This will facilitate a deeper comprehension of its wider biological functions in oncology and assess its viability as a candidate for targeted therapy.
This study employs bioinformatics approaches, integrating multiple public databases, to explore the expression patterns of C15ORF48 across pan-cancer and its potential mechanisms of action.
Materials and methods
Analysis of C15ORF48 expression levels in normal and cancerous tissues by Timer2.0 and GEPIA
We used the TIMER2.0 database to assess C15orf48 expression differences between normal and cancerous tissues. TIMER 2.0 is a widely used web server in tumor immunology research. It employs a range of computational approaches to quantify immune cell infiltration within TCGA tumor samples or transcriptomic datasets. The platform assists users in identifying associations between different cancer types, as well as between clinical and genomic features, enabling comprehensive analysis and visualization of tumor-infiltrating immune cells.
The TIMER2.0 (http://timer.cistrome.org/) database was used to analyze differential gene expression of C15ORF48 across cancer types and normal tissues via the Gene DE module, which is primarily applied to investigate the differential expression levels of specific genes across multiple cancer types and normal tissues in TCGA18,19,20.
To validate these findings, the GEPIA database was employed, integrating TCGA and GTEx data.
GEPIA(http://gepia.cancer-pku.cn/) is a comprehensive data analysis tool that integrates information from TCGA and GTEx databases, specifically intended for analyzing gene expression between tumor and normal tissues21. Its capabilities include patient survival evaluation, identification of genes with similar profiles, correlation assessments, and dimensionality reduction techniques, along with various additional functions.
On the GEPIA homepage, we entered “C15ORF48” into the “Enter gene name” input box and clicked the “go pia” button to initiate the search. This operation generated a “Gene Expression Profile” across all tumor samples and paired normal tissues, allowing us to conduct further detailed analysis.
Using the human protein atlas database to investigate the localization of C15ORF48 in lung
Protein expression and localization were examined using the Human Protein Atlas (https://www.proteinatlas.org/), a comprehensive database offering detailed data on protein expression levels and their localization within human tissues and cells, including data on normal tissues, cancer tissues, cell lines, and subcellular locations22,23,24,25,26,27,28. We navigated to the platform’s homepage. In the search bar, input “C15ORF48” and click “Search” to proceed to the next page. Then, selected the entry for the C15ORF48 molecule to view detailed information. C15ORF48 was searched, and lung tissue staining was reviewed under the “Tissue” tab. Subsequently, under the “LUNG -Antibody staining” section of the page, the corresponding image was located.
Relationship between C15ORF48 expression levels and immune infiltration
TIMER2.0’s “Immune” module analyzed associations between C15orf48 and immune alterations or clinical features. C15orf48 was input, and cells like CD8 + T cells, neutrophils, common lymphoid progenitors, and gamma-delta T cells were selected to evaluate infiltration correlations.
Association between C15ORF48 expression levels and patient survival
Survival impact was assessed via GEPIA’s “Survival” tab, with C15orf48 input, overall survival selected, median cutoff, hazard ratio/95% confidence interval enabled, months as axis units, and datasets like LUAD, LGG, LIHC, and SKCM chosen for plotting.
Investigating the association between C15orf48 and tumor mutations using cBioPortal
To further explore the connection between C15orf48 gene and tumors, the cBioPortal platform was employed to explore the association between C15orf48 and gene mutations. cBioPortal(https://www.cbioportal.org/) is a powerful online tool that enables the mining, transformation, and visualization of cancer genomics datasets29,30,31. “Quick Search” queried C15orf48 for alteration distributions. The “Mutations” tab added “Cancer Hotspots” and “Post-Translational Modifications (dbPTM)” annotations for locus mapping.
Prediction of C15orf48 phosphorylation sites using the PhosphoNET database
Post-translational modifications, particularly phosphorylation, represent a crucial mechanism for regulating protein function. To predict whether C15orf48 contains phosphorylation sites and to identify their locations, the PhosphoNET database is utilized. PhosphoNET is a comprehensive platform designed to explore and analyze protein phosphorylation sites and their potential biological roles32. Navigate to the PhosphoNET website (http://www.phosphonet.ca/), “C15orf48” was queried for results.
Forecast the three-dimensional configuration and pathogenic hotspots of C15orf48 by AlphaFold
To predict the three-dimensional structure and pathogenic hotspots of C15orf48, start by accessing the AlphaFold database (https://alphafold.com/). AlphaFold is an AI system capable of predicting a protein’s three-dimensional structure based on its amino acid sequence33,34,35. Access the AlphaFold database and enter “C15orf48” to query its predicted structure. From the results, select the entry with “Source Organism” as Homo sapiens to obtain the predicted structure and pathogenicity heatmap. Next, integrate phosphorylation sites from PhosphoNET and tumor hotspots from cBioPortal. From the pathogenicity heatmap, select key sites such as G25D, S28K, S29K, T38W, and K61N. Clicking “AlphaMissense Pathogenicity” will automatically highlight the corresponding amino acid locations on the 3D structure.
Results
The expression level of C15ORF48 is generally higher in cancerous tissues compared to normal tissues
As illustrated in the Figure S1A, the box plot enables a clear depiction of the median expression, range, and significant differences between cancer and normal samples. Overall, this diagram presents the distribution of mRNA expression levels of the C15ORF48 gene in cancer tissues and normal tissues across various cancer types. A total of 33 cancer types is included in the analysis, with 19 of them showing significant statistical differences. Notably, in these 19 cancer types, the expression level of the C15ORF48 gene is markedly higher in cancer tissues in comparison with the corresponding normal tissues in 14 of the cases. In summary, the expression of C15ORF48 is typically elevated in tumor tissues relative to normal counterparts.
In Figure S1B, A total of 33 cancer types is included. Among these, 23 cancer types exhibit statistically significant differences in gene expression, with 20 of them showing significantly higher C15ORF48 gene expression levels within tumor samples relative to matched normal tissues.
In Figure S1C, Among the 31 cancer types analyzed, a statistically significant difference was observed in several cases. Notably, in 26 of these cancer types, the level of gene expression of the C15ORF48 gene was markedly elevated in tumor tissues relative to their matched normal counterparts.
C15ORF48 exhibits low expression levels in lung tissue, characterized by weak staining signals overall
Figure S2 reveals that C15ORF48 shows weak expression in airway epithelial cells, with minimal or no detectable staining in the surrounding alveolar regions. Prior investigations have shown that C15ORF48 is predominantly expressed in the gastrointestinal tract, which aligns with these findings.
C15ORF48 expression shows a notable positive relationship with the infiltration levels of several immune cell types
Figure S3 illustrates the association between the C15ORF48 gene and infiltration of different immune cells. From the figure, it can be concluded that a significant association exists between C15ORF48 gene expression and the infiltration levels of various immune cell populations.
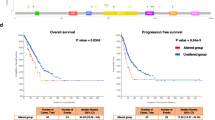
The expression level of C15ORF48 is negatively correlated with the survival rates of patients with LUAD, LGG, LIHC, and SKCM
Figure S4A demonstrates the correlation between C15ORF48 gene expression and overall survival in LUAD cases. The blue and red curves indicate the low and high C15ORF48 expression groups, respectively. Survival analysis revealed a statistically significant disparity between groups, with patients exhibiting elevated C15ORF48 expression demonstrating poorer survival outcomes. These findings indicate an inverse relationship between C15ORF48 expression and overall survival, though disease-free survival showed no statistically significant association. Additionally, the following Figure S4B、S4C、S4D indicate that C15ORF48 expression levels in cancers such as LGG, LIHC, and SKCM are also negatively correlated with patient survival. Whether this phenomenon reflects a general trend warrants further research and validation.
Frequency of C15orf48 alterations across various cancer types
The figure S5A demonstrates the distribution of different genetic alterations in the C15orf48 gene in multiple cancer types. The highest observed frequency of genetic alterations reaches approximately 4%. In total, 32 cancer types are represented, with genetic alterations detected in 22 of them. Notably, genetic alterations are present in both lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC).
Distribution and characteristics of C15orf48 mutation sites in cancer
In Figure S5B, key mutations include S29F in colon adenocarcinoma, T38I in melanoma, and K61N in rectal adenocarcinoma (full list in Supplementary Table S1).
These mutation sites may have significant impacts on protein function, and further research could help elucidate the specific roles these mutations play in cancer development. Epigenetic modifications also play a crucial role in gene expression and the progression of diseases. Key types of epigenetic modifications include acetylation, phosphorylation, and glycosylation. In the C15orf48 gene, acetylation is observed at position 9, while glycosylation occurs at positions 38, 53, and 57. Mutations at these sites could also contribute to disease and represent potential pathogenic targets for future studies.
Phosphorylation sites of C15orf48 and their research significance
From Figure S6, it can be observed that C15orf48 contains five potential phosphorylation sites: S28, S29, T38, T53, and T57. Among these, S28 and S29 have been experimentally validated in mammals, providing robust evidence for their phosphorylation. Although T38, T53, and T57 lack direct experimental validation, they have been predicted as likely phosphorylation sites, warranting further investigation. Additionally, analysis via cBioPortal reveals that T38, T53, and T57 are mutation hotspots in tumors and overlap with regulatory phosphorylation sites, suggesting their potential significance. These sites may serve as promising candidates for tumor screening and warrant detailed exploration in future studies.
The modeled 3D structure of C15orf48 and its potential pathogenic sites
From Figure S7A, it indicates that the majority of the predicted model is colored blue, indicating a high level of confidence in the predictions. From Fig. 7B, it can be observed that the red-marked sites represent amino acid residues with higher pathogenicity, including critical sites such as G25, S28, S29, T38, and K61. These sites are likely closely associated with the regulation of protein function and disease mechanisms, warranting further investigation. From Figure S7C, it can be seen that when glycine (G) at position 25 is replaced by aspartic acid (D), the pathogenicity score is 0.765, the pLDDT score is 97.99, and the AlphaMissense Pathogenicity score is 0.415. This indicates that the G25 mutation has a moderate level of pathogenic potential, while the high structural stability score suggests it may significantly impact protein function. From Figure S7D, it is evident that when serine (S) at position 28 is replaced by lysine (K), the pathogenicity score is 0.969, the pLDDT score is 97.48, and the AlphaMissense Pathogenicity score is 0.443. This result suggests that the S28 mutation exhibits a high pathogenic potential. Moreover, the predictive model indicates that this site is structurally stable, implying its potential role in protein activity. From Figure S7E, it can be seen that when serine (S) at position 29 is replaced by lysine (K), the pathogenicity score is 0.918, the pLDDT score is 95.54, and the AlphaMissense Pathogenicity score is 0.400. This indicates a significant pathogenic potential for the mutation. Combined with the stability prediction, S29 is likely to influence the functional domain of C15orf48. From Figure S7F, it can be observed that when threonine (T) at position 38 is replaced by tryptophan (W), the pathogenicity score is 0.597, the pLDDT score is 83.02, and the AlphaMissense Pathogenicity score is 0.280. This result suggests that the T38 mutation has a certain level of pathogenicity, and its structural changes may have potential impacts on the local structure or interaction interface of the protein.From Figure S7G, it can be seen that when threonine (T) at position 38 is again replaced by tryptophan (W), the pathogenicity score increases to 0.913, the pLDDT score is 89.75, and the AlphaMissense Pathogenicity score rises to 0.872. This high pathogenicity score Implies that T38 may play a critical role in disease occurrence under specific environmental or functional contexts.
Discussion
This study systematically applied bioinformatics tools to comprehensively analyze the expression patterns, genetic mutations, phosphorylation modifications of C15ORF48 across multiple cancer types, uncovering its potential biological functions and clinical relevance in cancer.
Our study reveals elevated C15ORF48 expression in cancer tissues versus normal tissues across multiple cancer types, suggesting its involvement in tumorigenesis and progression. These findings are consistent with the study by Chen et al., which reported elevated C15ORF48 expression in various cancers and its association with macrophage infiltration in thyroid cancer (THCA)14. Furthermore, our study observed that C15ORF48 is expressed at relatively low levels in lung tissues and is primarily localized to the cytoplasm. This result aligns with earlier research indicating that C15ORF48 is predominantly expressed in healthy gastrointestinal tissues7 suggesting a tissue-specific regulatory role for C15ORF48. In terms of the immune microenvironment, it reveals a significant positive correlation between C15ORF48 and the infiltration levels of various immune cells, such as CD8 + T cells, neutrophils, common lymphoid progenitors, and gamma-delta T cells. The phosphorylation sites S28.
C15ORF48 may regulate immune infiltration through cytokine signaling pathways or immune checkpoint modulation, potentially by influencing ligand-receptor interactions that alter the recruitment and function of immune cells in the tumor microenvironment. Studies on similar genes have demonstrated immune evasion mechanisms in cancer, where dysregulated cell-to-cell networks—driven by specific ligand-receptor pairs—enable tumors to evade CD8 + T-cell surveillance or enhance regulatory cell activity, as predicted in computational frameworks for multicellular interactions36. In this context, C15ORF48’s overexpression in tumors might parallel such evasion strategies, suggesting a role in checkpoint-like inhibition, though experimental validation in immune co-culture models is essential to confirm these inferences. This suggests that C15ORF48 may influence the tumor immune microenvironment by modulating immune cell infiltration or activity. These findings are in agreement with the study by Su-Su Zheng et al., which identified C15ORF48 as a potential prognostic target for hepatocellular carcinoma, with its high expression associated with advanced tumor staging and poorer survival37. Similarly, the study by Shaodi Wen et al. indicated that the C15orf48 gene might play a role in lung cancer immunotherapy and impact patient prognosis38. Our study further found that C15ORF48’s high expression in LUAD (lung adenocarcinoma), LGG (low-grade glioma), LIHC, and SKCM (skin cutaneous melanoma) is significantly negatively correlated with patient survival, reinforcing its value as a potential prognostic biomarker. These findings align with contemporary research emphasizing TME heterogeneity and biomarker utility in guiding immunotherapy. For example, IOBR-based analyses have revealed intricate antitumor immunity patterns, enabling refined TME characterization that complements our multi-dimensional integration of C15ORF48 data15. Similarly, dynamic network biomarkers have uncovered progression tipping points in thyroid cancers, paralleling our identification of mutation-phosphorylation overlaps as drivers of pan-cancer advancement16. Moreover, predictive models like iMLGAM highlight machine learning’s role in forecasting immunotherapy responses, which may extend our insights into C15ORF48’s immune-modulatory functions for personalized strategies17. In this context, our study innovatively combines expression profiling, genetic alterations, and post-translational modifications for C15ORF48, unlike prior single-aspect investigations, revealing novel hotspots (e.g., S29, T38) that may underpin tumorigenesis. Practically, this could inform prognostic tools and targeted therapies, such as inhibitors disrupting these sites to enhance immune infiltration, potentially improving outcomes in resistant tumors like lung adenocarcinoma—though rigorous validation in diverse cohorts is required.
Moreover, this study is the first to systematically integrate the expression patterns, genetic mutations, and phosphorylation modifications of C15orf48. We found that C15orf48 undergoes genetic alterations in 22 out of 32 cancer types, with multiple critical mutation sites (e.g., S29F, and T38I) potentially altering C15orf48’s structure and function, thereby driving tumorigenesis and progression. Notably, there is significant overlap between mutation sites, phosphorylation sites, and pathogenicity predictions, with hotspot regions such as positions S29 and T38 identified as critical for genetic mutation, phosphorylation modification, and pathogenicity. The phosphorylation sites S28 and T38 in C15ORF48 may have functional implications in cancer pathways, such as the MAPK/ERK cascade, where phosphorylation events typically regulate kinase activity and downstream signaling to influence cell proliferation, survival, and metastasis. For instance, databases like KinaseNET, which map human kinase networks, indicate that similar serine/threonine sites can be targeted by upstream kinases in the MAPK family, potentially leading to altered protein conformation and enhanced signal transduction that promotes tumor progression. In cancer contexts, aberrant phosphorylation within this pathway often activates transcription factors like c-Fos or Elk-1, driving oncogenic processes including immune evasion and angiogenesis, as observed in models where sustained ERK signaling suppresses apoptosis and facilitates matrix degradation39. Accordingly, phosphorylation at S28 and T38 could mimic these mechanisms in C15ORF48, possibly modulating inflammatory responses or immune cell recruitment in the tumor microenvironment, although direct kinase-substrate interactions and pathway-specific effects require empirical validation through site-directed mutagenesis and phosphoproteomic assays. These findings highlight these key sites as potential core regions for functional regulation and underscore C15ORF48’s translational potential as a therapeutic target in cancer, particularly given its associations with poor prognosis and immune infiltration, which may enable precision interventions akin to those for established biomarkers like PD-L1. For instance, while PD-L1 serves as a predictive marker for immune checkpoint inhibitors by facilitating T-cell exhaustion and tumor evasion, C15ORF48 could similarly inform targeted therapies aimed at disrupting phosphorylation-driven signaling or mutation hotspots to restore immune responsiveness and inhibit tumor progression. Ongoing clinical trials targeting immune-related genes, such as NCT03568097—a phase II study evaluating phased avelumab (an anti-PD-L1 antibody) combined with chemotherapy in extensive-stage small cell lung cancer—demonstrate the feasibility of such approaches, where modulating immune checkpoints improves outcomes in immune-dysregulated tumors40. In this framework, C15ORF48’s overexpression and genetic alterations suggest it may parallel PD-L1’s role in immunotherapy resistance, warranting preclinical models and basket trials to assess inhibitors targeting its key sites for enhanced therapeutic efficacy.
Despite these insights, this study has some limitations. The identified genetic mutations, phosphorylation modifications, and pathogenic sites have not yet been experimentally validated. Although these insights are derived from comprehensive database analyses, they require rigorous experimental validation to substantiate their biological relevance, such as through knockdown or overexpression assays in lung cancer cell lines to assess impacts on proliferation, migration, and immune infiltration. Future investigations may incorporate patient-derived xenograft (PDX) models to evaluate in vivo tumor dynamics and therapeutic responses, providing a more clinically translatable platform for confirming C15ORF48’s role in pan-cancer progression. Previous studies have successfully utilized xenograft models in cancer research; for example, one investigation employed a breast cancer xenograft model to demonstrate that volatile anesthetics like sevoflurane promote metastasis by enhancing angiogenesis, contrasting with intravenous agents like propofol that reduce vessel density41. Another study used colorectal cancer xenografts to show that BCAT2 deficiency in branched-chain amino acid catabolism accelerates tumor growth and metastasis via metabolic reprogramming42. Similarly, in non-small cell lung cancer xenografts, overexpression of hsa-miR-CHA2 suppressed tumor growth by targeting cyclin E1 and inducing G1/S arres43. These models highlight the utility of xenografts for mechanistic validation, though challenges like immune system differences necessitate complementary immunocompetent systems for comprehensive assessment. Future studies should integrate cell and animal models, along with large-scale clinical samples, to confirm the functional roles of these mutations and modifications and elucidate their specific mechanisms in cancer.
Our reliance on TCGA data for pan-cancer analysis aligns with a rich history of biomarker studies utilizing this resource, where Liu’s laboratory has been a pioneer since its early adoption, developing innovative bioinformatic strategies for identifying prognostic genes across tumor types. For instance, their work has systematically explored voltage-gated sodium channel β3 subunit (SCN3B) as a potential glioma biomarker, cyclin-dependent kinase 2 (CDK2) in glioma progression, aminoacyl tRNA synthetase complex interacting multifunctional protein 1 (AIMP1) for head-neck squamous cell carcinoma prognosis, cornichon family AMPA receptor auxiliary protein 4 (CNIH4) in head and neck squamous cell carcinoma, and RAD50 for breast cancer diagnosis and prognosis44,45,46,47,48. Although this body of work appears highly productive, the batch-like generation of similar analyses may suggest a templated approach that prioritizes volume over nuanced interpretation, potentially limiting depth in individual studies. Furthermore, while TCGA offers unparalleled advantages such as large-scale multi-omics data for cross-tumor comparisons and biomarker discovery, it is susceptible to technical biases (e.g., batch effects in sequencing) and biological limitations (e.g., bulk profiling masking intratumor heterogeneity), as detailed in recent critiques49,50. These factors warrant caution in over-relying on TCGA-derived findings, underscoring the need for orthogonal validation in diverse cohorts to mitigate such biases and enhance generalizability.
Although the expression and functional roles of C15ORF48 across various cancers have been comprehensively analyzed, its specific roles in certain cancer types remain underexplored. In particular, the detailed mechanisms by which C15ORF48 regulates the tumor immune microenvironment, metabolic pathways, and signal transduction warrant further investigation. Future studies may explore the establishment of specialized cell models tailored to this project, such as immortalized lung cancer cell lines overexpressing or knocking down C15ORF48, to dissect its roles in mutation-driven phosphorylation and immune modulation under controlled conditions. For instance, patient-derived cell lines could be engineered to mimic pan-cancer hotspots like S29 and T38, enabling high-throughput screening of inhibitors targeting these sites. As an example, one study successfully established breast phyllodes tumor cell lines via lentiviral immortalization with HPV-16 E6/E7 or SV40-T, preserving key morphological and functional features of the original tumors, including proliferation, migration, and invasion capacities, which facilitated mechanistic investigations. Such models highlight the potential for creating disease-specific platforms, though challenges like maintaining genetic fidelity and heterogeneity necessitate rigorous validation through comparative genomics and functional assays to ensure translational relevance51. Future studies should prioritize exploring the potential of key mutation and phosphorylation sites as molecular targets for therapeutic intervention, providing new directions for precision medicine.
In conclusion, C15ORF48’s elevated expression, immune infiltration correlations, and mutation-phosphorylation overlaps position it as a promising biomarker and therapeutic target for precision oncology, particularly in lung cancer.
Data availability
This study analyzed publicly available datasets, which can be accessed at the following location: (http://timer.cistrome.org/,http://gepia.cancer-pku.cn/, https://www.proteinatlas.org/,https://www.cbioportal.org/,http://www.phosphonet.ca/,https://alphafold.com/).
References
SiegelRL, GiaquintoAN & Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 74 (1), 12–49. https://doi.org/10.3322/caac.21820 (2024).
Kim, J. et al. Spatial profiling of non-small cell lung cancer provides insights into tumorigenesis and immunotherapy response. Commun. Biol. 7 (1), 930. https://doi.org/10.1038/s42003-024-06568-w (2024).
Fan, X. X. & Wu, Q. Decoding lung cancer at single-cell level. Front. Immunol. 13 https://doi.org/10.3389/fimmu.2022.883758 (2022).
Sonkin, D., Thomas, A. & Teicher, B. A. Cancer treatments: past, present, and future. Cancer Genet. 286–287, 18–24. https://doi.org/10.1016/j.cancergen.2024.06.002 (2024).
Joshi, R. M., Telang, B. & Soni, G. Overview of perspectives on cancer, newer therapies, and future directions. Oncol. Transl Med. 10 (3), 105–109. https://doi.org/10.1097/ot9.0000000000000039 (2024).
Steele, E. M. et al. Missed follow-up is associated with worse survival in stage I lung cancer: results from a large multi-site academic hospital system. Sci. Rep. 14, 17710. https://doi.org/10.1038/s41598-024-68351-5 (2024).
Zhou, J. et al. A novel gene, NMES1, downregulated in human esophageal squamous cell carcinoma. Int. J. Cancer. 101 (4), 311–316. https://doi.org/10.1002/ijc.10600 (2002).
C15orf48 protein expression summary - the human protein atlas. Accessed November 30 (2024). https://www.proteinatlas.org/ENSG00000166920-C15orf48
Lee, C. Q. E. et al. Coding and non-coding roles of MOCCI (C15ORF48) coordinate to regulate host inflammation and immunity. Nat. Commun. 12 (1), 2130. https://doi.org/10.1038/s41467-021-22397-5 (2021).
Spisák, S. et al. Genome-wide screening of genes regulated by DNA methylation in colon cancer development. PloS One. 7 (10), e46215. https://doi.org/10.1371/journal.pone.0046215 (2012).
Sova, P. et al. Discovery of novel methylation biomarkers in cervical carcinoma by global demethylation and microarray analysis. Cancer Epidemiol. Biomark. Prev. Publ Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol. 15 (1), 114–123. https://doi.org/10.1158/1055-9965.EPI-05-0323 (2006).
P W, H. L. CRISPR screening and cell line IC50 data reveal novel key genes for Trametinib resistance. Clin. Exp. Med. 25 (1). https://doi.org/10.1007/s10238-024-01538-2 (2024).
Su, A., Ra, S., Li, X., Zhou, J. & Binder, S. Differentiating cutaneous squamous cell carcinoma and pseudoepitheliomatous hyperplasia by multiplex qRT-PCR. Mod. Pathol. Off J. U S Can. Acad. Pathol. Inc. 26 (11), 1433–1437. https://doi.org/10.1038/modpathol.2013.82 (2013).
Li, C. et al. The prognostic and immune significance of C15orf48 in pan-cancer and its relationship with proliferation and apoptosis of thyroid carcinoma. Front. Immunol. 14, 1131870. https://doi.org/10.3389/fimmu.2023.1131870 (2023).
Fang, Y. et al. Systematic investigation of tumor microenvironment and antitumor immunity with IOBR. Med. Res. 1 (1), 136–140. https://doi.org/10.1002/mdr2.70001 (2025).
Zhang, H. et al. Optimized dynamic network biomarker Deciphers a high-resolution heterogeneity within thyroid cancer molecular subtypes. Med. Res. 1 (1), 10–31. https://doi.org/10.1002/mdr2.70004 (2025).
B Y, J F, L X, et al. iMLGAM: integrated machine learning and genetic algorithm-driven multiomics analysis for pan-cancer immunotherapy response prediction. iMeta 4 (2). https://doi.org/10.1002/imt2.70011 (2025).
Li, B. et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 17 (1), 174. https://doi.org/10.1186/s13059-016-1028-7 (2016).
Li, T. et al. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 77 (21), e108–e110. https://doi.org/10.1158/0008-5472.CAN-17-0307 (2017).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48 (W1), W509–W514. https://doi.org/10.1093/nar/gkaa407 (2020).
Tang, Z. et al. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 45 (W1), W98–W102. https://doi.org/10.1093/nar/gkx247 (2017).
Uhlen, M. et al. A genome-wide transcriptomic analysis of protein-coding genes in human blood cells. Science 366 (6472), eaax9198. https://doi.org/10.1126/science.aax9198 (2019).
Uhlen, M. et al. A pathology atlas of the human cancer transcriptome. Science 357 (6352), eaan2507. https://doi.org/10.1126/science.aan2507 (2017).
Karlsson, M. et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 7 (31), eabh2169. https://doi.org/10.1126/sciadv.abh2169 (2021).
A subcellular map of the human proteome - PubMed. Accessed December 9. (2024). https://pubmed.ncbi.nlm.nih.gov/28495876/
Sjöstedt, E. et al. An atlas of the protein-coding genes in the human, pig, and mouse brain. Science 367 (6482), eaay5947. https://doi.org/10.1126/science.aay5947 (2020).
Uhlén, M. et al. Proteomics. Tissue-based map of the human proteome. Science 347 (6220), 1260419. https://doi.org/10.1126/science.1260419 (2015).
The human secretome - PubMed. Accessed December 9. (2024). https://pubmed.ncbi.nlm.nih.gov/31772123/
Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal - PubMed. Accessed December 9. (2024). https://pubmed.ncbi.nlm.nih.gov/23550210/
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2 (5), 401–404. https://doi.org/10.1158/2159-8290.CD-12-0095 (2012).
Analysis and visualization of longitudinal genomic and clinical data from the AACR project GENIE biopharma collaborative in cBioPortal - PubMed. Accessed December 9, (2024). https://pubmed.ncbi.nlm.nih.gov/37668528/
Kinexus | PhosphoNET. Accessed December 10. (2024). http://www.phosphonet.ca/
Jumper, J. et al. Highly accurate protein structure prediction with alphafold. Nature 596 (7873), 583–589. https://doi.org/10.1038/s41586-021-03819-2 (2021).
Cheng, J. et al. Accurate proteome-wide missense variant effect prediction with alphamissense. Science 381 (6664), eadg7492. https://doi.org/10.1126/science.adg7492 (2023).
Varadi, M. et al. AlphaFold protein structure database in 2024: providing structure coverage for over 214 million protein sequences. Nucleic Acids Res. 52 (D1), D368–D375. https://doi.org/10.1093/nar/gkad1011 (2024).
Hou, R., Denisenko, E., Ong, H. T., Ramilowski, J. A. & Forrest, A. R. R. Predicting cell-to-cell communication networks using NATMI. Nat. Commun. 11, 5011. https://doi.org/10.1038/s41467-020-18873-z (2020).
Zheng, S. S., Wu, Y. F., Zhang, B. H., Huang, C. & Xue, T. C. A novel myeloid cell marker genes related signature can indicate immune infiltration and predict prognosis of hepatocellular carcinoma: integrated analysis of bulk and single-cell RNA sequencing. Front. Mol. Biosci. 10 https://doi.org/10.3389/fmolb.2023.1118377 (2023).
Wen, S. et al. Four differentially expressed genes can predict prognosis and microenvironment immune infiltration in lung cancer: A study based on data from the GEO. BMC Cancer. 22 (1), 193. https://doi.org/10.1186/s12885-022-09296-8 (2022).
Yj, G., Ww, P., Sb, L., Zf, S., Ll H., & Y. X. ERK/MAPK signalling pathway and tumorigenesis. Exp. Ther. Med. 19 (3). https://doi.org/10.3892/etm.2020.8454 (2020).
Linardou, H. et al. 1648P phased avelumab combined with chemotherapy as first-line treatment in extensive stage small cell lung cancer (PAVE): A phase II Hellenic cooperative oncology group study. Ann. Oncol. 32, S1163–S1164. https://doi.org/10.1016/j.annonc.2021.08.232 (2021).
Li, R., Huang, Y., Liu, H., Dilger, J. P. & Lin, J. Abstract 2162: comparing volatile and intravenous anesthetics in a mouse model of breast cancer metastasis. Cancer Res. 78 (13_Supplement), 2162. https://doi.org/10.1158/1538-7445.AM2018-2162 (2018).
Kang, Z. R. et al. Deficiency of BCAT2-mediated branched-chain amino acid catabolism promotes colorectal cancer development. Biochim. Biophys. Acta Mol. Basis Dis. 1870 (2), 166941. https://doi.org/10.1016/j.bbadis.2023.166941 (2024).
hsa-miR-CHA. 2, a novel microrna, exhibits anticancer effects by suppressing Cyclin E1 in human non-small cell lung cancer cells. Biochim. Biophys. Acta BBA - Mol. Basis Dis. 1870 (6), 167250. https://doi.org/10.1016/j.bbadis.2024.167250 (2024).
H L, J. W., Cl H., & Ap J. Is the voltage-gated sodium channel β3 subunit (SCN3B) a biomarker for glioma? Funct. Integr. Genomics. 24 (5). https://doi.org/10.1007/s10142-024-01443-7 (2024).
A comprehensive bioinformatic. Analysis of cyclin-dependent kinase 2 (CDK2) in glioma. Gene 822, 146325. https://doi.org/10.1016/j.gene.2022.146325 (2022).
Li, Y. & Liu, H. Clinical powers of aminoacyl tRNA synthetase complex interacting multifunctional protein 1 (AIMP1) for head-neck squamous cell carcinoma. Cancer Biomark. 34 (3), 359–374. https://doi.org/10.3233/CBM-210340 (2022).
Liu, H. & Li, Y. Potential roles of Cornichon family AMPA receptor auxiliary protein 4 (CNIH4) in head and neck squamous cell carcinoma. Cancer Biomark. 35 (4), 439–450. https://doi.org/10.3233/CBM-220143 (2022).
Chhatwal, K. S. & Liu, H. RAD50 is a potential biomarker for breast cancer diagnosis and prognosis. Published online September 14, 2024.09.07.611821 https://doi.org/10.1101/2024.09.07.611821 (2024).
Genetic expression in cancer research. Challenges and complexity. Gene Rep. 37, 102042. https://doi.org/10.1016/j.genrep.2024.102042 (2024).
Liu, H., Li, Y., Karsidag, M., Tu, T. & Wang, P. Technical and biological biases in bulk transcriptomic data mining for cancer research. J. Cancer. 16 (1), 34. https://doi.org/10.7150/jca.100922 (2025).
He, S. et al. Establishment of breast phyllodes tumor cell lines preserving the features of phyllodes tumors. BIO Integr. 4 (1). https://doi.org/10.15212/bioi-2022-0025 (2023).
Acknowledgements
We are grateful to the public databases, including GEPIA, Timer2.0, Human Protein Atlas, cBioPortal, PhosphoNET, and AlphaFold, for providing open access.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
JR, SX designed this study. HZ, ML, RZ, and WM extracted the information from the databases. JRanalyzed the data. SX supervised the entire study. JR wrote the manuscript. All authors revised the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ren, J., Zhu, H., Liu, M. et al. C15ORF48 serves as a potential biomarker and therapeutic target in pan-cancer with implications for lung cancer. Sci Rep 15, 32821 (2025). https://doi.org/10.1038/s41598-025-17966-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-17966-3