Abstract
BRAF mutations appear to varying degrees in human cancers. Proposed oxo-tetrahydro-pyrimidin-benzenesulfonamide hybrids target [αC-OUT/DFG-IN] conformation of BRAFV600Esimilar to second-generation FDA-approved drugs. Nine compounds (S1–S9) were synthesized and spectrally characterized using Mass, HRMS,1H, and13C NMR. All synthesized derivatives were tested for anti-proliferative activity against two cancer cell lines, and the percentage of BRAFV600E enzyme kinase inhibition was calculated using sorafenib as the standard. Molecular docking was performed for all compounds, while molecular dynamics simulations were conducted for the most active molecules, providing insights into their stability and interactions within the target binding site. The biological assay revealed that most compounds exhibited significant anticancer activity, with compound S4 demonstrating strong inhibition of the BRAFV600E kinase. Notably, S4 (91%) and S1 (87%) showed potent inhibitory effects, comparable to the reference drug, sorafenib (94%). Based on these promising results, S4 and S1 were selected for molecular dynamics simulations to elucidate their binding stability and conformational dynamics within the BRAFV600E active site. These findings highlight that these compounds may act as potential lead compounds for the development of BRAFV600E inhibitors.
Similar content being viewed by others
Introduction
Cancer is characterized by uncontrolled cell proliferation and metastasis1. In MAPK pathway, RAS, RAF, MEK & ERK are four major components. BRAF (RAS) plays a vital role in this signaling cascade. Mutation in MAPK signaling pathway is responsible for carcinogenesis2,3. Davies et al. initially identified somatic mutations in the BRAF gene in 2002. Subsequently, three different classes of BRAF mutants have been discovered4. Class I monomeric mutants (BRAFV600), class II BRAF homodimeric mutants (non-V600), and class III BRAF heterodimers (non-V600). Human cancers can have BRAF mutations to varied degrees; melanoma has about 70–90% of them, thyroid cancer has 30–50%, and ovarian cancer has 5–30%. 1–4% for non-small cell lung cancer, 1–3% for Langerhans cell histiocytosis, hairy cell leukaemia, and numerous other conditions5,6,7.
To date, three generations of BRAF inhibitors have been developed. Among them, only sorafenib (first generation), as well as dabrafenib, vemurafenib, and encorafenib (second generation), have received FDA approval. Other BRAF inhibitors are currently in various stages of clinical testing8,9,10.
The classification of BRAF inhibitors is fundamentally rooted in their capacity to engage distinct conformational states of the kinase domain, dictated by the spatial orientation of the DFG motif and αC-helix-critical structural elements governing ATP-binding pocket dynamics. Four principal binding modes have emerged:
-
(i)
Type I (αC-IN/DFG-IN): First-generation ATP-competitive inhibitors target the active kinase conformation, stabilizing BRAF in its catalytically competent state. While effective against monomeric BRAFV600E, these agents paradoxically activate wild-type BRAF in dimeric configurations, driving MAPK signaling in RAS-mutant cells and increasing risks of cutaneous squamous cell carcinomas.
-
(ii)
Type II (αC-IN/DFG-OUT): These inhibitors bind to a hydrophobic pocket adjacent to the ATP site, enforcing an inactive DFG-OUT conformation. These “paradox breakers” suppress both monomeric and dimeric BRAF signaling, mitigating off-target ERK activation while retaining efficacy against resistance mutations.
-
(iii)
Type III (αC-OUT/DFG-IN): These inhibitors exploit the αC-OUT conformation, selectively inhibiting BRAFV600E monomers without inducing RAF dimerization. Their shallow binding mode limits efficacy against dimer-dependent malignancies.
-
(iv)
Type IV (αC-OUT/DFG-OUT): A nascent class with limited clinical development, these compounds target rare “double-out” conformations, offering potential against dimer-stabilized BRAF mutants but challenged by poor pharmacokinetics. This structural taxonomy underscores a paradigm shift in kinase drug design-from initial ATP-mimetics to conformationally intelligent agents that decouple therapeutic efficacy from paradoxical pathway activation. Contemporary efforts focus on Type II/III hybrids to balance potency, selectivity, and resistance profiles, while Type IV inhibitors remain an underexplored frontier5,7,11,12.
Currently, FDA-approved first and second-generation BRAF kinase inhibitors resist BRAF dimerization. BRAF inhibitor-treated patients may develop secondary skin lesions like squamous cell carcinomas, keratoacanthomas, and hyperkeratosis. Patients taking BRAF inhibitors have also been reported to develop secondary melanomas, gastric and colonic polyps, and recurrences of prior cancers13,14. Because third-generation pan-RAF inhibitors were created as “paradox breakers”—that is, they activate BRAF in a particular way that stimulates dimerization and more strongly promotes paradoxical activation—they are especially intriguing. To address the issues of various BRAF mutations, dimerization, and paradoxical activation, numerous academic scientists and researchers began working on it. The biological evaluation of several BRAF inhibitors that contained different scaffolds, including pyrazine, imidazole, pyridine, pyrazole, pyrimidine, sulfonamide, quinoxaline, etc., as well as their hybrids, was published in this context15,16.
In this study, urea-based pyrimidine-cyclized sulfonamide derivatives were designed using structure-based and computational approaches. The compounds were subsequently synthesized, and their in vitro anti-proliferative and BRAFV600E kinase inhibitory activity were evaluated. Based on these finding a SAR was summarized.
Rational design
The design of target compounds has been made on the basis of different structural moieties present in first and second-generation FDA-approved BRAF inhibitors (Fig. 1).
The mobility of the DFG motif is critical for the selectivity of BRAF inhibitors. In order to design and develop BRAF inhibitors that take advantage of the characteristics of BRAF binding sites, conformational changes, and DFG rearrangement, a structure-based approach is highly advised. There are still many issues with the design and development of BRAF inhibitors because many of them are based on the DFG conformation and the αC-helix11,17,18. The designed compound has a binding affinity in the active core region of the ATP-binding pocket (Fig. 1). In this, R1-substituted sulfonamide interacts with the RAF selective pocket, phenyl ring interacts with the hydrophobic pocket, pyrimidine interacts with the ribose pocket, R2-substitution interact with adenine pocket, and [keto (-C = O)] interacts with other than ATP binding pocket which is a solvent assessable region. In addition, the developed pyrimidine-sulfonamide hybrids exhibited [αC-OUT/DFG-IN] conformation similar to the FDA-approved BRAFV600E inhibitors. The designed compounds interacted with the dimerization interface (DIF), making those potentially valuable agents against BRAFV600E resistance and malignancies caused by dimeric BRAF mutants.
Experimental
General
Sigma-Aldrich, Spectrochem Pvt. Ltd., and TCI India Chemicals Pvt. Ltd. were the suppliers of all reagents and solvents, which were used without additional purification. The Sartorius analytical balance (BSA224s-CW) was used for weighing. The organic compounds were dried using an ILMVAC rotary evaporator and the JSGW heating mantle for the reflux reaction. In a JSGW UV/fluorescent analysis cabinet and iodine chamber, petroleum ether/ethyl acetate, petroleum ether/acetone, or chloroform/methanol served as the mobile phase on pre-coated Merck TLC plates, and TLC was utilized to track the reactions’ progress. Compounds were purified using column chromatography (pore size 60 Å, particle size 60–120 mesh, Silica gel) to achieve the desired pure product. All melting points are uncorrected and were obtained using an open glass capillary tube and the Stuart melting point device (SMP-11). The structures of the final synthesized compounds were confirmed using1H-NMR13,C-NMR, Mass spectrometry. HRMS and deuterium exchange NMR was also done for representative compound.
The1H and13C nuclear magnetic resonance spectra for solutions in (DMSO-d6) were obtained using a JEOL, 600 MHz & 151 MHz spectrometer and are reported in parts per million (ppm), downfield from tetramethylsilane (TMS) as an internal standard. The mass spectrum of compounds was recorded using Shimadzu GC-MS and organic solvents.
General procedure for synthesis
Synthesis of phenyl sulfonyl derivatives (CPD 03)
Benzenesulfonyl chloride (CPD 3 A, 1 mmol) and amino acetophenone (CPD 3B, 1 mmol) were taken in RBF and dissolved by adding dichloromethane (DCM). In this, 3 to 5 ml (Et)3N was added drop by drop. The whole reaction was put on an ice bath at (0–5 °C) with continued stirring. After 6–8 h, the reaction was completed and confirmed by the TLC solvent system (pet ether: ethyl acetate) (7:3). The reaction was stopped, and the organic layer was separated using water: ethyl acetate extraction processes and rotavapor was used to evaporate the solvent and solid CPD 03 was obtained.
Synthesis of phenyl sulfonamide-chalcone derivatives (CPD 04)
A mixture of benzene sulfonyl derivatives (CPD 03, 1 mmol) and benzaldehyde (1 mmol) was added in RBF and dissolved by methanol. In this solution, 40% (NaOH) was added and stirred at room temperature for 8–10 h. The reaction was monitored by a TLC solvent system (chloroform: ethyl acetate) (3:7). After the competition, reaction was poured onto ice water, and the solid (CPD 04) obtained was collected by filtration.
Synthesis of oxo-tetrahydro-pyrimidin-benzenesulfonamide derivatives (S1-S9)
Sulfonamide-chalcone derivatives (CPD 04, 1 mmol) and urea (7 mmol) were taken in RBF for the final step. I2 pellets (2–3 equivalent) and 40% KOH were added and refluxed with stirring at 80–90 °C temperature for 8–10 h. TLC using chloroform: ethyl acetate solvent system monitored the reaction. After completion of reaction, it was cooled, and ethyl acetate and water were added to this organic layer, separated, rotated, and dried to get the final product (S1-S9). Column chromatography was used to purify the compounds. After purification, the spectral characterization was done on synthesized compounds (Fig. 2; Table 1)19,20.
Procedure for computational studies
Molecular docking
The three-dimensional high-resolution structure of the human BRAF kinase (PDB ID: 5JRQ, resolution: 2.29 Å) was retrieved from the RCSB Protein Data Bank21, as described in seminal study22. For molecular docking and subsequent molecular dynamics (MD) simulations, only the A chain of the protein was retained. Non-protein elements, including the ligand vemurafenib (VEM), water molecules, glycerol, dimethyl sulfoxide, and tetramethylammonium ions, were removed. Missing residues and atoms were reconstructed using UCSF Chimera’s implementation of the Šali and Blundell algorithm23, resulting in a model with a normalized Discrete Optimized Protein Energy (DOPE) score of −1.87.
To generate initial geometries for MD simulations, molecular docking experiments were performed. Ligand 3D coordinates were derived from SMILES representations using ChimeraX24. Ten conformers for each ligand were generated and energy-minimized using the MMFF94 force field25 via RDKit26. The lowest-energy conformer was then converted to pdbqt format using the Meeko toolkit27. The BRAF receptor structure was prepared for docking by retaining polar hydrogens and assigning Gasteiger charges using ChimeraX.
Molecular docking simulations were carried out with AutoDock Vina28, using a grid box centered at the crystallized ligand’s center of mass (coordinates: −11.46, −4.34, −29.45 Å). The grid box was cubic, with an edge length of 25 Å. Ligands were treated as flexible, while the protein was kept rigid. Docking parameters included an exhaustiveness value of 100 and a maximum of 100 poses per ligand. Conformations scoring within 4 kcal·mol-1 of the highest-ranked pose were retained, and the best-scoring pose, following visual inspection, was selected for further analysis.
The docking protocol was validated by re-docking vemurafenib into the BRAF binding site. The heavy-atom RMSD between the docked and crystallographic ligand structures was 3.00 Å, while the all-atom RMSD was 6.76 Å, primarily due to flexible groups such as the propyl chain and phenyl-OCH₃ moiety. Despite these deviations, the key ligand–receptor interactions were preserved. Detailed structural comparison and interaction analysis (Fig. S1–2, Table S1) support the reliability of the docking model as a starting point for MD simulations.
Molecular dynamics simulation
Using second-order Møller–Plesset perturbation theory (MP2) with the 6-31G basis set, the geometries of the top two compounds (S1 and S4) that showed the highest inhibition of BRAF protein in the BRAFV600E kinase assay, as well as the reference medication sorafenib, were optimized. Using the HF/6-31G method and the Merz–Singh–Kollman scheme29 the electrostatic potential was utilized to determine atomic charges at particular locations. The AMBER 22 Antechamber module30, was used for parameterization in molecular dynamics (MD) simulations, with general AMBER Force Field (GAFF2)31 for ligands and the AMBER ff19SB force field32 for the protein. To adjust the side chain protonation states of the residues to physiological pH (7.4), the PDB2PQR web server was utilized33. The geometries obtained by molecular docking experiments served as initial geometries of the ligand–protein complexes for the MD simulations. The parameterized protein-ligand complexes were then soaked in a truncated octahedral box with pre-equilibrated TIP3P water molecules. Periodic boundary conditions were applied, ensuring a minimum distance of 12 Å from any atom to the box boundary. This solvated system was neutralized by adding six chlorine anions, followed by additional Na+ and Cl− ions to achieve a salt concentration of 0.15 M, following the recommendations of Machado and Pantano34.
As confirmed in our earlier work, a minimization–heating–equilibration–production protocol was used for molecular dynamics simulation at physiological temperatures (310 K, 1 atm). 10,000 cycles of harmonic potential constraints (k = 10.0 kcal mol − 1Å−2) on all non-solvent molecules were used in the minimization process, which consisted of 4000 steepest descents followed by 6000 conjugate gradients. A second minimization, with no restrictions, followed a similar protocol. The systems were progressively heated from 0 K to 300 K over 500 ps without constraints after energy minimization, and then they were allowed to equilibrate for 500 ps. Lastly, at 310 K and 1 atm, a 300 ns MD production run was carried out with a 2 fs time step. A Langevin thermostat with a collision frequency of 1 ps − 1 was used to regulate the temperature. The SHAKE algorithm was used to limit the number of hydrogen atoms35, and long-range electrostatics were treated with the particle mesh Ewald method36, applying an 11 Å cutoff for non-bonded interactions. All directions were subject to periodic boundary conditions. The AMBER 22 software suite (https://ambermd.org) was utilized to run MD simulations on the Supek supercomputer located at the University Computing Center—SRCE at the University of Zagreb37 and ChimeraX (version 1.8) https://www.cgl.ucsf.edu/chimerax/download.html were used for illustrated the figure legends.
Free energy of binding calculations
The binding free energy was estimated using the molecular mechanics generalized Born surface area (MM/GBSA) method, a hybrid approach that combines molecular mechanics energies with continuum solvation models (generalized Born) to evaluate ligand–protein binding interactions. This method strikes a balance between computational efficiency and accuracy, lying between empirical scoring functions and rigorous alchemical perturbation techniques.
The MMPSBA.py script from the AmberTools package was utilized to implement the single-trajectory MM/GBSA protocol38. The binding free energy (∆Gbind) is calculated as:
Here, ∆EMM represents the change in molecular mechanics energy in the gas phase and is further broken down into internal (∆Einternal), electrostatic (∆Eelectrostatic), and van der Waals (∆EvdW) energy components:
The solvation free energy change (∆Gsol) is composed of polar (∆GGB) and non-polar (∆GSA) contributions:
The term T∆S accounts for the conformational entropy change upon binding.
For this study, 300 ns molecular dynamics trajectories were divided into six segments of 50 ns each. From each segment, 100 evenly spaced snapshots were extracted to compute ∆Gbind. The reported binding free energy represents the average value across all segments, along with the corresponding standard deviation. Additionally, residue-wise decomposition of MM/GBSA binding free energies was performed to identify critical interactions contributing to binding affinity39.
In vitro anti-proliferative assays
The anticancer activity of the synthesized derivatives was assessed against TPC-1 (thyroid cancer) and A375 (human malignant melanoma). The cell lines were obtained from National Centre for Cell Science (NCCS) Pune.
Cell cultures
DMEM supplemented with 10% heat-inactivated fetal bovine serum (FBS), 1% L-glutamine, and 50 mg/mL gentamycin sulphate were used to preserve the chosen cancer and normal cell lines (1 × 105 cells/well) at 37 °C in a CO2 incubator with humidified 5% CO2 and 95% air.
Preparation of samples
After dissolving the synthesized compounds in DMSO, stock solutions with a concentration of 1 mg/ml were made. The compounds were then diluted using 10% FBS and DMEM. A syringe filter (0.2 mM) was then used to filter and sterilize the solutions. Additional dilutions (1–100 mM) were made, sterilized, and filtered.
Cytotoxic activity measurement
The MTT assay was used to measure the compounds’ in vitro cytotoxic activities. After being diluted in culture medium, the synthesized compounds were added to the control wells and incubated for 48 h. Following the addition of 10 ml of MTT (5 mg/mL) to each well, the plates were incubated for the following four hours. Following the removal of the supernatant from each well, 100 milliliters of DMSO were used to dissolve the crystals that had formed. At 540 nm, the absorbance was measured. The reference medication used was sorafenib.
Statistical analysis
Cell viability (%) was calculated according the following formula:
Where OD represents optical density. The experiments for evaluation of the BRAFV600E inhibition and cytotoxicity were performed. All experiments were carried out three times with three replicates for each concentration tested. Determination of IC50 value was obtained by statistical analysis40,41,42.
BRAFV600E enzyme kinase assay
The kinase inhibitory activity of the BRAF enzyme was assessed using BRAFV600E kinase assay kit (Bioscience, Catalog# 48688). The BRAF Kit detects BRAF kinase activity with Kinase-Glo® MAX as a detection reagent. All samples and controls were tested in duplicate. Thawed 5X kinase buffer 1, ATP and 5X Raf substrate was taken and master mixture were prepared as positive control, test inhibitors and blank by using 5X kinase buffer 1 6 µL, ATP 1 µL, 5X Raf substrate 10µL, water 8 µL, inhibitor buffer (no inhibitor) 5 µL, 1X kinase buffer, BRAFV600E (2ng/µL) 20 µL total 50 µL. Added 5 µL inhibitor solution to each well labeled as test inhibitor. Added 5 µL of inhibitor-free solution (no inhibitor) in positive control and blank. To the blank wells, added 20 µL of 1X kinase buffer. The BRAFV600E enzyme thawed on ice. Once thawed, briefly spinned the tube containing the enzyme to recover the entire contents. To initiate the reaction, added 20 µL of diluted BRAFV600E enzyme to the “positive control” wells. Incubated at 30 °C for 45 min. Thawed kinase-Glo Max reagent. 50 µL of the kinase-Glo Max reagent was added to each well after 45 min. After covering the plate with aluminum foil, it was incubated for fifteen minutes at room temperature. A microplate reader was used to measure luminosity at 510 nm. The percentage of inhibition was computed43,44.
Results and discussion
Chemistry
An electrophile replaces an atom attached to an aromatic ring in the electrophilic aromatic substitution reaction, which was the initial step reaction in the synthesis. Among these reactions is the substitution of an electrophile for a hydrogen atom in a benzene ring.
Chalcone (β-unsaturated carbonyl compound) was synthesized in the second step.
When a strong base is present, a reaction between two esters or one ester and another carbonyl compound forms a carbon-carbon bond, producing a β-keto ester or a β-diketone. An enolate ion of the ketone is formed as a result of base-catalyzed Claisen-Schmidt condensation. As the nucleophile, this enolate ion attacks the aldehyde’s electrophilic carbon, turning it into an electron that absorbs the proton from the aqueous solution. Dehydration, the reaction’s last stage, enables the synthesis of chalcone.
The Michael addition reaction is the reaction used in the third step of the proposed compound’s synthesis. The Michael addition to the chalcones initiates the reaction, which is then followed by proton transfer, hydrolysis, Claisen addition cyclization, and spontaneous dehydration. The synthesized compounds were characterized by1H and13C NMR spectroscopy and Mass spectrometry. Presence of NH protons was confirmed by deuterium exchange experiment. The NH peak appeared as a singlet in the range of δ ppm 4.5 − 10.5. To further confirm the identity of one representative compound (S8), by deuterium exchange NMR.
Spectral characterization of synthesized compounds
4-Bromo-N-(4-(6-(4-chlorophenyl)−2-oxo-1,2,3,4-tetrahydropyrimidin-4-yl)phenyl)benzenesulfonamide (S1)
Dark yellow solid, yield: 55%, m.p.175–177 °C; 1H NMR (600 MHz, DMSO-d6) δ (ppm): 10.62 (s, NH), 9.81 (s, NH), 8.83 (s, NH), 7.75 (d, J = 8.9 Hz, 2 H, Ar-H), 7.66 (d, J = 8.8 Hz, 2 H, Ar-H), 7.51 (d, J = 8.4 Hz, 3 H, Ar-H), 7.39 (d, J = 8.6 Hz, 3 H, Ar-H), 7.11 (d, J = 9.0 Hz, 2 H, Ar-H), 6.16 (d, J = 6.2 Hz, 1 H), 5.18 (d, J= 4.2 Hz, 1 H).13C NMR (151 MHz, DMSO-d6) δ (ppm): 186.24 (C = O), 154.51, 150.07, 143.95, 140.47, 135.39, 131.74 (2 C), 129.38 (2 C), 129.12 (2 C), 128.70 (2 C), 127.17 (2 C), 123.73, 113.83 (2 C), 112.66, 97.27, 64.70. MS (EI) m/z: Calculated for C22H17BrClN3O3S: 517.00; Found: 517.10 [M]+.
4-Bromo-N-(4-(6-(4-bromophenyl)−2-oxo-1,2,3,4-tetrahydropyrimidin-4-yl)phenyl) benzenesulfonamide (S2)
Yellow solid, yield: 52%, m.p.176–178 °C; 1H NMR (600 MHz, DMSO-d6) δ (ppm): 9.53 (s, NH), 9.03 (s, NH), 7.44 (d, J = 8.7 Hz, 2 H, Ar-H), 7.31 (d, J = 8.7 Hz, 4 H, Ar-H), 7.17 (d, J = 8.9 Hz, 4 H, Ar-H), 6.51 (d, J = 8.9 Hz, 2 H, Ar-H), 5.32 (s, NH), 5.13 (d, J = 6.9 Hz, 1 H), 5.06 (d, J= 5.3 Hz, 1 H). 13C NMR (151 MHz, DMSO-d6) δ (ppm): 176.75 (C = O), 151.10, 148.69, 136.51, 132.46 (2 C), 131.73, 130.51 (2 C), 128.17 (4 C), 128.14, 126.83 (2 C), 123.40, 121.61, 114.84 (2 C), 98.08, 55.40. MS (EI) m/z: Calculated for C22H17Br2N3O3S: 563.26; Found: 563.05 [M]+.
4-Bromo-N-(4-(6-(2,4-dichlorophenyl)−2-oxo-1,2,3,4-tetrahydropyrimidin-4-yl)phenyl)benzenesulfonamide (S3)
Dark yellow solid, yield: 56%, m.p.180–181 °C; 1H NMR (600 MHz, DMSO-d6) δ (ppm): δ 10.49 (s, NH), 9.75 (s, NH), 9.07 (s, NH), 7.76 (d, J = 7.6 Hz, 1 H, Ar-H), 7.59 (s, 1 H, Ar-H), 7.53 (d, J = 7.9 Hz, 1 H, Ar-H), 7.36 (d, J = 8.6 Hz, 2 H, Ar-H), 7.30 (d, J = 7.4 Hz, 2 H, Ar-H), 7.20 (d, J = 8.8 Hz, 2 H, Ar-H), 7.07 (d, J = 8.6 Hz, 2 H, Ar-H), 5.26 (d, J = 4.8 Hz, 1 H), 5.06 (d, J= 4.6 Hz, 1 H). 13C NMR (151 MHz, DMSO-d6) δ (ppm): 196.80 (C = O), 149.87, 143.76, 138.85, 135.19, 132.92, 132.27, 130.19, 129.01, 128.91 (2 C), 128.50 (2 C), 126.95 (2 C), 120.51, 119.82, 118.58, 113.63 (2 C), 97.05, 54.19. HRMS: Calculated for C22H16BrCl2N3O3S [M + H]: 550.9523; Found: 550.9522.
4-Bromo-N-(4-(6-(2-nitrophenyl)−2-oxo-1,2,3,4-tetrahydropyrimidin-4-yl)phenyl)benzenesulfonamide (S4)
Dark brown solid, yield: 55%, m.p.183–185 °C; 1H NMR (600 MHz, DMSO-d6) δ (ppm): 9.57 (s, NH), 9.05 (s, NH), 7.46 (dd, J = 8.2, 7.8 Hz, 2 H, Ar-H), 7.34 (d, J = 7.8 Hz, 1 H, Ar-H), 7.30 (d, J = 7.9 Hz, 1 H, Ar-H), 7.17 (d, J = 8.9 Hz, 4 H, Ar-H), 6.51 (d, J = 8.9 Hz, 4 H, Ar-H), 5.33 (s, NH), 5.16 (d, J = 5.5 Hz, 1 H), 5.06 (d, J= 5.5 Hz, 1 H). 13C NMR (151 MHz, DMSO-d6) δ (ppm): 175.42 (C = O), 149.86, 143.76, 140.23, 135.19, 132.28, 131.51, 130.87, 130.51, 129.01, 128.91 (2 C), 128.50 (2 C), 126.95 (2 C), 120.52, 113.64 (2 C), 112.79, 97.06, 71.47. MS (EI) m/z: Calculated for C22H17BrN4O5S: 529.37; Found: 529.60 [M]+.
4-Bromo-N-(4-(6-(4-nitrophenyl)−2-oxo-1,2,3,4-tetrahydropyrimidin-4-yl)phenyl)benzenesulfonamide (S5)
Brown solid, yield: 57%, m.p.181–183 °C; 1H NMR (600 MHz, DMSO-d6) δ (ppm): 9.60 (s, NH), 9.07 (s, NH), 7.48 (d, J = 8.2 Hz, 2 H, Ar-H), 7.33 (dd, J = 7.8 Hz, 3.5 Hz, 2 H, Ar-H), 7.18 (d, J = 8.9 Hz, 4 H, Ar-H), 6.52 (d, J = 9.0 Hz, 4 H, Ar-H), 5.18 (s, NH), 5.16 (d, J = 6.7 Hz, 1 H), 5.07 (d, J= 5.7 Hz, 1 H). 13C NMR (151 MHz, DMSO-d6) δ (ppm): 176.15 (C = O), 143.99, 140.49, 139.25, 134.93, 134.04, 133.13, 130.36 (2 C), 129.66 (2 C), 129.26 (2 C), 127.89 (2 C), 127.64 (2 C), 120.18 (2 C), 101.09, 80.19, 54.88. MS (EI) m/z: Calculated for C22H17BrN4O5S: 529.37; Found: 529.40 [M]+.
N-(4-(6-(4-Chlorophenyl)−2-oxo-1,2,3,4-tetrahydropyrimidin-4-yl)phenyl)−4-nitrobenzenesulfonamide (S6)
Turmeric yellow solid, yield: 56%, m.p.180–182 °C; 1H NMR (600 MHz, DMSO-d6) δ (ppm): 9.52 (s, NH), 9.03 (s, NH), 7.43 (d, J = 8.8 Hz, 3 H, Ar-H), 7.30 (d, J = 8.6 Hz, 3 H, Ar-H), 7.16 (d, J = 8.9 Hz, 3 H, Ar-H), 6.50 (d, J = 8.9 Hz, 3 H, Ar-H), 5.32 (s, NH), 5.12 (d, J = 5.5 Hz, 1 H), 5.05 (d, J= 6.6 Hz, 1 H). 13C NMR (151 MHz, DMSO-d6) δ (ppm): 196.80 (C = O), 149.86, 144.17, 138.96, 138.18, 135.19, 134.20, 132.93, 131.90 (2 C), 131.83 (2 C), 129.06, 128.85 (2 C), 127.30 (2 C), 119.79 (2 C), 113.62 (2 C), 97.00. MS (EI) m/z: Calculated for C22H17ClN4O5S: 484.06; Found: 484.20 [M]+.
N-(4-(6-(2,4-Dichlorophenyl)−2-oxo-1,2,3,4-tetrahydropyrimidin-4-yl)phenyl)−4-nitrobenzenesulfonamide (S7)
Dark yellow solid, yield: 54%, m.p.182–184 °C; 1H NMR (600 MHz, DMSO-d6) δ (ppm): 9.69 (s, NH), 8.97 (s, NH), 7.58 (s, 1 H, Ar-H), 7.50 (d, J = 8.8 Hz, 1 H, Ar-H), 7.29 (d, J = 8.7 Hz, 1 H, Ar-H), 7.10 (d, J = 9.0 Hz, 4 H, Ar-H), 6.47 (d, J = 9.6 Hz, 4 H, Ar-H), 5.33 (s, NH), 5.32 (d, J = 6.9 Hz, 1 H), 5.10 (d, J= 6.8 Hz, 1 H). 13C NMR (151 MHz, DMSO-d6) δ (ppm): 186.41 (C = O), 154.30, 147.09, 143.67, 141.05, 133.14, 132.43 (2 C), 131.30, 129.95, 129.82, 129.21, 124.69, 123.25, 118.90, 118.10, 116.98, 116.20, 113.26 (2 C), 108.20, 64.69. MS (EI) m/z: Calculated for C22H16Cl2N4O5S: 518.02; Found: 517.95 [M]+.
4-Nitro-N-(4-(6-(4-nitrophenyl)−2-oxo-1,2,3,4-tetrahydropyrimidin-4-yl)phenyl)benzenesulfonamide (S8)
Dark brown solid, yield: 51%, m.p.185–186 °C; 1H NMR (600 MHz, DMSO-d6) δ (ppm): 9.65 (s, NH), 9.16 (s, NH), 7.78 (d, J = 8.9 Hz, 2 H, Ar-H), 7.70 (d, J = 8.8 Hz, 2 H, Ar-H), 7.37 (d, J = 8.7 Hz, 2 H, Ar-H), 7.29 (d, J = 8.9 Hz, 2 H, Ar-H), 6.66 (dd, J = 8.5, 8.9 Hz, 4 H, Ar-H), 5.45 (s, NH), 5.24 (d, J = 5.4 Hz, 1 H), 5.17 (d, J= 8.5 Hz, 1 H). 13C NMR (151 MHz, DMSO-d6) δ (ppm): 187.26 (C = O), 149.95, 149.37, 144.49, 141.93, 136.44, 134.00, 129.81, 128.99 (2 C), 128.21 (2 C), 126.33 (2 C), 124.75 (2 C), 118.63 (2 C), 113.17 (2 C), 92.04, 71.03. MS (EI) m/z: Calculated for C22H17N5O7S: 495.09; Found: 495.0 [M]+.
4-Nitro-N-(4-(6-(2-nitrophenyl)−2-oxo-1,2,3,4-tetrahydropyrimidin-4-yl)phenyl)benzenesulfonamide (S9)
Dark brown solid, yield: 52%, m.p.184–186 °C; 1H NMR (600 MHz, DMSO-d6) δ (ppm): 9.51 (s, NH), 9.02 (s, NH), 7.77 (m, 2 H, Ar-H), 7.42 (d, J = 8.8 Hz, 4 H, Ar-H), 7.29 (d, J = 8.8 Hz, 4 H, Ar-H), 7.15 (d, J = 8.9 Hz, 1 H, Ar-H), 6.49 (d, J = 8.9 Hz, 1 H, Ar-H), 5.30 (s, NH), 5.11 (d, J = 5.8 Hz, 1 H), 5.04 (d, J= 5.4 Hz, 1 H). 13C NMR (151 MHz, DMSO-d6) δ (ppm): 196.77 (C = O), 153.57, 151.56, 143.14, 142.17, 138.81, 132.87 (2 C), 132.54, 130.90, 130.69, 130.14 (2 C), 128.97 (2 C), 127.48, 118.54 (2 C), 116.74, 116.17, 113.91, 112.97. MS (EI) m/z: Calculated for C22H17N5O7S: 495.09; Found: 495.05 [M]+.
Molecular Docking and molecular dynamics simulations
Molecular docking studies were conducted on a series of nine synthesized compounds and the reference drug, sorafenib, against the BRAFV600E protein. Docking studies, utilizing AutoDock Vina, were performed to predict binding affinities. Table 2 summarizes the docking results, including binding energies.
In vitro kinase assays demonstrated a range of inhibitory activities among the tested compounds, with potencies varying from 65 to 94% inhibition. Compounds S4 (91%) and S1 (87%) exhibited the most potent inhibition, comparable to the reference drug sorafenib (94%). Based on these promising results, compounds S4 and S1 were selected for subsequent molecular dynamics simulations to comprehensively investigate their binding stability and conformational dynamics within the BRAFV600E active site.
Interestingly, despite the significant variation in experimental inhibitory activities, the docking scores, representing predicted binding free energies, exhibited minimal differences across the compounds, falling within a narrow range of 2 kcal·mol-1. This discrepancy between predicted binding affinities and experimental inhibition data suggests that the docking methodology, while providing a valuable initial assessment, may not fully capture the intricate interactions and dynamics that contribute to the observed inhibitory potencies. Factors such as solvent effects, protein flexibility, and ligand-induced conformational changes, which are not explicitly considered in the rigid-receptor docking approach employed in this study, likely play crucial roles in determining the observed differences in inhibitory activity45.
Lipinski’s rule of five (Ro5), a well-established guideline for drug-likeness, defines criteria for successful oral drugs, including a molecular weight below 500 Da, fewer than 10 hydrogen bond acceptors (HBA), fewer than 5 hydrogen bond donors (HBD), and a calculated logP value below 5. These criteria aim to improve drug absorption and bioavailability, addressing historical challenges in drug development.
A bioavailability radar plot (Fig. 3) was employed to assess the drug-likeness of the identified compounds. Analysis of the radar plot revealed that only S6 fully adhered to all criteria of Lipinski’s rule of five. Others, while adhering to the HBA and HBD count limits, their molecular weights slightly exceeded the 500 Da threshold. Notably, Reese et al.46 demonstrated that a significant proportion (~ 38%) of FDA-approved oral drugs deviate from the original Ro5 criteria, with molecular weight being the most frequent violation. Furthermore, Khanna and Ranganathan47 observed that 16% of drugs in the DrugBank database exceed the 500 Da molecular weight limit, with an average molecular weight of 355 ± 259 Da. These findings suggest that the identified compounds, despite slightly exceeding the 500 Da threshold, fall within the typical molecular weight range observed for approved drugs, indicating their potential as viable drug candidates. All compounds exhibited a moderate level of flexibility with between five and seven rotatable bonds. Compound S4 exhibited a calculated logP value below 5.0. The molecular weight, HBA count, HBD count, calculated logP, and number of rotable bonds (ROT) for all compounds are summarized in Table S2.
The bioavailability radar plot shows the drug-likeness of the compounds. The shaded area represents the optimal range for key physicochemical properties according to Lipinski’s rule of five: molecular weight, hydrogen bond acceptors (HBA), hydrogen bond donors (HBD), calculated logP, and rotatable bonds (ROT).
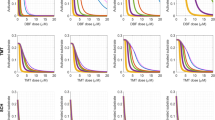
Building upon the docking protocol, validated the top-ranked docking pose for each compound, S1, S4, and the reference drug, SOR were selected as the initial structures for subsequent molecular dynamics simulations. To ensure reproducibility, three independent 300 ns MD simulations were performed for each protein-ligand complex. The stability of the protein-ligand complexes was assessed by monitoring the root-mean-square deviation (RMSD, Fig. 4), radius of gyration (Rg), and solvent-accessible surface area (SASA) relative to the initial structure. Furthermore, the flexibility of key residues within the binding site was evaluated by calculating the root-mean-square fluctuation (RMSF) values.
Root mean square deviation (RMSD) of the protein-ligand complexes over 300 ns molecular dynamics simulations. RMSD values were calculated for Cα atoms relative to the starting conformation of each simulation. Three independent simulations were performed for each complex: S1 (BRAF: S1), S4 (BRAF: S4), and SOR (BRAF: SOR).
Root mean square deviation is a critical metric used to assess the stability of molecular systems during molecular dynamics simulations. It quantifies the average positional deviation of atoms over time relative to a reference structure, typically the starting conformation. RMSD provides insights into the structural stability and flexibility of protein-ligand complexes during simulations. Low and stable RMSD values typically indicate that the system has reached equilibrium and maintains structural integrity, while large or fluctuating RMSD values may suggest flexibility, conformational changes, or instability in the system.
In this study, RMSD was calculated for three protein-ligand complexes: BRAF: S1, BRAF: S4, and BRAF: sorafenib (BRAF: SOR) over 300 ns of MD simulations, with three independent replicates performed for each complex. The results are displayed in Fig. 4.
The RMSD for the BRAF: S1 complex stabilized around 2 Å after an initial equilibration phase of approximately 10–20 ns. All three replicates exhibited similar trends with minor fluctuations, suggesting that the BRAF: S1 complex maintains a relatively stable conformation throughout the simulation. The consistent RMSD values across replicates indicateed good reproducibility and limited flexibility in the binding mode of the ligand within the protein’s active site. The BRAF: S4 complex displayed higher RMSD values, stabilizing around 3 Å. Notably, one replicate showed substantial fluctuations during the simulation, particularly after 150 ns. These variations highlighted the importance of performing multiple replicates to capture the full range of conformational dynamics.
The BRAF: SOR complex showed intermediate stability, with RMSD values stabilizing between 2 and 2.5 Å. The three replicates demonstrated consistent trends, with one replicate exhibiting slightly higher RMSD values and transient fluctuations. The data suggested that SOR formed a moderately stable complex with the BRAF protein, retaining its binding pose while allowing some degree of flexibility.
In the BRAF: S1 complex, ligand RMSD values fluctuated between 1.0 and 2.5 Å across the three replicates. Replicate 2 exhibited the largest deviations, with RMSD occasionally approaching 3 Å after 150 ns. Visual inspection of the trajectory showed that bromophenyl sulfonyl moiety of ligand (exposed to solvent), rotated around N-S bond. In contrast, replicates 1 and 3 showed comparatively stable profiles after 100 ns, suggesting the ligand maintains a relatively consistent binding mode with minor positional adjustments. The BRAF: S4 complex displayed the highest degree of variability among the three systems. RMSD frequently exceeded 2.5 Å in simulations 2 and 3, with peaks surpassing 3 Å. Just like in BRAF: S1 complex, bromophenyl sulfonyl moiety is responsible for increased structural deviations. The BRAF: SOR complex demonstrated the most stable RMSD profiles. Across all three replicates, RMSD values consistently ranged from 1.0 to 2.0 Å, with minimal fluctuations.
The radius of gyration (Rg, Fig. S3), a key indicator of protein structural compactness48 and the spatial distribution of mass around the protein’s centre of mass49, exhibited minimal fluctuations across the simulations, suggesting a stable and well-defined tertiary structure for all three protein-ligand complexes. This observation is further supported by low standard deviations in Rg values (Table 3). Concurrent with Rg, the solvent-accessible surface area (SASA, Fig. S4), a measure of protein-solvent interactions, displayed consistent trends across the simulations. While subtle variations were observed, the BRAF: SOR complex exhibited the highest mean SASA and standard deviation. These findings are corroborated by minimal alterations in the secondary structure of the BRAF protein throughout the simulations (Fig. S5). Detailed statistical parameters extracted from the molecular dynamics simulations are summarized in Table 3.
Root-mean-square fluctuations (RMSF) analysis (Fig. S6) revealed dynamic regions within the BRAF: S1, BRAF: S4, and BRAF: SOR complexes during 300 ns molecular dynamics simulations. While most residues exhibited limited fluctuations (< 3 Å), indicative of overall protein stability, several regions displayed significant flexibility (Fig. 5). Notably, the N- and C-termini, the loops connecting helices H4-H5 (His542-Thr546), H7-H8 (Ser602-Ser614), H9-H10 (Gln628-Asp629), and H10-H11 (Asn658-Asn661) demonstrated increased mobility. Helix numbering is based on (Fig. S7).
The loops connecting helices H4-H5 and H10-H11, which are solvent-exposed and lack direct ligand interactions, may exhibit inherent flexibility. Furthermore, the loops connecting helices H7 and H8, and H9 and H10, characterized by high flexibility, lack experimental structural data, likely due to its intrinsic disorder and increased dynamics. Within these regions, only a segment of the Ser602-Ser614 loop directly interacts with the ligand within the binding pocket. This interaction may influence the accessibility of the unoccupied binding site, potentially contributing to increased ligand binding affinity. The loop’s inherent flexibility could facilitate ligand accommodation and stabilize ligand-protein interactions by enabling local conformational adjustments.
Analysis of hydrogen bond (H-bond) formation across triplicate simulations revealed distinct interaction profiles for each complex (Fig. S8). The BRAF: S4 complex exhibited the highest average H-bond count (1.83), with instances reaching up to six H-bonds, particularly evident in the second replicate. This robust H-bonding pattern suggests strong and frequent interactions between the ligand and the binding site, likely contributing to its enhanced stability. Conversely, the BRAF: SOR complex displayed the lowest average H-bond count (0.98), indicating less frequent and potentially weaker interactions. The BRAF: S1 complex showed an intermediate average H-bond count (1.33) with relatively stable H-bond formation throughout the simulations, typically ranging between 1 and 2. This consistent interaction pattern suggests moderate and sustained binding interactions within the BRAF: S1 complex, contributing to its observed conformational stability.
The protein-ligand fingerprint plots provide a detailed temporal profile of various interactions between the ligand and its binding site residues during the 300 ns molecular dynamics simulations (Fig. 6, Fig. S9-11). Key interaction types include hydrophobic contacts, hydrogen bond acceptors and donors, van der Waals (vdW) contacts, and π-π or π-cation stacking. When analysing fingerprint plots of top hit candidate, S4, in BRAF: S4 complex one can see that hydrogen bond donors and acceptors involving residues such as Ser535 and Glu611 are intermittent, indicating a dynamic exchange of hydrogen bonds. Residues such as Leu505, Val471, and Phe610 demonstrated sustained hydrophobic interactions across the simulation, suggesting their critical role in maintaining the ligand’s stable positioning within the binding site. The Vander wall interactions, prominently observed with residues such as Val482 and Ala543, provide additional stabilizing forces, indicative of close packing within the binding site. Residues like Phe583 and Tyr538 exhibited occasional π-stacking or π-cation interactions. For BRAF: S1 complex, hydrophobic interactions were consistently observed throughout the simulation with residues such as Val471, Val482, and Phe583. Hydrogen bond donors and acceptors, particularly involving Ser465, Ser535, and Ser544, appeared intermittently, indicating their role in stabilizing transient interactions. Additionally, vdW contacts were highly prevalent, especially with residues like Leu514, Thr529, and Gly534, which indicate a robust non-specific binding mode contributing to the overall ligand stability. π-cation interactions, though less frequent, were evident with aromatic residues such as Trp531 and Phe583, highlighting their potential role in ligand anchoring.
BRAF-S4 (left) and BRAF-S1 (right) interaction fingerprints during the 300 ns molecular dynamics simulation of the ligand-protein complex for triplicates 2 (left) and 3 (right). The interactions, plotted against simulation time, include hydrophobic contacts (green), hydrogen bond acceptors (blue), hydrogen bond donors (cyan), van der Waals contacts (yellow), π-cation interactions (purple), and π-stacking (magenta). Key interacting residues are labeled on the y-axis, illustrating the dynamic nature of binding interactions and the persistence of critical contacts throughout the trajectory.
The consistent hydrophobic and vdW interactions indicate a stable core interaction network, while the dynamic hydrogen bonding and stacking interactions reflect the adaptability required for ligand accommodation. Ligand interaction networks for S1, S4, and SOR bound to BRAF protein are presented in Figs. S12-14.
Free energy of binding calculations
Binding affinities of S1, S4, and SOR to the BRAF protein were assessed using MM/GBSA calculations (Table 4). Key energy components analyzed included van der Waals interactions (ΔEvdW), electrostatic interactions (ΔEele), polar and non-polar solvation energies (ΔGsolele and ΔGsolnp), binding free energy without entropic contribution (ΔGbind, w/o T), and the total binding free energy (ΔGbind). Entropic contributions were evaluated at a temperature of 310 K. S4 exhibited the most favorable binding affinity (ΔGbind = −18.1 ± 5.3 kcal mol−1), followed by S1 (−15.9 ± 4.1 kcal mol−1) and SOR (−14.6 ± 5.6 kcal mol−1). Van der Waals interactions (ΔEvdW) were the dominant contributors to binding of all ligands, with S4 displayed the most significant contribution (−59.0 ± 4.0 kcal mol−1), followed by SOR (−52.0 ± 4.8 kcal mol−1) and S1 (−51.4 ± 4.9 kcal mol−1). Electrostatic interactions (ΔEele) also played a role, with S4 demonstrating the most favorable ΔEele. Solvation energy (ΔGsol), encompassing both electrostatic and non-polar components, contributed significantly to the overall binding free energy. S4 incurred the largest solvation penalty (+ 34.4 kcal mol−1). Entropy contributions (TΔS) were unfavorable for all ligands, reflecting a decrease in conformational freedom upon binding. S4 exhibited the largest entropic penalty (−25.9 ± 1.5 kcal mol−1). Despite the significant solvation and entropic penalties, S4 emerged as the most potent binder due to its substantial favorable van der Waals and electrostatic interactions.
The per-residue free energy decomposition from MM/GBSA calculations highlights key residues contributing to the binding of ligands S1, S4, and SOR to the BRAF protein, revealing distinct trends in residue type and interaction mechanisms (Table 5). Across all ligands, Phe583 consistently exhibited the most favorable contributions, stabilizing the ligands through van der Waals, hydrophobic and occasional π-π stacking interactions, as suggested by the fingerprint analyses. Other significant contributors varied depending on the ligand. For S1, Trp531 and Ile463 were key, along with Cys532 and Val471, indicating a mix of π-π stacking, hydrophobic contacts, and hydrogen bonding. For S4, Thr529, Cys532, and Val471 were prominent. In the case of SOR, Ile463, Val471, and Leu514 were notable, with His539 emerging as a unique contributor.
The protein-ligand fingerprint analyses align well with these findings, emphasizing the importance of hydrophobic contacts, π-π stacking, and hydrogen bonding. Residues such as Phe583, Trp531, and Ile463 were identified as key interaction hotspots in both analyses. Figure 7. Visualizes key residues for each complex, identified as having per-residue contributions to binding energy lower than − 1.5 kcal mol−1. These residues were determined from the final geometry of each trajectory.
The 3D structures of the designed molecules in complex with the BRAF protein, captured from the final frame of 300 ns molecular dynamics simulations. Only protein residues contributing more than − 1.5 kcal·mol−1 to the binding free energy, as determined by MM/GBSA decomposition analysis, are highlighted for clarity. [software ChimeraX, (version 1.8)].
Pharmacological assay
Anti-proliferative assays
The cytotoxicity of the newly synthesized oxo-tetrahydro-pyrimidin-benzenesulfonamide derivatives (S1–S9) was evaluated using the MTT assay against A375 (human malignant melanoma) and TPC-1 (thyroid cancer) cell lines. Sorafenib, a known BRAF inhibitor, was used as a reference drug and served as a cytotoxicity control. Cell viability was assessed by plotting survival curves as a function of drug concentration. The anticancer potency of each compound was determined based on the half-maximal inhibitory concentration (IC₅₀), which represents the concentration required to inhibit 50% of cell viability. The IC₅₀ values for all derivatives are summarized in Table 6.
Against the A375 cell line, the IC₅₀ values of the synthesized derivatives ranged from 7.14 ± 0.18 µM to 9.75 ± 0.16 µM, while the reference drug sorafenib exhibited an IC₅₀ of 0.80 ± 0.22 µM. The A375 cell line is known to exhibit high BRAF mutation expression among various cancer cell lines. However, none of the synthesized compounds demonstrated inhibition higher than sorafenib. Seven compounds with IC₅₀ values exceeding 5.0 µM exhibited only mild inhibitory activity. Notably, compounds S3 and S6 had significantly higher IC₅₀ values and were classified as inactive (NA). Despite this, all compounds displayed potent antitumor activity, with IC₅₀ values below 10 µM.
Against the TPC-1 cell line, the IC₅₀ values of the synthesized derivatives ranged from 6.45 ± 0.28 µM to 9.39 ± 0.30 µM, while sorafenib demonstrated an IC₅₀ of 0.62 ± 0.22 µM. Similar to the A375 results, none of the synthesized compounds exhibited inhibitory potency comparable to sorafenib. Seven compounds with IC₅₀ values exceeding 5.0 µM showed mild inhibitory activity. Compounds S6 and S8 had the highest IC₅₀ values and were classified as inactive (NA). However, the majority of the compounds demonstrated potent antitumor activity, with IC₅₀ values below 10 µM.
BRAFV600E enzyme kinase assay
Using sorafenib as the reference standard, all target compounds were examined in vitro for their ability to inhibit the BRAFV600E kinase. All of the tested compounds successfully inhibited the BRAFV600E enzyme’s kinase activity, as shown in Table 6, with percentage inhibition at the nanomolar level at 500 nM, which is equivalent to the reference drug sorafenib. Among the screened compounds, compound S4 was revealed as an excellent BRAFV600E inhibitor. Sorafenib has 94.81% BRAFV600E inhibitory activity; compared to Sorafenib, compound S4 has 91.20% BRAFV600E inhibitory activity.
Compounds S1 and S9 showed 87.27% and 83.69% BRAFV600E inhibitory activity close to sorafenib. Four compounds, S8, S7, S5, and S6, showed (70–80) % BRAFV600E inhibitory activity, respectively 79.75, 78.27, 76.92, and 71.04%. Two compounds were having 60–70% BRAFV600E inhibitory activity; in this, compound S2 had 69.02% BRAFV600E inhibitory activity, and compound S3 had 65.29% BRAFV600E inhibitory activity. All synthesized compounds had more than 60% BRAFV600E inhibitory activity. Compound S4 was the most potent compound compared to sorafenib, having good BRAFV600E kinase inhibitory activity.
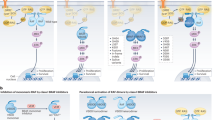
SAR of Oxo-tetrahydro-pyrimidin-benzene-sulfonamide hybrids
Based on computational and pharmacological assays such as anti-proliferative activity against two cancer cell lines and BRAFV600E kinase inhibition, a generalized structure–activity relationship (SAR) is summarized in Fig. 8.
The synthesized compounds exhibited binding affinity within the active core region of the ATP-binding pocket (Fig. 8). Specifically, the R-substituted sulfonamide moiety interacted with the RAF-selective pocket, the phenyl ring occupied the hydrophobic pocket, the pyrimidine ring interacted with the ribose pocket, the R1-substitution targeted the adenine pocket, and the carbonyl group (C = O) extended into a solvent-accessible region outside the ATP-binding site.
Among the synthesized compounds, the 4-chloro derivative (S1) demonstrated potent anti-proliferative activity against BRAFV600E kinase. The 4-bromo derivative (S2) exhibited comparable anti-proliferative activity against both A375 and TPC-1 cell lines. Similarly, the 2-nitro (S4) and 4-nitro (S5) derivatives showed significant activity against the A375 cell line. The 2,4-dichloro derivative (S7) also displayed potent anti-proliferative effects against both A375 and TPC-1 cell lines, while the 2-nitro derivative (S9) showed notable BRAFV600E kinase inhibitory activity.
In contrast, compounds such as the 2,4-dichloro (S3), 4-chloro (S6), and 4-nitro (S8) derivatives exhibited lower anti-proliferative and BRAFV600E kinase inhibitory activities.
Conclusion
Among the designed and synthesized oxo-tetrahydro-pyrimidine–benzene-sulfonamide hybrids, computational studies revealed that seven compounds exhibited superior docking scores, while two showed scores comparable to the reference drug, sorafenib. MM/GBSA binding free energy calculations, along with RMSD, RMSF, and SASA analyses, provided a comprehensive understanding of the binding dynamics of these ligands within the BRAF protein binding pocket.
Compound S1 demonstrated strong binding affinity and higher stability compared to other candidates, as indicated by favorable binding free energy value and lower RMSD and RMSF values. In contrast, compound S4 displayed higher RMSD but lower RMSF value than sorafenib, suggesting distinct binding behavior. These insights are essential for guiding the rational design of BRAF inhibitors.
Notably, compound S4 exhibited the most potent BRAFV600E kinase inhibitory activity among the tested compounds. These findings highlight the critical influence of electron-withdrawing substituents on anticancer activity. Overall, most synthesized derivatives demonstrated promising anticancer potential and S4, along with related analogs, can be considered potential lead compounds for BRAFV600E inhibition.
Data availability
The Amber trajectories for the BRAF: S1, BRAF: S4, and BRAF: SOR (Sorafenib) protein-ligand complexes over 300 ns of molecular dynamics simulations are openly available in the FULIR DATA repository at https://urn.nsk.hr/urn: nbn: hr:241:411570 (accessed on 18 March 2025).
Abbreviations
- BRAF:
-
v-RAF murine sarcoma viral homolog B1
- DMEM:
-
Dulbecco’s modified eagle’s medium
- FDA:
-
Food and drug administration
- MAPK:
-
Mitogen-activated protein kinase
- MD:
-
Molecular dynamics
- MM/GBSA:
-
Molecular mechanics/generalized born surface area
- RMSD:
-
Root mean squire deviation
- RMSE:
-
Root mean squire error
- RMSF:
-
Root mean squared fluctuations
- RoG:
-
Radius of gyration
- SASA:
-
Solvent accessible surface area
References
Singh, A., Roghini, S. & Cancer Unraveling the complexities of uncontrolled growth and metastasis. Int. J. Pharm. Sci. 2, 59–73 (2023).
Asati, V., Mahapatra, D. K. & Bharti, S. K. PI3K/Akt/mTOR and ras/raf/mek/erk signaling pathways inhibitors as anticancer agents: structural and Pharmacological perspectives. Eur. J. Med. Chem. 109, 314–341 (2016).
Santarpia, L., Lippman, S. M. & El-Naggar, A. K. Targeting the MAPK–RAS–RAF signaling pathway in cancer therapy. Expert Opin. Ther. Targets. 16, 103–119 (2012).
Tuveson, D. A., Weber, B. L. & Herlyn, M. BRAF as a potential therapeutic target in melanoma and other malignancies. Cancer Cell. 4, 95–98 (2003).
Singh, A. K. et al. Challenges and opportunities in the crusade of BRAF inhibitors: from 2002 to 2022. ACS Omega. 8, 27819–27844 (2023).
Ram, T. et al. MEK inhibitors in cancer treatment: structural insights, regulation, recent advances and future perspectives. RSC Med. Chem. 14, 1837–1857 (2023).
Singh, A. K., Kumar, A., Thareja, S. & Kumar, P. Current insights into the role of BRAF inhibitors in treatment of melanoma. Anti-Cancer Agents Med. Chem. 23, 278–297 (2023).
Koelblinger, P., Thuerigen, O. & Dummer, R. Development of Encorafenib for BRAF-mutated advanced melanoma. Curr. Opin. Oncol. 30, 125–133 (2018).
Sun, J., Zager, J. S. & Eroglu, Z. Encorafenib/binimetinib for the treatment of BRAF-mutant advanced, unresectable, or metastatic melanoma: design, development, and potential place in therapy. OncoTarg Ther, 9081–9089 (2018).
Broman, K. K., Dossett, L. A., Sun, J., Eroglu, Z. & Zager, J. S. Update on BRAF and MEK Inhibition for treatment of melanoma in metastatic, unresectable, and adjuvant settings. Expert Opin. Drug Saf. 18, 381–392 (2019).
Röck, R. et al. BRAF inhibitors promote intermediate BRAF (V600E) conformations and binary interactions with activated RAS. Sci. Adv. 5, eaav8463 (2019).
Clayton, J. et al. Mechanism of dimer selectivity and binding cooperativity of BRAF inhibitors. eLife 13, RP95334 (2025).
Dcruz, A. C. et al. BRAF gene as a potential target to attenuate drug resistance and treat cancer. Gene Rep. 30, 101740 (2023).
Lee, J. J., Kroshinsky, D. & Hoang, M. P. Cutaneous reactions to targeted therapy. Am. J. Dermatopathol. 39, 67–82 (2017).
Wang, P. F., Qiu, H. Y. & Zhu, H. L. A patent review of BRAF inhibitors: 2013–2018. Expert Opin. Ther. Pat. 29, 595–603 (2019).
Maji, L. et al. An updated literature on BRAF inhibitors (2018–2023). Mol. Divers. 28, 2689–2730 (2024).
Pratap Reddy Gajulapalli, V. Development of Kinase-Centric Drugs: A Computational Perspective.. ChemMedChem18, e202200693 (2023).
Khan, P. S. et al. Recent advances in B-RAF inhibitors as anticancer agents. Bioorg. Chem. 120, 105597 (2022).
Parai, M. K., Panda, G., Srivastava, K. & Puri, S. K. Design, synthesis and antimalarial activity of benzene and isoquinoline sulfonamide derivatives. Bioorg. Med. Chem. Lett. 18, 776–781 (2008).
Ghorab, M. M., Ragab, F. A., Heiba, H. I., El-Gazzar, M. G. & Zahran, S. S. Synthesis, anticancer and radiosensitizing evaluation of some novel sulfonamide derivatives. Eur. J. Med. Chem. 92, 682–692 (2015).
Berman, H. M. et al. The protein data bank. Nucleic Acids Res. 28, 235–242 (2000).
22 Grasso, M. et al. Chemically linked Vemurafenib inhibitors promote an inactive BRAFV600E conformation. ACS Chem. Biol. 11, 2876–2888 (2016).
Šali, A. & Blundell, T. L. Comparative protein modelling by satisfaction of Spatial restraints. J. Mol. Biol. 234, 779–815 (1993).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Tosco, P., Stiefl, N. & Landrum, G. Bringing the MMFF force field to the rdkit: implementation and validation. J. Cheminf. 6, 1–4 (2014).
Landrum, G. Rdkit Documentation. Release 1, 4 (2013).
Bhattacharya, P. & Mandal, A. Identification of Amentoflavone as a potent SARS-CoV-2 Mpro inhibitor: a combination of computational studies and in vitro biological evaluation. J Biomol. Struct. Dyn 1–19 (2024).
Morris, G. M. et al. AutoDock4 and AutoDockTools4: automated Docking with selective receptor flexibility. J. Comput. Chem. 30, 2785–2791 (2009).
Besler, B. H., Merz Jr, K. M. & Kollman, P. A. Atomic charges derived from semiempirical methods. J. Comput. Chem. 11, 431–439 (1990).
Wang, J., Wang, W., Kollman, P. A. & Case, D. A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graphics Modell. 25, 247–260 (2006).
Wang, J., Wolf, R. M., Caldwell, J. W., Kollman, P. A. & Case, D. A. Development and testing of a general amber force field. J. Comput. Chem. 25, 1157–1174 (2004).
Tian, C. et al. ff19SB: amino-acid-specific protein backbone parameters trained against quantum mechanics energy surfaces in solution. J. Chem. Theory Comput. 16, 528–552 (2019).
Dolinsky, T. J. et al. PDB2PQR: expanding and upgrading automated Preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 (2007).
Machado, M. R. & Pantano, S. Split the charge difference in two! A rule of thumb for adding proper amounts of ions in MD simulations. J. Chem. Theory Comput. 16, 1367–1372 (2020).
Andersen, H. C. & Rattle A velocity version of the shake algorithm for molecular dynamics calculations. J. Comput. Phy. 52, 24–34 (1983).
Darden, T., York, D. & Pedersen, L. Particle mesh ewald: an N log (N) method for Ewald sums in large systems. J. Chem. Phy. 98, 10089–10089 (1993).
Cruzeiro, V. W. D., Feliciano, G. T. & Roitberg, A. E. Exploring coupled redox and pH processes with a force-field-based approach: applications to five different systems. J. Am. Chem. Soc. 142, 3823–3835 (2020).
Genheden, S. & Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discovery. 10, 449–461 (2015).
Rastelli, G., Rio, A. D., Degliesposti, G. & Sgobba, M. Fast and accurate predictions of binding free energies using MM-PBSA and MM‐GBSA. J. Comput. Chem. 31, 797–810 (2010).
Jiao, Y. et al. Design, synthesis and evaluation of novel 2-(1H-imidazol-2-yl) pyridine Sorafenib derivatives as potential BRAF inhibitors and anti-tumor agents. Eur. J. Med. Chem. 90, 170–183 (2015).
Pathak, P. et al. Quinazoline based 1, 3, 5-triazine derivatives as cancer inhibitors by impeding the phosphorylated RET tyrosine kinase pathway: design, synthesis, docking, and QSAR study. Arch. Pharm. 352, 1900053 (2019).
Ram, T. et al. Design, one-pot synthesis, computational and biological evaluation of diaryl benzimidazole derivatives as MEK inhibitors. J. Biomol. Struct. Dyn. 42, 11812–11827 (2024).
Khormi, A. Y. et al. Novel thiazole derivatives incorporating phenyl sulphonyl moiety as potent BRAFV600E kinase inhibitors targeting melanoma. RSC Adv. 12, 27355–27369 (2022).
Tobeigei, F. H. et al. Computational high-throughput screening and in vitro approaches identify CB-006-3; A novel PI3K-BRAFV600E dual targeted inhibitor against melanoma. Oncol. Res. 29, 305 (2022).
Chen, Y. C. Beware of docking! Trends Pharmacol. Sci. 36, 78–95 (2015).
Reese, T. C. et al. Evaluating physiochemical properties of FDA-approved orally administered drugs. Expert Opin. Drug Discovery. 19, 225–238 (2024).
Khanna, V. & Ranganathan, S. In BMC Bioinf. 1–18 (Springer).
Sneha, P. & Doss, C. G. P. Molecular dynamics: new frontier in personalized medicine. Adv. Protein Chem. Struct. Biol. 102, 181–224 (2016).
Lobanov, M. Y., Bogatyreva, N. & Galzitskaya, O. Radius of gyration as an indicator of protein structure compactness. Mol. Biol. 42, 623–628 (2008).
Acknowledgements
Authors are thankful to Central University of Punjab, Bathinda, DST-FIST India. JN thanks the Croatian Science Foundation (grant ref. IP-2022-4658) for support, and the University of Zagreb, University Computing Center – SRCE, for granting access to the Supek supercomputer. PK is also thankful to ANRF SERB (CRG/2023/001580), ICMR (IIRPSG-2024–01–01573), and DST-PURSE (SR/PURSE/2023/220) for financial/infrastructure support.
Funding
The APC was funded by King Abdullah University of Science and Technology, Thuwal, Jeddah, Saudi Arabia.
Author information
Authors and Affiliations
Contributions
Conceptualization: Pradeep Kumar; spectral characterization, computational methodology, pharmacological assay and their data interpretation: Ankit Kumar Singh, Jurica Novak, Prateek Pathak; Data curation and Figure drawing: Ankit Kumar Singh, Adarsh Kumar, Harshwardhan Singh, and Jurica Novak. The interpretation of the results, writing, and reviewing of the manuscript: Ankit Kumar Singh, Prateek Pathak, Jurica Novak, Amita Verma, Habibullah Khalilullah, Mariusz Jaremko, Abdul-Hamid Emwas and Pradeep Kumar.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Singh, A.K., Kumar, A., Singh, H. et al. Targeted design, synthesis, molecular dynamics, ADME and in –vitro anticancer assessment of oxo-tetrahydro-pyrimidin-benzenesulfonamide hybrids as potential BRAFV600E inhibitors. Sci Rep 15, 35088 (2025). https://doi.org/10.1038/s41598-025-18835-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-18835-9