Abstract
Gestational diabetes mellitus (GDM), one of the prevalent pregnancy-related metabolic disorders, have shown immediate or long-term adverse health outcomes for maternal and fetal health. Therefore, it is crucial to understand the ongoing cellular and molecular changes in GDM patients for characterizing novel biomarkers for diagnosis and therapeutic purposes. In the current study, we analyzed 3 transcriptomic datasets, characterized 449 unique upregulated and 785 downregulated DEGs, and performed several analyses. Gene ontology shows enrichment of migration, development, and immune-related processes in GDM patients. KEGG pathway shows enrichment of pathways like “type 1 diabetes mellitus” and “graft versus host disease”. Disease ontology shows enrichment of “female reproductive system disease,” “anemia,” etc. Integration of methylation and transcriptomic data identified 11 genes (RASSF2, WSCD1, TNFAIP3, TPST1, UBASH3B, ZFP36, CRISPLD2, IGFBP7, TNS3, TPM2, and VTRNA1-2), as potential novel diagnostic biomarkers and therapeutic targets. Furthermore, immune cell-type infiltration analysis shows higher memory B-cells and lower M1 macrophages and CD8 T-cells. Protein-protein interaction analysis followed by ROC analysis in an independent dataset identified 7 hub genes (POLR2G, VWF, COL5A1, COL6A1, CD44, COL3A1, and COL1A1) with high diagnostic potential. Overall, we obtained 18 genes that could serve as novel diagnostic biomarkers and therapeutic targets in GDM patients.
Similar content being viewed by others
Introduction
Gestational Diabetes Mellitus (GDM) is one of the most prevalent pregnancy-related metabolic disorders, referring to a diabetogenic state affecting both the mother and the fetus, often leading to severe pregnancy outcome1. With increasing prevalence and incidence rates, it is becoming of utmost importance to develop novel diagnostic and treatment methods2. The current diagnostic procedure in pregnancies mostly relies on the patients’ oral glucose tolerance, with the International Association of Diabetes and Pregnancy Study Groups (IADPSG) method being the most prevalent one. The blood HbA1c (glycosylated hemoglobin) level measurement is one of the other diagnostic procedures for predicting GDM3. The HbA1c levels above 5.7% are referred to as being at risk for GDM. The current prevalence rate of GDM stands at 16.9%4 worldwide, which has been increasing with every passing year. The prevalence rate of GDM not only varies from one country to another but also between different regions of the same country depending on different factors, like ethnicity, food habits, body mass index, age, and pre-existing clinicopathological conditions. The prevalence rate of GDM according to IADPSG criteria is often observed to be 19.6% in high-income countries513.4% in low-and-middle-income countries6and 10.2% in low-income countries7 thereby showing the drastic reduction in the prevalence rates from high-income countries to low-income countries. The prevalence rates also significantly vary between people staying in urban and rural areas. According to a study by Seshiah et al., the prevalence rate in India according to the IADPSG criteria is 16.2%, with 17.8% occurring in urban areas, 13.8% in semi-urban areas, and 9.9% in rural areas8. This suggests that individuals living in urban environments are more susceptible to developing GDM compared to those living in rural areas.
The current treatment procedures for GDM by clinicians throughout the globe vary depending on the plasma glucose levels of the patient. Based on the treatment procedures, GDM can be broadly divided into two categories, GDMA1 and GDMA29. GDMA1 refers to the patients who are having mild elevations in blood sugar levels in pregnancy, which can be managed by using a dietary modification10. Such patients are recommended for medical nutritional therapy for the management of GDM. Patients belonging to the GDMA2 category generally have highly elevated blood glucose levels, and these cannot be managed by medical nutritional therapy. In such cases, management of GDM requires the use of oral drugs with patients with very highly elevated blood glucose levels, often requiring subcutaneous insulin injections. The oral drugs that are generally used for the management of GDMA2 include metformin and glyburide (a second-generation sulfonylurea), with metformin being the most commonly used drug11. Metformin generally acts by increasing the gastrointestinal glucose uptake12 by inhibiting the hepatic gluconeogenesis13mitochondrial respiration14fructose-1,6-bisphosphatase14 and activating the adenosine monophosphate-activated protein kinase13.
Though we have treatments against GDM, there are certain limitations associated with it. For example, existing treatment shows individual variability in treatment response, long-term risk for mother and baby, etc. Furthermore, current treatment primarily manages hyperglycemia but does not address underlying mechanisms like β-cell dysfunction, inflammation, and placental abnormalities, which contribute to GDM progression and its long-term complications, including Type 2 diabetes (T2D). The application of multi-omics has helped identify new biomarkers and molecular subtypes in various diseases. Therefore, there is a pressing need to identify new biomarkers and therapeutic targets by analyzing various high-throughput data to address the above challenges for GDM patients.
Methodology
Dataset creation
We downloaded gene expression data from three different transcriptome datasets from the Gene Expression Omnibus (GEO) database15where mothers have been diagnosed with gestational diabetes, i.e., GSE87295, GSE103552, and GSE70493, along with the normal counterparts. To get the differentially expressed genes (DEGs) from the above datasets, we used the ‘DESeq2‘16 and ‘limma‘17 tools. In the case of GSE103552, we directly used the results provided by the authors. As the datasets were obtained from different sequencing platforms, we processed the datasets using the raw reads to obtain the DEGs. This approach allows for ensuring statistical robustness and eliminating any batch effects. We further selected up- and downregulated genes based on LogFC values ( > = 0.5) in the case of GSE87295 and GSE103552. In the case of GSE70493, LogFC values considered were > = 0.1 due to the lower range of LogFC throughout the data. Next, we took the union of all the up- and downregulated genes for further analyses.
Gene enrichment analyses
We did gene enrichment analyses to obtain enriched biological processes, cellular components, and molecular functions associated with the up- and downregulated genes. We used the “clusterProfiler 4.0”18 R package for the analyses. First, we used the ‘enrichGO’ function to characterize the processes, ‘org.Hs.eg.db’ as the human database, minGSSize (minimum gene size) as 10, and maxGSSize (maximum gene size) as 500. Next, we removed the redundant terms using the ‘simplify’ function with a cutoff value of 0.8, ‘p.adjust’ as a statistical measure, ‘min’ as select_fun, and “Wang” as a measure. We used Enrichr integrated Kyoto Encyclopedia of Genes and Genomes’ (KEGG) module to perform the KEGG pathway19,20,21 and disease ontology analysis associated with the up- and down-regulated genes.
Integration of epigenomic data
We downloaded the dataset provided by Lu et al.22where he profiled DNA methylation in GDM patients and matched controls. We downloaded the processed data and obtained the list of hyper- and hypomethylated genes as per delta values, which is defined as the difference between the mean methylation value obtained in GDM patients and the mean methylation value obtained in controls. If the delta value is positive, the gene is called hypermethylated, and if it is negative, it is called hypomethylated.
Immune cell infiltration analysis
We used CIBERSORTx23 to estimate the infiltration of 22 different immune cells in GDM and normal samples. Gene expression of case and control was used separately as an input file. We used the default ‘LM22’ gene signature, ‘B-mode’ as the batch correction mode since the datasets belong to different platforms, and ‘100’ for permutations for significance. We further computed the Spearman’s correlation among the enriched immune cell types in GDM patients.
Protein-Protein interaction (PPI) network and hub genes characterization
We further investigated if there is an existence of correlations among DEGs proteins; we performed a PPI analysis using the STRING database24. In the SEARCH tab, upregulated and downregulated genes were given as input. The species used was Homo sapiens, and a PPI score of 0.7 was used to build the PPI network. We further visualized this network in Cytoscape25 and constructed the subnetwork using the ‘cytoHubba’ plugin. We computed the node score using the degree centrality, which represents the topological feature of the network. Nodes with high scores indicated that the proteins played a crucial role in the obtained network. The top 10 nodes with the highest scores were listed and used for therapeutic analyses.
Receiver operating characteristic curve analysis
The receiver operating characteristic (ROC) curve and areas under curve (AUC) analyses were used to evaluate the characterized hub genes as potential diagnostic biomarkers in GDM patients. ‘pROC’26 R package was used for the analysis.
Results
DEGs characterization
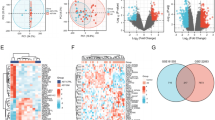
We processed the datasets and characterized the statistically significant up- and downregulated DEGs. For the dataset GSE87295, we obtained 237 upregulated DEGs (LogFC > = 0.5, FDR < = 0.05) and 441 downregulated DEGs (LogFC >= -0.5, FDR < = 0.05). In the case of GSE70493, we obtained 129 upregulated DEGs (LogFC > = 0.1, FDR < = 0.05) and 308 downregulated DEGs (LogFC >= -0.1, FDR < = 0.05). In the case of GSE103552, we used the authors provided data, i.e., 121 upregulated DEGs (LogFC > = 0.5, FDR < = 0.25) and 90 downregulated DEGs (LogFC >= -0.5, FDR < = 0.25). Figure 1A-C presents a volcano plot that illustrates the differential gene expression results for all three datasets. We combined the characterized DEGs (up and downregulated) to see if there are common genes among the three datasets; however, we observed only 1 downregulated gene as common among the three datasets (Fig. 1D & E). Therefore, we combined the genes from all three datasets and took the union, resulting in 472 unique upregulated genes and 806 unique downregulated genes. The complete results of differential expression analysis and the list of up- and down-regulated genes (for each dataset and unique) are provided in Supplementary Tables S1-S4.
Gene enrichment analysis
We performed gene enrichment analysis using the list of unique up- and downregulated genes to compute significant enriched biological processes, cellular components, and molecular functions. We observed that upregulated genes were largely enriched for processes associated with migration and development, for example, “ameboidal-type cell migration”, “Endothelial Development”, “Regulation of Angiogenesis”, “Regulation of Blood Vessel Endothelial Cell Migration”, etc. (Fig. 2A), whereas downregulated genes were enriched for processes such as “Extracellular Matrix Organization”, “Collagen Fibril Organization”, “MHC Class II Protein Complex Assembly”, etc. [Fig. 2B]. Our findings were supported by the previous studies; for example, Kwon et al. show that impaired angiogenic function of fetal endothelial progenitor cells are associated with PCHD10 in GDM27. Likewise, McElwain et al. have reported the endothelial dysfunction instigated by various oxidative stress, pro-inflammatory cytokines, etc., in pre-eclampsia and GDM28. Supplementary Table S5-S6 provides a complete list of enriched processes associated with up- and down-regulated genes respectively.
When we looked at the cellular components, we observed upregulated genes were associated with “secretory granule membrane”, “respiratory chain complex”, etc. [Fig. 2C], whereas downregulated genes were enriched for components such as “collagen-containing extracellular matrix”, “immunoglobulin complex”, “endoplasmic reticulum lumen”, etc. [Fig. 2D]. Previous studies29,30 supported these findings. Supplementary Tables S7 and S8 provide a full list of cellular components associated with upregulated and downregulated genes, respectively. In the case of molecular functions, only downregulated genes show significant enrichment, including functions such as “antigen binding”, “extracellular matrix structural constituent”, etc. [Supplementary Figure S1]. A complete list of enriched molecular functions associated with downregulated genes is provided in Supplementary Table S9.
KEGG pathway analysis19,20,21 shows enrichment of only three pathways associated with upregulated genes, i.e., “N neutrophil extracellular trap formation”, “Systemic lupus erythematosus” and “Alcoholism” (Fig. 3A), whereas the number of pathways was associated with downregulated genes, including “Type I diabetes mellitus”, “Graft-versus-host disease”, “Autoimmune thyroid disease”, etc. (Fig. 3B). Complete details of KEGG pathways19,20,21 are provided in Supplementary Tables S10 and S11. Next, we analyzed the enriched disease ontology associated with the DEGs (up and down) and observed that upregulated genes were associated with diseases such as “Kuhnt-Junius degeneration”, “female reproductive system disease”, “esophageal cancer”, etc. (Fig. 3C), whereas downregulated genes were associated with diseases such as “Ehlers-Danlos syndrome”, “collagen disease”, “osteoarthritis”, etc. (Fig. 3D). We also observed “gestational diabetes” as an enriched disease ontology with downregulated genes. See Supplementary Tables S12 and S13 for complete details.
DNA methylation analysis reveals potential therapeutic targets
We further integrated the DNA methylation pattern with the transcriptome to characterize genes that could be potential and novel therapeutic targets, as transcriptomic and epigenomic collectively provide a comprehensive picture of a gene’s molecular and cellular function. We looked at the gene methylation data from Lu et al.‘s datasets on GDM and normal placenta and obtained the list of genes that were hyper- and hypomethylated. We compared the hypo- and hypermethylated gene sets along with the up- and downregulated DEGs, characterized based on gene expression. In the case of upregulated DEGs, RASSF2 and WSCD1 were found to be hypomethylated; however, none of the genes show hypermethylation. We further investigated the functional role of the above two genes and found that RASSF2 encodes a protein that contains a Ras-associated domain. Its role has been previously seen in diseases such as colorectal cancer, but its role in GDM is yet to be explored. Likewise, WSCD1 (WSC domain containing 1) is typically associated with cell wall integrity signaling and cellular responses to environmental stress. Its role has been shown previously in GDM patients by Pinney et al.31. Furthermore, high expression of WSCD1 could possibly be associated as a response to the stress caused due to the use of endocrine disruptors, which is one of the major factors associated with ~ 50% of the GDM and other pregnancy-related issues in the women32,33. When we looked at the list of downregulated DEGs along with sets of genes that were hypo- or hypermethylated, we found that 4 genes (TNFAIP3, TPST1, UBASH3B, and ZFP36) were hypermethylated and 5 genes (CRISPLD2, IGFBP7, TNS3, TPM2, and VTRNA1-2) were hypomethylated.
To further validate our findings, we plotted the gene expression patterns of the above genes in all three GDM datasets we used in this study. As shown in Fig. 4 (A-C), RASSF2 and WSCD1, as per expectation, show higher gene expression in the case compared to the control, as it has been shown that hypomethylation in the promoter region removes the transcriptional repression34. Likewise, CRISPLD2, IGFBP7, TNS3, TPM2, and VTRNA1-2 show lower gene expression in the case compared to the controls, which can be explained by the fact that hypermethylation in the promoter region typically silences gene expression by preventing TF binding. Lastly, for the genes that were hypomethylated, i.e., TNFAIP3, TPST1, UBASH3B, and ZFP36, they show overall lower gene expression in the case compared to the control, with a few exceptions where the case shows higher expression. This observation can be explained by the fact that an additional regulatory mechanism, for example, transcriptional repression by other factors such as proteins, miRNA, or chromatin modifications, might be playing a role. Overall, the integration of epigenetic information along with gene expression provides a clear insight about disease pathogenesis and potential novel therapeutic targets.
Immune infiltration analysis in the GDM patients
We performed immune cell infiltration analysis using the CIBERSORTx algorithm for the GDM patients and normal population to assess the enrichment of 22 different immune cell types. Supplementary Figures S2A and S2B show the enrichment of various immune cell types in normal and GDM patients, respectively. The Mann Whitney statistical test shows that GDM patients have a higher enrichment of memory B-cells, while M1 macrophages and CD8 T-cells are more enriched in the normal population [Fig. 5A]. This observation can be explained by the fact that GDM patients exhibit chronic low-grade inflammation, and as memory B-cells play a crucial role in innate immunity and elevated inflammation, it is possible that due to increased inflammation and autoantibody production, we are observing high enrichment of memory B-cells in GDM patients compared to normal. We further looked at the correlation among the various immune cells using Spearman correlation analysis and observed that many cell types were positively correlated and some of them showed negative correlation. For example, memory B-cells show positive correlation with M1 macrophages, follicular helper T-cells, and regulatory T-cells, whereas negative correlation with NK resting cells and mast cells. Similarly, CD8 T-cells show a positive correlation with regulatory T-cells, monocytes, and resting NK cells, whereas there is a negative correlation with neutrophils, activated mast cells, and naïve CD4 T-cells [Fig. 5B].
Immune cell enrichment analysis. (A) Box plot with mann-whitney statistical analysis representing comparison among 22 immune cell types in GDM patient and control. P-value < 0.05 was considered as statistical measure. (B) Heatmap showing the correlation among the 22 immune cell types enriched in GDM patients.
The PPI network analysis
We used STRING and Cytoscape to perform PPI analysis and hub gene identification. First, we downloaded the STRING interaction network from the STRING database and imported it into Cytoscape to construct and visualize the network. As shown in Supplementary Figure S3A & S3B, we observed clusters of networks associated with the up- and downregulated genes, respectively. We further used the ‘cytoHubba’ plugin to compute the hub genes from the above complex network, which shows a strong correlation with the disease biology. We used ‘Degree’ as a measure to compute the node score, and based on the score, we selected the top 10 hub genes. In the case of upregulated genes, the top 10 genes based on the node score include H4C6, KDR, H3C13, UQCR10, VWF, H3C12, H2BC5, H2AC8, H2AC6, and POLR2G, where the node score ranges between 8 and 13 (Fig. 6A and Table 1). Likewise, in the case of downregulated genes, the top 10 genes based on node score include FN1, COL1A1, IL1B, COL1A2, CD44, COL3A1, CDK1, COL6A1, COL6A2, and COL5A1, where the node score ranges between 24 and 54 (Fig. 6B & Table 2).
Hub Gene Characterization. Top10 hub genes characterized from protein-protein interaction network created using (A) upregulated genes and (B) downregulated genes. Cytoscape tool was implemented to obtain hub genes. (C) Correlation plot showing correlation among hub genes computed using gene expression. Spearman correlation was computed.
We further assess the correlation between the gene expression level of hub genes. Only 12 (3 up- and 9 downregulated) of the 20 hub DEGs had expression data that was present in all three datasets used in the study (see Method). Figure 6C reveals an overall high correlation among the expression levels of hub genes, except POLR2G and COL3A1. One possible reason for this observation could be due to distinct role of the two genes, where COL3A1 is associated with extracellular matrix (ECM) organization whereas POLR2G, a subunit of RNA polymerase II, playing a central role in transcription regulation and hence not directly associating with ECM remodeling.
In the case of upregulated genes (KDR, POLR2G, and VWF), we observed mixed results. For instance, KDR showed high correlation with the genes, whereas POLR2G showed poor correlation, and VWF showed moderate correlation with the other genes. Interestingly, we observed that different collagen genes showed different correlation patterns with other genes. For example, COL1A1 shows a high correlation of 0.78 with KDR, whereas there is a poor correlation of 0.34 with POLR2G; even among themselves, collagen genes show variability in gene expression. For example, COL1A2 shows a strong correlation of 0.79 with COL3A1, whereas there is a moderate correlation of 0.59 with COL5A1. This observation suggests tissue- or condition-specific regulation.
ROC curve analysis reveals hub genes as potential biomarker
We carried out ROC analysis to predict the prognostic potential biomarker property of the hub genes for the GDM. We performed this analysis on an independent dataset that has not been used in this study to maintain the statistical significance and robustness of the study and avoid any bias. We downloaded the gene expression of GDM and the normal population from the GEO database with ID number GSE4952435. In our new independent dataset, we observed data for only 11 genes (3 upregulated and 8 downregulated) out of 12 hub genes; hence, we performed the analysis for these 11 genes. As shown in Fig. 7, out of 3 upregulated genes (Fig. 7A-C), VWF and POLR2G achieved the AUC of 0.78, whereas KDR showed the AUC of 0.56. In the case of 8 downregulated genes (Fig. 7D-K), IL1B and COL1A2 achieved the AUC of 0.56, COL6A2 achieved the AUC of 0.67, COL5A1 and COL6A1 achieved the AUC of 0.78, CD44 obtained the AUC of 0.89, and COL3A1 and COL1A1 showed the maximum AUC of 1. As the AUC ranges between 0.56 and 1, we implemented the cutoff of 0.75 to call a gene as a potential biomarker. In total, 7 genes—POLR2G, VWF, COL5A1, COL6A1, CD44, COL3A1, and COL1A1—were found to have AUC values of 0.75 or above.
Discussion
GDM, defined as glucose intolerance, is the most prevalent pregnancy-associated metabolic disorder. It is first evident at some point during pregnancy, and approximately 13% of pregnancies worldwide get complicated due to GDM. GDM has a complicated etiology that includes both environmental and genetic components36,37. Numerous studies have demonstrated the serious negative health effects of GDM on both the mother and her children, both in the short and long term. Women with GDM are prone to both immediate and long-term health issues such as T2D, cardiovascular diseases, etc38. Additionally, GDM causes a large financial strain on healthcare resources and society39. Currently, the onset and progression of GDM are uncertain and complex, and pathogenesis remains uncertain. It is therefore essential to identify novel molecular biomarkers for the diagnosis and treatment of GDM patients to provide them with tailored clinical intervention to alleviate the prognosis of not only the pregnancy but also fetal health both during the pre- and post-natal stages of its growth.
In the current analysis, we analyzed several GDM-associated transcriptomic datasets and characterized several DEGs associated with the disease pathophysiology. First, we did gene enrichment analyses and discovered that genes that were upregulated are mostly linked to processes that involve cell migration and development, while genes that were downregulated are mostly linked to immune system processes, specifically antigen processing and presentation. Previous studies27,28 also supported these findings. Next, we looked at the enriched KEGG pathways19,20,21which depicted that DEGs were enriched for “type 1 diabetes mellitus”, “graft versus host disease”, “neutrophil extracellular trap formation”, etc., and had a keystone role in GDM, as shown in previous studies29. The disease ontology analysis revealed enrichment of the disease like “female reproductive system”, “musculoskeletal system cancer”, “arteriosclerotic cardiovascular disease”, “anemia”, etc.
One interesting observation we made was the enrichment of the biological process “multicellular organismal-level iron ion homeostasis” and the disease “anemia.”. These two ontologies hinted towards the relationship of GDM with another disease prevalent in pregnant women, known as “iron-deficient gestational anemia (IDGA).”. GDM, one of the most common pregnancy-related clinicopathological conditions, is often related to the most common pregnancy complication of iron-deficient gestational anemia (IDGA)40. IDGA refers to reduced hemoglobin (Hb) levels, generally classified into three categories of mild anemia (9 to 10.9 g/dL), moderate anemia (7 to 8.9 g/dL), and severe anemia (< 7 g/dL) by the World Health Organization41. The relationship between IDGA and GDM is quite complex, as several studies noted that the incidence of GDM among IDGA patients is quite low, showing their inverse relationship42. This may be due to the reduced Hb levels in anemic patients, which reduces the glycosylated Hb levels as well, thereby reducing the prevalence of diabetes43; contrastingly, in GDM patients, the prevalence of IDGA is seen to be higher, mostly because of the increased demand for Hb to support the macrosomic fetus and the maternal health44. We also observed a process, “multicellular organismal-level iron ion homeostasis,” supporting the relationship between GDM and IDGA.
We would also like to highlight another interesting observation we made during our study. Recently, Hivert et al. characterized gene IGFBP1 (Insulin-like Growth Factor Binding Protein 1) as a circulating biomarker in the placenta during early pregnancy45. IGFBP1 binds to insulin growth factor 1 (IGF1) with high affinity and is associated with the increased risk of insulin resistance and gestational diabetes. Interestingly, in our analysis we found IGFBP7 (Insulin-like Growth Factor Binding Protein 7), a member of the same family, i.e., IGFBP (Insulin-like Growth Factor Binding Protein), to be differentially expressed in GDM patients. IGFBP7, in contrast to IGFBP1, binds with lower affinity to IGF1 and is associated with multiple other roles independent of IGF, including implantation, angiogenesis, trophoblast invasion, etc., suggesting a stronger link with placental biology. Previous studies have shown high expression of IGFBP7 in the placenta post-implantation and remodeling of placental tissue during pregnancy46,47,48. Notably, some studies also reported lower expression reflecting variability based on gestational timing, obesity status, and treatment49. Our findings therefore complement the IGFBP1 literature by identifying IGFBP7-centered placental pathways as a mechanistically unique axis pertinent to GDM pathophysiology and risk stratification.
The integration of DNA methylation data with gene expression data allowed us to characterize the potential novel prognostic biomarkers associated with the etiopathology of the GDM patients. We observed a methylation-expression relationship for 11 genes. Methylation (hypo and hyper) in the promoter region is associated with TF binding and gene regulation. We analyzed the same relationship and observed that two genes show hypomethylation in the promoter region as well as higher expression in GDM patients compared to control, whereas 5 genes show hypermethylation in the promoter region and downregulation in gene expression. Interestingly, 4 genes that were downregulated show hypomethylation, which could be due to additional regulatory mechanisms such as transcriptional repression by other factors such as proteins, miRNA, or chromatin modifications. The gene expression comparison of these 11 genes across multiple datasets, including GDM patients and healthy individuals, further validated the above observation. However, a big problem with combining epigenomic data and transcriptomic data is that they come from different types of tissues; the transcriptomic data comes from primary umbilical cord cells and nearby cells, while the epigenomic data comes from placental tissue. Such differences may lead to tissue-specific biases and may confound integration.
Next, we performed immune cell enrichment analysis to characterize the immune cell types enriched in GDM patients. Previous studies have established the relationship between immune response and GDM50. They have shown the presence of dysregulated immune systems and low-grade inflammation in the GDM population51,52. We perform a similar analysis and obtain a significant infiltration of memory B cells in GDM patients and M1 macrophages and CD8 T cells in the normal population. As the above finding is based on the deconvolution principle, we compared our findings with the recent study that performed single-cell analysis. Overall, we found mixed observations; similar to CIBERSORTx, single-cell analysis also shows the lower M1 macrophage gene signature expression; however, signals for increased memory B cells and reduced CD8 T cells were less consistent across single-cell studies53,54,55. This heterogeneity could be due to differences in sampling site, gestational age, and methodological limits of bulk RNA-Seq-based deconvolution versus single-cell technique. We further analyzed the correlation among the enriched immune cell types and found strong positive as well as negative correlations.
PPI analysis using the STRING database revealed a strong interaction network, and subsequent analysis using a cystoscope characterized the putative top 10 hub genes (upregulated and downregulated) with a high node score. We further looked at the correlation among the gene expression of these hub genes in all three datasets (only 12 genes were common) and observed a strong correlation with a Spearman correlation value of up to 0.96. We further established the therapeutic potential of 11 out of the 12 above-mentioned hub genes, as data was present for only 11 of them, by performing ROC curve analysis on an independent dataset. We observed that based on gene expression; these 11 genes can classify GDM patients with control with high accuracy thereby providing a unique insight into the etiopathology of GDM. This performance can be further improved in future by developing a model with combined information of genes on a larger dataset to avoid overfitting.
In the last, we propose 7 potential biomarkers i.e. POLR2G, VWF, COL5A1, COL6A1, CD44, COL3A1, and COL1A1. Genes such as POLR2G, VWF, and CD44 have been detected in circulating blood56,57 and can be evaluated as biomarkers in serum or plasma, whereas collagen is predominantly expressed in tissues. However, it’s possible to find their fragments or post-translationally modified peptides in circulation or in extracellular vesicles, making these genes clinically relevant and a potential non-invasive biomarker. There is a need to prove this by performing various in-vitro and in-vivo experiments.
Data availability
We used open-source multiple transcriptomic and epigenetic (methylation) datasets to characterize novel diagnostic and prognostic biomarkers. IDs of these datasets are provided in the manuscript. The transcriptome datasets used are GSE87295 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse87295), GSE103552 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse103552), and GSE70493 ([https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70493](https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE70493) ). The epigenomic dataset used is GSE49524 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE49524).
References
Mitra, T. et al. Prospecting of exosomal-miRNA signatures as prognostic marker for gestational diabetes mellitus and other adverse pregnancy outcomes. Front. Endocrinol. 14 https://doi.org/10.3389/fendo.2023.1097337 (2023).
Modzelewski, R., Stefanowicz-Rutkowska, M. M., Matuszewski, W. & Bandurska-Stankiewicz, E. M. Gestational diabetes Mellitus—Recent literature review. J. Clin. Med. 11 (19), 5736. https://doi.org/10.3390/jcm11195736 (2022).
Sherwani, S. I., Khan, H. A., Ekhzaimy, A., Masood, A. & Sakharkar, M. K. Significance of HbA1c test in diagnosis and prognosis of diabetic patients. Biomark. Insights. 11, 95–104. https://doi.org/10.4137/BMI.S38440 (2016).
Guariguata, L., Linnenkamp, U., Beagley, J., Whiting, D. R. & Cho, N. H. Global estimates of the prevalence of hyperglycaemia in pregnancy. Diabetes Res. Clin. Pract. 103 (2), 176–185. https://doi.org/10.1016/j.diabres.2013.11.003 (2014).
Alsaedi, S. A., Altalhi, A. A., Nabrawi, M. F., Aldainy, A. A. & Wali, R. M. Prevalence and risk factors of gestational diabetes mellitus among pregnant patients visiting National guard primary health care centers in Saudi Arabia. Saudi Med. J. 41 (2), 144–150. https://doi.org/10.15537/smj.2020.2.24842 (2020).
Universal screening of gestational diabetes mellitus in Upper Egypt. Prospective cohort study of the prevalence, risk factors, and short-term outcomes. Urol. Nephrol. Open. Access. J. 11 (Issue 2). https://doi.org/10.15406/unoaj.2023.11.00337 (2023).
Kahimakazi, I. et al. Prevalence of gestational diabetes mellitus and associated factors among women receiving antenatal care at a tertiary hospital in South-Western Uganda. Pan Afr. Med. J. 46 (50). https://doi.org/10.11604/pamj.2023.46.50.38355 (2023).
Seshiah, V. et al. Prevalence of gestational diabetes mellitus in South India (Tamil Nadu)--a community based study. J. Assoc. Physicians India. 56, 329–333 (2008).
Ogamba, I. et al. Risk assessment of gestational diabetes mellitus type A2 in socioeconomically disadvantaged women. Am. J. Obstet. Gynecol. 226 (1), S497. https://doi.org/10.1016/j.ajog.2021.11.822 (2022).
Zhou, X. et al. Analysis Glycemic Variability in Pregnant Women with Various Type of Hyperglycemia. https://www.authorea.com/users/804116/articles/1191731-analysis-glycemic-variability-in-pregnant-women-with-various-type-of-hyperglycemia Accessed March 6, (2025).
Ryu, R. J., Hays, K. E. & Hebert, M. F. Gestational diabetes mellitus management with oral hypoglycemic agents. Semin Perinatol. 38 (8), 508–515. https://doi.org/10.1053/j.semperi.2014.08.012 (2014).
Pernicova, I. & Korbonits, M. Metformin—mode of action and clinical implications for diabetes and cancer. Nat. Rev. Endocrinol. 10 (3), 143–156. https://doi.org/10.1038/nrendo.2013.256 (2014).
LaMoia, T. E. & Shulman, G. I. Cellular and molecular mechanisms of Metformin action. Endocr. Rev. 42 (1), 77–96. https://doi.org/10.1210/endrev/bnaa023 (2021).
Rena, G., Hardie, D. G. & Pearson, E. R. The mechanisms of action of Metformin. Diabetologia 60 (9), 1577–1585. https://doi.org/10.1007/s00125-017-4342-z (2017).
Barrett, T. et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 41 (D1), D991–D995. https://doi.org/10.1093/nar/gks1193 (2013).
Chaudhuri, D. et al. Corticosteroids in COVID-19 and non-COVID-19 ARDS: a systematic review and meta-analysis. Intensive Care Med. 47 (5), 521–537. https://doi.org/10.1007/s00134-021-06394-2 (2021).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47. https://doi.org/10.1093/nar/gkv007 (2015).
Wu, T. et al. ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innov. Camb. Mass. 2 (3), 100141. https://doi.org/10.1016/j.xinn.2021.100141 (2021).
Kanehisa, M. & Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. https://doi.org/10.1093/nar/28.1.27 Accessed August 21, (2025).
Kanehisa, M. Toward Understanding the origin and evolution of cellular organisms. https://doi.org/10.1002/pro.3715
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51 (D1), D587–D592. https://doi.org/10.1093/nar/gkac963 (2022).
Lu, S. et al. DNA methylation and expression profiles of placenta and umbilical cord blood reveal the characteristics of gestational diabetes mellitus patients and offspring. Clin. Epigenetics. 14 (1), 69. https://doi.org/10.1186/s13148-022-01289-5 (2022).
Newman, A. M. et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 37 (7), 773–782. https://doi.org/10.1038/s41587-019-0114-2 (2019).
Szklarczyk, D. et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51 (D1), D638–D646. https://doi.org/10.1093/nar/gkac1000 (2023).
Otasek, D., Morris, J. H., Bouças, J., Pico, A. R. & Demchak, B. Cytoscape automation: empowering workflow-based network analysis. Genome Biol. 20 (1), 185. https://doi.org/10.1186/s13059-019-1758-4 (2019).
Robin, X. et al. pROC: an open-source package for R and S + to analyze and compare ROC curves. BMC Bioinform. 12, 77. https://doi.org/10.1186/1471-2105-12-77 (2011).
Kwon, H. et al. Impaired angiogenic function of fetal endothelial progenitor cells via PCDH10 in gestational diabetes mellitus. Int. J. Mol. Sci. 24 (22), 16082. https://doi.org/10.3390/ijms242216082 (2023).
McElwain, C. J., Tuboly, E., McCarthy, F. P. & McCarthy, C. M. Mechanisms of endothelial dysfunction in Pre-eclampsia and gestational diabetes mellitus: windows into future cardiometabolic health. Front. Endocrinol. 11, 655. https://doi.org/10.3389/fendo.2020.00655 (2020).
Chen, J. L., Dai, H. F., Kan, X. C., Wu, J. & Chen, H. W. The integrated bioinformatic analysis identifies immune microenvironment-related potential biomarkers for patients with gestational diabetes mellitus. Front. Immunol. 15, 1296855. https://doi.org/10.3389/fimmu.2024.1296855 (2024).
Radaelli, T., Varastehpour, A., Catalano, P. & Hauguel-de Mouzon, S. Gestational diabetes induces placental genes for chronic stress and inflammatory pathways. Diabetes 52 (12), 2951–2958. https://doi.org/10.2337/diabetes.52.12.2951 (2003).
Pinney, S. E. et al. Exposure to gestational diabetes enriches Immune-Related pathways in the transcriptome and methylome of human amniocytes. J. Clin. Endocrinol. Metab. 105 (10), 3250–3264. https://doi.org/10.1210/clinem/dgaa466 (2020).
Mitra, T. et al. Endocrine disrupting chemicals: gestational diabetes and beyond. Diabetol. Metab. Syndr. 16 (1), 95. https://doi.org/10.1186/s13098-024-01317-9 (2024).
Bellavia, A. et al. Association of self-reported personal care product use with blood glucose levels measured during pregnancy among women from a fertility clinic. Sci. Total Environ. 695, 133855. https://doi.org/10.1016/j.scitotenv.2019.133855 (2019).
Ehrlich, M. DNA hypomethylation in cancer cells. Epigenomics 1 (2), 239–259. https://doi.org/10.2217/epi.09.33 (2009).
Ambra, R. et al. Transcriptome analysis of human primary endothelial cells (HUVEC) from umbilical cords of gestational diabetic mothers reveals candidate sites for an epigenetic modulation of specific gene expression. Genomics 103 (5–6), 337–348. https://doi.org/10.1016/j.ygeno.2014.03.003 (2014).
Szmuilowicz, E. D., Josefson, J. L. & Metzger, B. E. Gestational diabetes mellitus. Endocrinol. Metab. Clin. North. Am. 48 (3), 479–493. https://doi.org/10.1016/j.ecl.2019.05.001 (2019).
Johns, E. C., Denison, F. C., Norman, J. E. & Reynolds, R. M. Gestational diabetes mellitus: mechanisms, treatment, and complications. Trends Endocrinol. Metab. TEM. 29 (11), 743–754. https://doi.org/10.1016/j.tem.2018.09.004 (2018).
de la Torre, N. G. et al. Effectiveness of following mediterranean diet recommendations in the real world in the incidence of gestational diabetes mellitus (GDM) and adverse Maternal-Foetal outcomes: A prospective, universal, interventional study with a single group. The St Carlos study. Nutrients 11 (6), 1210. https://doi.org/10.3390/nu11061210 (2019).
Ovesen, P. G., Jensen, D. M., Damm, P., Rasmussen, S. & Kesmodel, U. S. Maternal and neonatal outcomes in pregnancies complicated by gestational diabetes. A nation-wide study. J. Matern-Fetal Neonatal Med. Off J. Eur. Assoc. Perinat. Med. Fed. Asia Ocean. Perinat. Soc. Int. Soc. Perinat. Obstet. 28 (14), 1720–1724. https://doi.org/10.3109/14767058.2014.966677 (2015).
Kumari, R. S. et al. Potential role of SLC40A1 and HMOX1 in Establishing relation between gestational anemia and gestational diabetes mellitus. Indian Heart J. 76, S60. https://doi.org/10.1016/j.ihj.2024.11.120 (2024).
Okia, C. C. et al. Prevalence, morphological classification, and factors associated with anemia among pregnant women accessing antenatal clinic at Itojo hospital, South Western Uganda. J. Blood Med. 10, 351–357. https://doi.org/10.2147/JBM.S216613 (2019).
Impact of Iron Deficiency Anemia on Prevalence of Gestational Diabetes Mellitus | Diabetes Care | American Diabetes Association. https://diabetesjournals.org/care/article/27/3/650/23077/Impact-of-Iron-Deficiency-Anemia-on-Prevalence-of Accessed March 7. (2025).
Guo, W., Zhou, Q., Jia, Y. & Xu, J. Increased levels of glycated hemoglobin A1c and iron deficiency anemia: A review. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 25, 8371–8378. https://doi.org/10.12659/MSM.916719 (2019).
Hayashi, I., Sakane, N., Suganuma, A. & Nagai, N. Association of a pro-inflammatory diet and gestational diabetes mellitus with maternal anemia and hemoglobin levels during pregnancy: a prospective observational case-control study. Nutr. Res. 115, 38–46. https://doi.org/10.1016/j.nutres.2023.05.003 (2023).
Hivert, M. F. et al. Placental IGFBP1 levels during early pregnancy and the risk of insulin resistance and gestational diabetes. Nat. Med. 30 (6), 1689–1695. https://doi.org/10.1038/s41591-024-02936-5 (2024).
Chen, L., Hui, L. & Li, J. The multifaceted role of insulin-like growth factor binding protein 7. Front. Cell. Dev. Biol. 12 https://doi.org/10.3389/fcell.2024.1420862 (2024).
Anderlová, K. et al. Association between gestational diabetes mellitus and bioavailability of insulin-like growth factors and role of their binding proteins. Growth Horm. IGF Res. 67, 101511. https://doi.org/10.1016/j.ghir.2022.101511 (2022).
Wu, P. L. et al. IGFBP7 enhances trophoblast invasion via IGF-1R/c-Jun signaling in unexplained recurrent spontaneous abortion. Published Online November. 1 https://doi.org/10.1530/REP-21-0501 (2022).
Lipska, K. J. & Montori, V. M. In type 1 diabetes, intensive insulin therapy for 6.5 y reduced mortality at 27 y compared with usual care. Ann. Intern. Med. 162 (10), JC12. https://doi.org/10.7326/ACPJC-2015-162-10-012 (2015).
Sharma, S., Banerjee, S., Krueger, P. M. & Blois, S. M. Immunobiology of gestational diabetes mellitus in Post-Medawar era. Front. Immunol. 12, 758267. https://doi.org/10.3389/fimmu.2021.758267 (2021).
Zhang, Z. et al. Association of Circulating galectin-3 with gestational diabetes mellitus, progesterone, and insulin resistance. J. Diabetes. 13 (1), 54–62. https://doi.org/10.1111/1753-0407.13088 (2021).
Cao, W. et al. Maternal lipids, BMI and IL-17/IL-35 imbalance in concurrent gestational diabetes mellitus and preeclampsia. Exp. Ther. Med. 16 (1), 427–435. https://doi.org/10.3892/etm.2018.6144 (2018).
Yang, Y. et al. Transcriptomic profiling of human placenta in gestational diabetes mellitus at the Single-Cell level. Front. Endocrinol. 12, 679582. https://doi.org/10.3389/fendo.2021.679582 (2021).
Kang, Y. E. et al. Increased Pro-Inflammatory Senescent T Cells, and Immune-Check Point Molecules in the Placentas of Patients With Gestational Diabetes Mellitus. J. Korean Med. Sci. 37(48). https://doi.org/10.3346/jkms.2022.37.e338 (2022).
Li, H., Huang, Q., Liu, Y. & Garmire, L. X. Single cell transcriptome research in human placenta. Published Online Dec. 1 https://doi.org/10.1530/REP-20-0231 (2020).
Chen, Y. et al. Non-Invasive detection of fetal vascular endothelial function in gestational diabetes mellitus. Front. Endocrinol. 12, 763683. https://doi.org/10.3389/fendo.2021.763683 (2021).
Gao, M. et al. Serum CD44 levels in early pregnancy and its genetic variants for increased risk of gestational diabetes mellitus in Chinese pregnant women. J. Proteom. 307, 105268. https://doi.org/10.1016/j.jprot.2024.105268 (2024).
Acknowledgements
All contributors have been given authorship.
Funding
Open access funding provided by SRM Institute of Science and Technology for SRMIST – Medical & Health Sciences.
Author information
Authors and Affiliations
Contributions
T.M., P.A., and R.J. designed the experimental program; P.A. performed the software analysis; T.M., D.V.Y. and P.A. wrote the original draft of the manuscript; S.K.R., P.A. and R.J. contributed to the reviewing and editing of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
No ethical declaration is required for this article.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mitra, T., Yadav, D.V., Kumari R., S. et al. Integrating transcriptomic and epigenomic data to identify potential biomarkers in gestational diabetes mellitus patients. Sci Rep 15, 32768 (2025). https://doi.org/10.1038/s41598-025-18018-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-18018-6