Abstract
In this study, a dataset for solubility of raloxifene and CO2 density was analyzed using different regression models to reveal the correlation between inputs and drug solubility via supercritical processing. The models were developed and analyzed for their accuracy in predicting the process variables. Three models of Elastic Net Regression (ENR), Orthogonal Matching Pursuit (OMP), and Gaussian Process Regression (GPR) are optimized for the dataset of drug. For CO2 density estimation, GPR turned out to be the best R2, which confirmed a robust correlation. Therefore, the GPR model can be considered the most accurate and reliable for predicting CO2 density in this context. Regarding solubility, both the ENR and OMP models demonstrated similar performance with high R2 of 0.89062 and 0.89125, respectively. However, GPR outperformed the others with an impressive R2 of 0.97755, a lower RMSE of 3.3221E-01, and a relatively lower AARD% of 7.08009E + 00. Hence, the GPR model is recommended for accurate solubility predictions of raloxifene. Overall, the GPR model consistently demonstrated excellent predictive performance for both CO2 density and solubility, making it the preferred choice for modeling these relationships.
Similar content being viewed by others
Introduction
Drug solubility can be estimated by computational models such as thermodynamic or data-driven models. In pharmaceutical industry, it is important to determine the solubility of medicines at different solvents, as the solubility prediction is used for design of formulations with enhanced solubility as well as analysis of crystallization process for separation of solid (drug) from solvent. Solubility plays a crucial role in crystallization of pharmaceutical compounds as it is needed to find the metastable zone and supersaturation conditions for driving the process1,2. Equation of state (EoS) is one of the major approaches in thermodynamic modeling of drugs solubility in supercritical solvents. Zhang et al.3 developed PC-SAFT equation of state to correlate solubility data of anticancer drugs in supercritical CO2. They used measured data for optimization of temperature-independent parameters of the model which showed great prediction accuracy. Also, other EoS-based models have been proposed for estimation of solubility data which are based on the thermodynamic nature4,5. For modeling solubility of drugs, usually a solid-liquid equilibrium is assumed for building thermodynamic models.
Measured solubility data are needed for building correlative models such as thermodynamic models, while it is complicated to obtain measured data for large number of APIs (active pharmaceutical ingredients). On the other hand, building and optimization of thermodynamic models for large number of APIs is time consuming and thus other modeling techniques should be explored for correlation of drug solubility dataset. Data-driven models are great alternatives to thermodynamic models in correlation of solubility data, and have been used with reliability in drug solubility estimation6. The data are generated via solubility measurements and then used for fitting the data-driven models. The trained and tested models can be used for estimation of drug solubility values at wide conditions including pressure, temperature, and composition7,8. Better accuracy has been obtained for data-driven models in comparison to thermodynamic models which shows the usefulness of these models in pharmaceutical modeling9.
Machine learning (ML) models are considered as data-driven approach which can be used in correlating drug solubility dataset10. This approach has applications in a wide range of fields and industries. Regression models are an integral part of ML for quantitative analysis of large dataset. In regression, the goal is to learn a mapping function that can predict the target variable using the features11,12,13. Elastic Net Regression (ENR), Orthogonal Matching Pursuit (OMP), and Gaussian Process Regression (GPR) are three powerful regressive methods for correlation of API solubility data.
The ML models used in this study are all justified for their robustness in correlation of drug dataset. In fact, ENR combines the benefits of Lasso and Ridge regression, offering a comprehensive framework for variable selection and regularization. The model’s flexibility, interpretability, and ability to handle multicollinearity contribute to its wide applicability and relevance in diverse domains14. GPR is a Bayesian approach that provides not only point predictions but also a measure of uncertainty by estimating the conditional probability distribution, enabling robust and flexible modelling in a wide range of applications15. Also, in OMP regression, the algorithm commences with a null set of chosen inputs and proceeds to incrementally incorporate the feature that shows the greatest correlation with the residual16,17.
In this study, Elastic Net Regression, Orthogonal Matching Pursuit, and Gaussian Process Regression models are used with Grey Wolf Optimization (GWO) as hyperparameters tuner to determine solubility of raloxifene and CO2 density under supercritical state. The modeling strategy offers a unique platform for the first time in correlating raloxifene solubility with the aim of increasing fitting precision compared to the previous ML models. The models are then compared to reveal their accuracy in handling the dataset.
Dataset
We analyzed a supercritical processing using machine learning, and the dataset of this research which is taken from18, consists of measurements of solubility of raloxifene, temperature, pressure, and supercritical CO2 density. Raloxifene was considered as a case study because its water solubility is poor and the method of supercritical can be assessed for application of this drug to nanonize the drug particles for more aqueous solubility. All parameters were considered for building ML models in two steps, i.e., training and validation. The variables were selected to ensure that the solvent (CO2) is in the supercritical state which is at the pressure of 7.38 MPa and temperature of 304.1 K for CO2. Here is a breakdown of the different columns in the dataset:
-
Temperature (T): This column represents the temperature values measured in Kelvin (K). The dataset includes temperature values of 313 K, 318 K, 328 K, and 343 K.
-
Pressure (P): This column represents the pressure values measured in bar. The dataset includes pressure values of 100 bar, 120 bar, 140 bar, 165 bar, 185 bar, 205 bar, and 240 bar. All values are above the solvent’s supercritical pressure.
-
Solubility of Raloxifene (y): This column represents the solubility of raloxifene. The solubility values are given as numerical values for each combination of temperature and pressure.
-
Supercritical CO2 Density: This column represents the density of carbon dioxide (CO2). The density values are also provided as numerical values corresponding to each combination of temperature and pressure. Density is selected as the output because the solvent is compressible, and density changes can have major influence on the variations of raloxifene solubility.
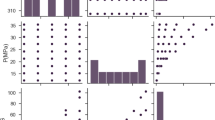
Figure 1 illustrated the frequencies of input and output variables in histogram plots using raw data. As seen, the distribution of density is more skewed compared to the drug solubility data. While low solubility points have higher frequency, the solvent density distribution shows higher frequency for large numbers which is due to increasing pressure in the process which significantly impacted the solvent density as it is a compressible solvent in the process.
Methodology
Grey Wolf optimization (GWO)
GWO is a metaheuristic technique taking cues from the hunting strategies exhibited by grey wolves. The algorithm simulates the hunting dynamics of a pack consisting of four different categories of wolves, namely alpha, beta, delta, and omega. Each wolf in the pack is associated with a vector of decision variables representing its position within the search space. The search process in GWO is guided by the positions of these wolves, which are progressively optimized according to their own positions and the positions of other wolves in the pack19,20.
The updating equation for the position of each wolf is given by21:
In the above equation, \(\:\overrightarrow{C}\) and \(\:\overrightarrow{A}\) denote coefficient vectors, \(\:\overrightarrow{{r}_{1}}\) stands for a random vector, \(\:\overrightarrow{D}\) indicates the distance vector, and \(\:\overrightarrow{{x}_{i}}\left(t\right)\) stands for the position of the i-th wolf at t-th iteration.
Elastic net regression (ENR)
Considering the training samples \(\:X=\left[{x}_{1},{x}_{2},\dots\:,{x}_{N}\right]\in\:{R}^{N\times\:D}\), where \(\:N\) denotes the quantity of samples and \(\:D\) is the dimensionality, the Elastic Net (EN) regression algorithm aims to find the optimal coefficients that minimize the sum of squared errors, while simultaneously promoting sparsity. EN optimization problem is formulated as22:
where \(\:{\upbeta\:}\) is the coefficient vector, \(\:{y}_{i}\) is the target output associated with the i-th sample, \(\:{x}_{i}\) is the corresponding feature vector, \(\:{\left|\cdot\:\right|}_{1}\) denotes the \(\:{\text{l}}_{}norm\left(L1norm\right),{\left|\cdot\:\right|}_{}\) denotes the \(\:{\text{l}}_{2}\) norm (Euclidean norm), \(\:{{\uplambda\:}}_{1}\) controls the amount of L1 regularization, and \(\:{{\uplambda\:}}_{2}\) controls the amount of L2 regularization23.
Orthogonal matching pursuit (OMP)
OMP combines the inherent strengths of the OMP algorithm with an additional regularization term, resulting in enhanced performance and interpretability24. Given a set of training samples \(\:X=\left[{x}_{1},{x}_{2},\dots\:,{x}_{N}\right]\in\:{R}^{N\times\:D}\), where \(\:N\) shows the total count of samples and \(\:D\) is the dimensionality, the OMP regression algorithm aims to find the sparsest representation of each input sample in terms of a learned dictionary25. The dictionary matrix \(\:D\in\:{R}^{D\times\:K}\), with \(\:K\) being the number of dictionary atoms, is constructed by selecting a subset of \(\:K\) atoms from a larger candidate pool26.
To estimate the coefficients, OMP regression solves the following optimization problem for each sample27:
where \(\:{{\upalpha\:}}_{i}\) is the coefficient vector for the i-th sample, \(\:{y}_{i}\) is the target output associated with \(\:{x}_{,}{\left|\cdot\:\right|}_{2}^{}\) denotes the squared Euclidean norm, \(\:{\left|\cdot\:\right|}_{0}\) stands for the \(\:{\text{l}}_{0}\) “pseudo-norm” that counts the number of nonzero entries in \(\:{{\upalpha\:}}_{i}\), and \(\:{\uplambda\:}\) controls the trade-off between fitting the data and promoting sparsity25.
Gaussian process regression (GPR)
In ML, GPR is applied for estimating the conditional probability distribution of a continuous response variable, denoted as y, given a set of predictor variables, denoted as x. The key concept underlying GPR is the Gaussian processes (GPs), which represent a set of random variables following a Gaussian distribution. In the context of GPR, GPs are employed to model the unknown function that establishes the interrelationship between the input variables and the output targets15,28.
In the GPR model, the response variable y is patterned after a Gaussian process with a covariance function k(x, x’) and a mean function m(x). This is expressed as \(\:y\left(x\right)\sim\:\mathcal{G}\mathcal{P}\left(m\left(x\right),k\left(x,{x}^{{\prime\:}}\right)\right)\) in which, m(x) denotes the expected value of the response given the predictors, and k(x, x’) captures the similarity between responses at different predictor values. Deciding on the covariance function such as the squared exponential or Matern functions, determines the level of similarity between responses and is typically selected from a family of parametric or non-parametric functions29,30,31.
The GPR algorithm involves the following steps32,33:
-
Choose a mean and a covariance function for the Gaussian process.
-
Maximize the marginal likelihood of the training data to estimate the covariance function hyperparameters.
-
Given the observed training data, compute the posterior variance and mean of the Gaussian process at the predictor values in the test data.
-
Predict the response of the new measurement as the posterior mean of the Gaussian process.
Results and discussion
Solvent density
The models described in this research were fitted and developed via Python version 3.8 software, as the open-source software which was downloaded from: https://www.python.org.
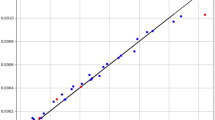
Table 1 shows the statistical evaluations associated with the prediction of supercritical solvent density via the three different methods. Comparative analysis was carried out using several statistical parameters as listed in Table 1. The results indicate that the GPR model achieved the highest R2 score (0.98578), illustrating a great correlation. This accuracy is attributed to the proper selection of features and optimizing the hyperparameters using GWO33. It also indicated the lowest RMSE (26.255) and AARD% (4.83286), demonstrating accurate and reliable predictions. Therefore, the GPR model was the best model for CO2 density prediction. Figure 2 displays the residuals of GPR for CO2 density correlation. Also, Figs. 3 and 4 show the direct relationship between density and pressure and its inverse relationship with temperature. The predictive function of the GPR model for this output is also shown in 3D in Fig. 5. Contour plot of CO2 density is shown in Fig. 6 using the GPR model. As the density is changed with pressure (see Fig. 3), it is expected that the solubility to be increased with pressure which is a great advantage of supercritical CO2 as the solvent. This has been also reported in other works for drug solubility estimation via machine learning6,33,34,35,36,37.
3D surface of supercritical CO2 density simulated using GPR model. Drawn by Python 3.8, which can be accessed from the link: https://www.python.org.
Solubility analysis
The analysis of solubility prediction using the three models is listed in Table 2. Similar to the density correlation, four major criteria are presented and applied for comparative evaluation of ML models33. Both the ENR and OMP models exhibited similar performance metrics, achieving high R2 and relatively small RMSE. However, GPR outperformed them with the highest R2 of 0.97755. The GPR model also demonstrated the lowest RMSE (0.33221) and AARD% (7.08009). Consequently, the GPR model is chosen as the most robust model for solubility prediction.
The results of ML model (GPR) in this study are greater than the thermodynamic models developed for raloxifene as reported by18. Table 3 compares the performance of GPR model developed in this research for raloxifene with the thermodynamic models which are based on Equation of State model of Peng Robinson as well as semi empirical correlation which is Mendez-Santiago-Teja (MST)18.
Figure 7 shows the residuals of GPR for drug solubility correlation. Also, Figs. 8 and 9 show the increase in solubility with the increase in both input parameters. The final predictive function of the GPR model for this output is also shown in 3D in Fig. 10. Also, contour plot of solubility of drug is shown in Fig. 11. The dual effect of T on raloxifene solubility is related to the solvent compressibility. At higher T values, the density of supercritical CO2 is reduced, however the solubility of raloxifene is increased due to the higher interactions between the drug molecules and solvent which dominates the density reduction with increasing temperature10. These solubility changes were also reported in other studies with similar trends6,33,35,36,37.
3D surface of raloxifene solubility constructed via GPR model. Drawn by Python 3.8, which can be accessed from the link: https://www.python.org.
To validate the generalizability of the Gaussian Process Regression model trained on raloxifene data, an external dataset consisting of 15 additional drugs with diverse molecular structures and physicochemical properties was analyzed. For each compound, solubility predictions were compared with experimental data, and model performance was evaluated using the R², RMSE, and AARD%. Data was collected from published sources for different drugs38,39,40,41,42. As shown in Table 4, the GPR model consistently achieved R² values above 0.91 and AARD% below 10% for all drugs, demonstrating high predictive accuracy and robustness. The strong performance across compounds such as sunitinib malate, lansoprazole, and buprenorphine HCl highlights the versatility of the model, confirming its suitability as a generalized solubility prediction tool for pharmaceutical process design under supercritical CO₂ conditions.
Conclusion
Three regression models of ENR, OMP, and GPR were tuned and fitted to predict raloxifene solubility and CO2 density via P and T. These models were optimized using the Grey Wolf Optimization algorithm to obtain their optimal hyperparameters. Based on the evaluation metrics, three models performed well in predicting the solubility and CO2 density. The GPR model showed the highest accuracy for CO2 density and solubility. It also showed the RMSE and AARD% values among the models. The ENR and OMP models also yielded satisfactory results, with decent R-squared and reasonably low errors. Overall, the results confirmed the validity of machine learning regression models in predicting the solubility and CO2 density for raloxifene drug. The accurate predictions obtained from these models can contribute to deeper knowledge of the drug’s behavior and aid in the optimization of pharmaceutical processes. Further research can focus on exploring additional features and applying these models to larger datasets to enhance their predictive capabilities in pharmaceutical applications.
Data availability
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
References
Thakur, A. K. et al. A critical review on thermodynamic and hydrodynamic modeling and simulation of liquid antisolvent crystallization of pharmaceutical compounds. J. Mol. Liq. 362, 119663 (2022).
Yu, Z. Q., Zhang, F. K. & Tan, R. B. H. Liquid–liquid phase separation in pharmaceutical crystallization. Chem. Eng. Res. Des. 174, 19–29 (2021).
Zhang, C. et al. Thermodynamic modeling of anticancer drugs solubilities in supercritical CO2 using the PC-SAFT equation of state. Fluid. Phase. Equilibria. 587, 114202 (2025).
Faraz, O. et al. Thermodynamic modeling of pharmaceuticals solubility in pure, mixed and supercritical solvents. J. Mol. Liq. 353, 118809 (2022).
Ardestani, N. S., Majd, N. Y. & Amani, M. Experimental measurement and thermodynamic modeling of capecitabine (an anticancer Drug) solubility in supercritical carbon dioxide in a ternary system: effect of different cosolvents. J. Chem. Eng. Data. 65 (10), 4762–4779 (2020).
Aldawsari, M. F., Mahdi, W. A. & Alamoudi, J. A. Data-driven models and comparison for correlation of pharmaceutical solubility in supercritical solvent based on pressure and temperature as inputs. Case Stud. Therm. Eng. 49, 103236 (2023).
Li, M. et al. Optimization of drug solubility inside the supercritical CO2 system via numerical simulation based on artificial intelligence approach. Sci. Rep. 14 (1), 22779 (2024).
Ghazwani, M. & Begum, M. Y. Computational intelligence modeling of hyoscine drug solubility and solvent density in supercritical processing: gradient boosting, extra trees, and random forest models. Sci. Rep. 13 (1), 10046 (2023).
He, L. et al. Theoretical understanding of pharmaceutics solubility in supercritical CO2: Thermodynamic modeling and machine learning study. J. Supercrit. Fluids. 223, 106605 (2025).
Alotaibi, H. F. et al. Computational machine learning Estimation of Digitoxin solubility in supercritical solvent at different temperatures utilizing ensemble methods. Sci. Rep. 15 (1), 29248 (2025).
Huang, J. C. et al. Application and comparison of several machine learning algorithms and their integration models in regression problems. Neural Comput. Appl. 32, 5461–5469 (2020).
Alpaydin, E. Introduction To Machine Learning (MIT Press, 2020).
Jin, H. et al. Development of machine learning-based solubility models for Estimation of hydrogen solubility in oil: models assessment and validation. Case Stud. Therm. Eng. 51, 103622 (2023).
Algamal, Z. Y. & Lee, M. H. High dimensional logistic regression model using adjusted elastic net penalty. Pakistan Journal of Statistics and Operation Research 11(4), 667-676 (2015).
Schulz, E., Speekenbrink, M. & Krause, A. A tutorial on Gaussian process regression: modelling, exploring, and exploiting functions. J. Math. Psychol. 85, 1–16 (2018).
Pati, Y. C., Rezaiifar, R. & Krishnaprasad, P. S. Orthogonal matching pursuit: Recursive function approximation with applications to wavelet decomposition. in Proceedings of 27th Asilomar conference on signals, systems and computers. IEEE. (1993).
Yang, F. et al. Artificial intelligence for computation and development of nanodrug solubility in supercritical solvent: analysis of temperature and pressure influence. J. Mol. Liq. 414, 126095 (2024).
Notej, B. et al. Increasing solubility of phenytoin and raloxifene drugs: application of supercritical CO2 technology. Journal of Molecular Liquids, 2023: p. 121246.
Mirjalili, S., Mirjalili, S. M. & Lewis, A. Grey Wolf optimizer. Adv. Eng. Softw. 69, 46–61 (2014).
Dereli, S. A new modified grey Wolf optimization algorithm proposal for a fundamental engineering problem in robotics. Neural Comput. Appl. 33 (21), 14119–14131 (2021).
Li, Z. et al. A novel discrete grey wolf optimizer for solving the bounded knapsack problem. in Computational Intelligence and Intelligent Systems: 10th International Symposium, ISICA 2018, Jiujiang, China, October 13–14, Revised Selected Papers 10. 2019. Springer. (2018).
Heiss, F., Hetzenecker, S. & Osterhaus, M. Nonparametric Estimation of the random coefficients model: an elastic net approach. J. Econ. 229 (2), 299-321 (2022).
Zou, H. & Hastie, T. Regularization and variable selection via the elastic net. J. Royal Stat. Society: Ser. B (statistical methodology). 67 (2), 301–320 (2005).
Tropp, J. A. & Gilbert, A. C. Signal recovery from random measurements via orthogonal matching pursuit. IEEE Trans. Inf. Theory. 53 (12), 4655–4666 (2007).
Li, M. et al. Employment of artificial intelligence approach for optimizing the solubility of drug in the supercritical CO2 system. Case Stud. Therm. Eng. 57, 104326 (2024).
Goyal, P. & Singh, B. Sparse signal recovery through regularized orthogonal matching pursuit for WSNs applications. in 6th International Conference on Signal Processing and Integrated Networks (SPIN). 2019. IEEE. 2019. IEEE. (2019).
Needell, D. & Vershynin, R. Uniform uncertainty principle and signal recovery via regularized orthogonal matching pursuit. Found. Comput. Math. 9, 317–334 (2009).
Bernardo, J. et al. Regression and classification using Gaussian process priors. Bayesian Stat. 6, 475 (1998).
Pustokhina, I. et al. Developing a robust model based on the Gaussian process regression approach to predict biodiesel properties. Int. J. Chem. Eng. 2021, 1–12 (2021).
Rasmussen, C. E. & Nickisch, H. Gaussian processes for machine learning (GPML) toolbox. J. Mach. Learn. Res. 11, 3011–3015 (2010).
Shi, J. Q. & Choi, T. Gaussian Process Regression Analysis for Functional Data (CRC, 2011).
Ruiz, A. V. & Olariu, C. A general algorithm for exploration with gaussian processes in complex, unknown environments. in IEEE International Conference on Robotics and Automation (ICRA). 2015. IEEE. 2015. IEEE. (2015).
Ghazwani, M. et al. Development of advanced model for Understanding the behavior of drug solubility in green solvents: machine learning modeling for small-molecule API solubility prediction. J. Mol. Liq. 386, 122446 (2023).
Ghazwani, M., Yasmin, M. & Begum Machine learning aided drug development: assessing improvement of drug efficiency by correlation of solubility in supercritical solvent for nanomedicine Preparation. J. Mol. Liq. 387, 122511 (2023).
Wu, S. et al. Intelligence modeling of nanomedicine manufacture by supercritical processing in Estimation of solubility of drug in supercritical CO2. Sci. Rep. 15 (1), 23193 (2025).
Liu, Y. et al. Machine learning based modeling for Estimation of drug solubility in supercritical fluid by adjusting important parameters. Chemometr. Intell. Lab. Syst. 254, 105241 (2024).
Abouzied, A. S. et al. Assessment of solid-dosage drug Nanonization by theoretical advanced models: modeling of solubility variations using hybrid machine learning models. Case Stud. Therm. Eng. 47, 103101 (2023).
Luo, B. et al. Experimental validation and modeling study on the drug solubility in supercritical solvent: case study on exemestane drug. J. Mol. Liq. 377, 121517 (2023).
Sodeifian, G. et al. Measurement, correlation and thermodynamic modeling of the solubility of ketotifen fumarate (KTF) in supercritical carbon dioxide: evaluation of PCP-SAFT equation of state. Fluid. Phase. Equilibria. 458, 102–114 (2018).
Sodeifian, G. et al. Solubility of buprenorphine hydrochloride in supercritical carbon dioxide: study on experimental measuring and thermodynamic modeling. Arab. J. Chem. 16 (10), 105196 (2023).
Sodeifian, G., Sajadian, S. A. & Derakhsheshpour, R. Experimental measurement and thermodynamic modeling of Lansoprazole solubility in supercritical carbon dioxide: application of SAFT-VR EoS. Fluid. Phase. Equilibria 507, 112422 (2020).
Saadati Ardestani, N. & Amani, M. Supercritical solvent impregnation of sodium valproate nanoparticles on polymers: characterization and optimization of the operational parameters. J. CO2 Utilization. 64, 102159 (2022).
Acknowledgements
This research was funded by Taif University, Saudi Arabia, Project No. (TU-DSPP-2024-82).
Author information
Authors and Affiliations
Contributions
H.O.A.: Supervision, Funding, Method development, Validation, Writing, Investigation. Y.S.A.: Writing, Software, Conceptualization, Resources, Analysis. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alsaab, H.O., Althobaiti, Y.S. Intelligence modeling of solubility of raloxifene and density of solvent for green supercritical processing of medicines for enhanced solubility. Sci Rep 15, 34615 (2025). https://doi.org/10.1038/s41598-025-18223-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-18223-3