Abstract
Uterine corpus endometrial carcinoma (UCEC) is a significant cause of cancer-related mortality among women worldwide. Prior research has demonstrated an association between cyclin-dependent kinase inhibitor 2 A (CDKN2A) and various tumors. As a member of the INK4 family, CDKN2A is involved in cell cycle regulation by controlling CDKs. In the present study, bioinformatics was used to analyze public datasets. The expression levels, signaling pathways, and copy number variations of CDKN2A in UCEC were explored, along with its immune cell subset associations. CDKN2A expression was found to be elevated in UCEC, particularly in the signaling pathways involved in cell proliferation and inflammation. Analysis of somatic copy number alterations in the TCGA (The Cancer Genome Atlas)-UCEC dataset revealed a connection between CDKN2A and drug metabolism in UCEC. Assessment of the relationship between CDKN2A and genes involved in immunotherapy for UCEC patients showed a negative correlation between CDKN2A and CD8+ T cell activity, as well as IL-2 and TP53. Collectively, these insights suggest that CDKN2A may be a potential biomarker for prognosis and treatment strategies in UCEC.
Similar content being viewed by others
Introduction
Endometrial cancer is a significant health issue for women. In China, the number of new malignant endometrial cancer cases in 2022 was approximately 80,000, with about 15,000 deaths. This disease has a high incidence and mortality rate among female patients with tumors1. Morphologically, endometrial cancer is categorized into nine subtypes, including endometrioid, serous, clear cell, dedifferentiated carcinoma, and others2. According to The Cancer Genome Atlas (TCGA), endometrial cancer can be divided into four molecular subtypes: non-specific molecular profile, MMR (Mismatch Repair) deficiency, POLE (Polymerase ɛ) mutation, and p53 abnormality3. These new molecular classifications provide valuable tools for diagnosing endometrial cancer, although some challenges remain4. The integration of molecular and histological classifications has led to diverse therapeutic approaches, introducing greater complexity in diagnostics and treatment. Developing a prognostic system that includes molecular classifications enhances risk stratification, potentially benefiting certain cases5. While molecular classification is useful for diagnosing uterine corpus endometrial carcinoma (UCEC), it is important to recognize that endometrial cancer lacks a definitive early biomarker. Thus, molecular classifications should be viewed as supplementary to histomorphometry classification and not serve as a complete replacement6. In addition, unresolved issues with molecular classification necessitate the identification of new biomarkers for early UCEC diagnosis and the prioritization of target discoveries for UCEC therapy.
CDKN2A is located on chromosome 9p21 and is a member of the INK4 family of cyclin-dependent kinase inhibitors. It is expressed in various human cells and encodes several proteins, including p14ARF and p16INK4a, which regulate cell cycle and inhibit tumor development and progression7. CDKN2A deficiency correlates with worse outcomes and poorer prognosis in non-small cell lung cancer, while high CDKN2A expression is closely related to immune infiltration in hepatocellular carcinoma. CDKN2A is a potential biomarker and therapeutic target in both renal cell carcinoma and triple-negative breast cancer8,9,10,11. CDKN2A abnormalities, such as aberrant mutations, deletions, and copy number variations (CNVs), have varied effects on the cell, and specific CDKN2A mechanisms in UCEC should be further explored12.
The roles of CDKN2A in various aspects of UCEC were investigated in the present study. Bioinformatics and public datasets were utilized to evaluate its potential to offer novel predictive or therapeutic insights for UCEC patients. CDKN2A expression levels were analyzed in different cancer stages and histological subtypes, noting an upregulation at both transcriptional and protein levels, which correlated with poor prognosis. CDKN2A was also found to be related to immune-related pathways. Thus, the relationship between CDKN2A and immune therapy was investigated. In addition, CDKN2A somatic copy number alterations (SCNAs) were examined in UCEC samples, revealing significant enrichment of CDKN2A-associated SCNA genes in drug metabolism and suggesting a tight link between CDKN2A and tumor therapy.
Results
Screening process for target gene CDKN2A relates with cell growth in UCEC
Genes identified via intersection analysis are statistically robust and biologically relevant, potentially engaging in disease’s molecular underpinnings, like signal transduction, cell cycle governance, and apoptosis. An intersection of two gene sets from GEPIA2 (Gene Expression Profiling Interactive Analysis) was performed in the present study. One was associated with endometrial cancer prognosis and included 500 genes (p-value < 0.05), and the other was correlated with differential gene expression between endometrial cancer and normal tissue, comprising 7,131 genes (p-value < 0.05). The study aimed to identify key genes that are significantly associated with tumor prognosis and play a crucial role in tumorigenesis, progression, and therapeutic response.
The intersection included 115 genes. A Venn diagram was created using DAVID online (Fig. 1A). The functions and pathways of these genes were then categorized using gene ontology (GO) (Fig. 1B) and Kyoto Encyclopedia of Genes and Genomes (KEGG) (Fig. 1C) analyses.
Screening process for target genes and CDKN2A related to cell growth in UCEC. (A) Venn diagram for genes associated with survival and differential expression. (B, C) GO and KEGG enrichment analysis show different functions of 115 intersecting genes. Genes associated with “growth” and cancer-related genes were selected (red box).
Then, the set of genes related to “growth” from the GO analysis was intersected with the set of cancer-related genes from the KEGG analysis. The genes identified using this intersection were apoptosis factors BAX and CDKN2A. BAX functions as an anti- or pro-apoptotic regulator in various cellular activities, and CDKN2A plays a key role in cell cycle regulation, with somatic mutations frequently observed in most human cancers. Since CDKN2A is a cell cycle-related regulator, it was selected as the focus of the present research due to its alterations in UCEC.
CDKN2A transcription level is upregulated in UCEC associated with poor prognosis
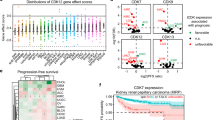
First, CDKN2A transcription levels were evaluated across 24 different cancer types, including UCEC (Fig. 2A). Subsequently, CDKN2A transcription levels were examined in 35 normal and 546 primary tumor tissues from UCEC patients. A differential mRNA expression was observed between normal and tumor tissues, with elevated CDKN2A mRNA levels in the tumor samples (Fig. 2B).
CDKN2A is upregulated in UCEC patients and associated with poor prognosis. (A) CDKN2A expression in pan-cancer. Blue represents normal, red shows tumor. (B) CDKN2A expression between normal and primary tumor tissue. (C) CDKN2A expression in different cancer stages. (D) CDKN2A expression in different histological subtypes. (E) Overall survival rate of low and high CDKN2A expression groups. 174 tumor samples from GTEx were analyzed. Log-rank p = 0.0055. Group cutoff: median. Hazard ratio (HR) = 2.9, p = 0.0078. HR was calculated based on Cox PH model. 95% confidence interval is shown as dotted line. *: p < 0.05; **: p < 0.01; ***: p < 0.001.
Furthermore, elevated CDKN2A transcription levels were noted in tumor tissues across various cancer stages (Fig. 2C) and different histological subtypes (Fig. 2D), which correlated with tumor progression. This suggests that increased CDKN2A mRNA levels are proportional to tumor severity. Concurrently, an increased CDKN2A transcription level was observed in serous and mixed endometrial cancers, suggesting variability in its expression across different UCEC types. Tumor progression and elevated CDKN2A transcription in tumor tissue may be associated with decreased patients’ survival rates (Fig. 2E).
CDKN2A protein expression is upregulated in UCEC
CDKN2A protein levels were analyzed using the Clinical Proteomic Tumor Analysis Consortium (CPTAC) dataset and The University of Alabama at Birmingham Cancer (UALCAN) data analysis portal. CDKN2A protein expression exhibited a positive correlation with UCEC, as evidenced by data from 31 normal and 100 UCEC samples (Fig. 3A). Further analysis of CDKN2A protein expression across different UCEC stages (Fig. 3B), grades (Fig. 3C), and histological subtypes (Fig. 3D) revealed significant upregulation in the early stages and across different histological subtypes.
CDKN2A protein expression is upregulated in UCEC patients. (A) CDKN2A protein expression increases significantly in UCEC samples. Z-values represent standard deviations from the median across samples. CDKN2A protein expression level in different UCEC (B) stages, (C) grades, and (D) histological subtypes. (E) Statistical analysis of all CDKN2A IHC images from the Human Protein Atlas. (F) IHC image of CDKN2A in normal and UCEC tissue. *: p < 0.05; **: p < 0.01; ***: p < 0.001.
To enhance the visualization of CDKN2A protein expression, immunohistochemistry (IHC) UCEC images were collected from the Human Protein Atlas (HPA) and thoroughly analyzed (Fig. 3E). The majority of IHC images of normal endometrial tissues within the HPA indicated “not detected,” with only a few displaying moderate or low expression. Conversely, the expressions in tumor tissues varied. Seven showed strong expression, fourteen were moderate, ten were weak, and only three were unexpressed (Fig. 3E). Patient information, expression status, and IHC images for both normal and strongly expressed samples were assembled (Fig. 3F), facilitating the visualization of elevated CDKN2A expression levels in UCEC.
According to the protein expression analysis, CDKN2A expression was already significant at stage 1 and grade 1 of endometrial cancer, indicating that the increase in CDKN2A expression occurs early in the disease process. For patients, IHC should be one of the most intuitive and convenient methods to diagnose tumors. Therefore, early disease detection using IHC could be highly significant.
Two CDKN2A signaling pathways involved in UCEC
To further elucidate the role of CDKN2A in UCEC pathogenesis, the CPTAC-UCEC dataset was utilized to explore the potential signaling pathways associated with CDKN2A. The CPTAC-UCEC dataset consists of 99 UCEC cases divided into “pathway altered” and “others” based on whether the pathway is altered. The CDKN2A protein expression levels in these two groups were compared to those in 31 normal adjacent cases. The findings indicated that the CDKN2A protein levels were closely correlated with various signaling pathways, such as Hippo, Wnt, p53/Rb, mTOR, RTK, MYC/MYCN, chromatin modifiers, and NRF2 (Fig. 4). Notably, CDKN2A protein expression was closely linked to significant changes in the p53/Rb and NRF2 pathways related to inflammation (indicated in the red box), suggesting CDKN2A’s involvement in the p53 pathway and tumor immunity.
Alteration in p53/Rb- and NRF2-related pathways associated with CDKN2A protein expression. UALCAN analysis of CDKN2A proteomic expression based on Hippo, Wnt, p53/Rb, mTOR, RTK, MYC/MYCN, SWI/SNF complex, chromatin modifier, and NRF2 status. CDKN2A protein expression was determined using mass spectrometry. Z-values represent standard deviations from the median across UCEC samples. A total of 99 CPTAC-UCEC cases were used. *: p < 0.05; **: p < 0.01; ***: p < 0.001.
CDKN2A has interactions with immune therapy in UCEC.
CDKN2A protein expression was observed to be associated with alterations in the p53/Rb and NRF2 inflammation-related pathways (Fig. 4). Considering that the p53 and NRF2 pathways are closely linked to immunotherapy, the association between CDKN2A and immune therapies was investigated.
To explore the correlation between CDKN2A expression and immune cells in UCEC patients, TCGA-UCEC data (n = 545) were processed using TIMER (Tumor Immune Estimation Resource). The results showed that CDKN2A was more negatively correlated with CD8+ T cell activity (p = 2.43e-08) and partially correlated with CD4+ T cells activity (p = 0.0111) (Fig. 5A). A significant correlation was noted between CDKN2A expression and UCEC patients’ survival (p = 7.88e-05) (Fig. 5B). Among the different immune cell types, CD8+ T cells (log-rank p = 0.022) and B cells (log-rank p = 0.019) were shown to correlate with UCEC patients’ survival rate (Fig. 5B). The relationship between CDKN2A, interleukin-2 (IL2), interferon alpha 1 (IFNA1), tumor necrosis factor (TNF), and TP53 were also investigated (Fig. 5C). CDKN2A expression was found to be associated with IL2, IFNA1, TNF, and TP53, with respective correlation coefficients of -0.155 (log-rank p = 2.83e-04), 0.157 (log-rank p = 2.3e-04), 0.144 (log-rank p = 7.54e-04), and − 0.178 (log-rank p = 2.87e-05).
Interaction between CDKN2A and immune therapy in UCEC. (A) CDKN2A correlation with immune cells in the immune system shows a purity-corrected fraction of Spearman rho value and statistical significance. Log2(TPM) is the log2 of the transcript count per million. (B) Clinical relevance of B cells, CD8+T cells, CD4+T cells, macrophages, neutrophils, dendritic cells, and CDKN2A expression with UCEC samples from TCGA. Group cutoff: median. (C–F) IL2, IFNA1, TNF, and TP53 correlation. A total of 545 TCGA-UCEC cases were used.
CDKN2A somatic copy number alteration relates to drug metabolism in UCEC
SCNA is a prevalent characteristic of human cancer and plays a crucial role in tumorigenesis by influencing gene expression. Abnormal DNA CNV constitutes a significant molecular mechanism in various conditions, including cancer, hereditary diseases, and cardiovascular diseases. In UCEC, copy number alterations significantly influence prognosis.
In LinkedOmics, CDKN2A-SCNA data from the TCGA-UCEC dataset (n = 539) was integrated with RNA-seq data from the same cohort for analysis. As shown in the volcano plot in Fig. 6A, the genes on the right (dark red dots) have a significant positive correlation with CDKN2A-SCNA, while the genes on the left (dark green dots) have a significant negative correlation. The expression of the top 50 correlated genes with the most significant p-value were represented using a heatmap (Fig. 6B). Enrichment analysis of the obtained up- and down-regulated genes showed that multiple biological functions were associated with CDKN2A SCNA (Fig. 6C). Drug metabolism analysis revealed the most significant correlation (Fig. 6D). Therefore, because the down-regulated genes associated with CDKN2A-SCNA were correlated with drug metabolism, CDKN2A drug sensitivity was analyzed in both the GDSC (Genomics of Drug Sensitivity in Cancer) and CTRP (The Cancer Therapeutics Response Portal) databases to investigate the relationship between CDKN2A and different drugs (Fig. 6E). Drug sensitivity analysis demonstrated that CDKN2A is related to the sensitivity of numerous drugs. Cerulenin, manumycin A, and PRIMA-1 significantly negatively correlated with CDKN2A. Conversely, Nutlin-3a (-) and PD-0332991 show a significant positive correlation with CDKN2A.
Somatic copy number alteration in CDKN2A is related to drug metabolism in UCEC. (A) Differential genes from RNA-seq and CDKN2A mutant-related SCNA data for TCGA-UCEC differential genes. P-value obtained from Pearson correlation test. (B) Heatmap of differential genes. (C) Top 25 KEGG pathways for differential genes. (D) GSEA plot of CDKN2A-related differential genes. FDR (false discovery rate) < 0.05, t-test corrected for multiple testing. (E) Correlation between CDKN2A expression and sensitivity of GDSC and CTRP drugs (top 30) in pan-cancer. Orange shows positive correlation; purple represents negative correlation.
Discussion
The CDKN2A gene is located at human chromosome band 9p21 and is expressed in a variety of human cells. It encodes several proteins, including p14ARF and p16INK4a, which are known to regulate the cell cycle and inhibit tumor development and progression7. In normal tissue, p16INK4a binds to CDK4/6 with Cyclin D to prevent the cell from entering the S phase and initiating DNA synthesis. It also dephosphorylates pRb (Retinoblastoma protein) to inhibit cell proliferation. However, CDKN2A alterations can impair p16INK4a, increase pRb phosphorylation, and promote tumorigenesis13,14. In normal tissue, p14ARF typically forms a trisomy complex with MDM2 and p53 15. However, p14ARF loses its ability to bind to MDM2 (Mouse double minute 2) and cannot form a complex when abnormalities occur in CDKN2A. Consequently, MDM2 promotes p53 degradation, leading to tumorigenesis. Therefore, CDKN2A is crucial for maintaining tissue homeostasis and a balanced interplay between tumor suppression and senescence.
CDKN2A expression was found to be upregulated in UCEC in the present study, and this upregulation was closely associated with a reduction in patients’ survival time. Additionally, CDKN2A expression was significantly elevated at the early stage of tumor development, with corresponding increases in protein expression. The IHC statistical data from the HPA indicated that CDKN2A protein expression was higher in UCEC compared to the level in normal endometrial tissue.
Tumor development is a complex process that involves the interaction of multiple signaling pathways. In the present proteomics study, CDKN2A was linked to the p53/Rb and NRF2 pathways. Previous studies have shown that the role of NRF2 in immune metabolism and redox balance is significant for immunotherapy16. Consequently, CDKN2A was found to be correlated with CD8+ T cells, which are vital for immune defense against infections and cancer17. Observations in Fig. 5 indicate that a decrease in CD8+ T cell activity correlates with increased CDKN2A expression, suggesting a negative relationship between CDKN2A and CD8+ T cells. The specific molecular mechanisms behind this correlation remain unclear. In addition, the evolutionarily conserved p53 signaling pathway is pivotal in either controlling cell death or maintaining cellular homeostasis through various regulations, and it plays a crucial role in immune therapy18. IL2, IFNA1, and TNF also have important roles in tumor immunotherapy19,20,21.
In the study, CDKN2A expression was associated with the p53/Rb and NRF2 inflammatory pathways, as evidenced by proteomic mass spectrometry data. This discovery prompted further investigation into the potential connections between CDKN2A and immune therapy.
CNV is the duplication or deletion events of the genome22. Uterine serous carcinoma is a subtype of UCEC that is linked to TP53 misexpression, has high mortality and recurrence, and is often CNV-high, marked by cell cycle deregulation and CDKN2A misexpression23. CNV-low-UCEC has a moderate prognosis, with few TP53 mutations but many in the Wnt pathway and in genes like PTEN3,24,25. CNV-high-UCEC is characterized by frequent mutations in TP53, PIK3CA, and PPP2R1A and is associated with poor prognosis25,26. The TCGA database shows that CNV-high-UCEC is heterogeneous and covers nearly all subtypes3,27, with most patients having poor outcomes. Previous studies have indicated that CNV influences drug response and personalized therapy, while integrating mRNA and CNV omics data improves drug sensitivity prediction28,29. Given the link between CNV and UCEC prognosis, TCGA UCEC data were utilized in order to combine SCNA data for CDKN2A with RNA-seq data. GSEA was then conducted on differentially expressed genes. The results showed that down-regulated genes were significantly enriched in drug metabolism in the analysis of SCNA differential genes associated with CDKN2A. Thus, the relationship between CDKN2A and drug sensitivity was evaluated in a variety of cell lines. We found that PRIMA-1, Nutlin-3a, and PD-0332991 interact with the CDKN2A-encoded proteins p16INK4a and p14ARF in the process of regulating the cell cycle and tumor growth30,31,32, while the relationship between Cerulenin and Manumycin A with CDKN2A requires further study.
Next, we plan to delve deeper into the role of CDKN2A in drug metabolism. Molecular assessments of UCEC cases will be collected and performed, then we’ll categorize them into two groups depending on whether CDKN2A exhibits SCNA. Following the sequencing process and other experiments, the variations in gene expression will be analyzed to further investigate the implications of CDKN2A-SCNA.
Using GTEx UCEC data, the present study identified 115 genes associated with prognosis by intersecting the top 500 survival-related genes with those genes expressed differentially. CDKN2A was highlighted via the GO and KEGG analyses, which showed its increased mRNA and protein levels in UCEC and correlation with poor survival. Changes in CDKN2A protein levels were linked to alterations in the p53/Rb and NRF2 pathways, suggesting its relevance in immune therapy. In addition, GSEA of CNV-related downregulated genes in TCGA-UCEC cases indicated significant enrichment in drug metabolism. Elevated CDKN2A expression was closely tied to UCEC prognosisand various treatment strategies, including drug sensitivity, positioning it as a potential biomarker and therapeutic target for UCEC. Further research is needed to explore its mechanisms.
Methods
Gene of interest filtration
GEPIA2 33 (GEPIA 2 (cancer-pku.cn)) is an interactive web application for gene expression analysis based on 8,587 normal and 9,736 tumor samples from the GTEx34 (https://gtexportal.org/home/). There are 174 UCEC cases (with 174 tumor samples paired with 13 normal adjacent tissue samples) in the GTEx. Based on the GTEx-UCEC cases, the top 500 most significant survival-related genes related to UCEC were collected. The median of expression served as the group cutoff. The genes met the predefined statistical significance criteria (p-value < 0.05) and a minimum fold change threshold. A total of 7,131 differential expression genes were used based on the requirements of adjusted p-value < 0.05 and |log2 fold change| > 1.
At https://bioinformatics.psb.ugent.be/webtools/Venn/, The two gene types were intersected to generate a Venn plot and obtain 115 genes with intersections.
Integrated bioinformatics analysis
DAVID35 (https://bioinformatics.psb.ugent.be/webtools/Venn/) is an online analysis tool with integrated discovery and annotation capabilities that provides typical batch annotation and GO terminology augmentation analysis to highlight the most relevant content with a list of GO terms associated with a given gene.
Function enrichment analysis based on the GO36 and KEGG37,38,39 pathway map of 115 correlative genes was carried out using the online OMICSHARE cloud platform tool, which is an open data analysis platform that was used to visualize the enrichment analysis (http://www.omicshare.com/tools/Home/Soft/pathwaygsea).
Investigation of expression
UALCAN is an interactive and convenient web resource for comprehensively analyzing cancer OMICS data40. UALCAN was utilized to examine gene expression across various forms of cancer (http://ualcan.path.uab.edu). Initially, CDKN2A transcription was investigated in 24 different cancer types using data from TCGA41. To explore the mRNA expression levels of CDKN2A in UCEC patients, the TCGA-UCEC dataset encompassing 546 cases was subsequently employed. Further details regarding this dataset are accessible on the TCGA portal via the following link: https://portal.gdc.cancer.gov/. Graphs and plots were generated to delineate the expression profiles for outcome visualization.
UALCAN was used to investigate CDKN2A protein expression in UCEC tissue and to explore the potential signaling pathways involved in CDKN2A activation. The CPTAC-UCEC dataset from the Clinical Proteomic Tumor Analysis Consortium (CPTAC)42 was utilized. This dataset comprises 100 cases (with 100 tumor samples paired with 31 normal adjacent tissue samples) and is accessible at https://pdc.cancer.gov/pdc/. CDKN2A protein expression in UCEC tissue was quantified using tissue mass spectrometry with isobaric labels (tandem mass tags − 10). The method protocol can be found in43. This dataset was also used to investigate the expression profile of CDKN2A based on various pathways (HIP, mTOR, MYC/MYCN, RTK, SWI-SNF complex, Wnt, p53/Rb-related, chromatin modifier, and NRF2 pathways). Z-values represent standard deviations from the median across samples for the given cancer type. Log2 spectral count ratio values from CPTAC were first normalized within each sample profile and then across samples.
Survival rate analysis
Kaplan-Meier (KM) plotter was used to determine the association between target gene expression and OS in UCEC patients. A total of 174 cases (174 tumor samples paired with 13 normal adjacent tissue samples) were analyzed. The correlation between CDKN2A expression and clinical prognosis of UCEC patients was explored based on the different expression levels in GEPIA2 33. Median expression served as the group cutoff. The hazard ratio (HR) was calculated based on the Cox proportional hazards model with 95% confidence interval (CI) (dotted line). Log-rank p-values were also computed.
TIMER analysis
The TIMER (https://cistrome.shinyapps.io/timer/) database was employed to investigate the correlation between CDKN2A expression and immune cell infiltrates, which offers a comprehensive resource for the analysis of immune infiltrates across various cancer types44. TIMER provides estimates of immune cell abundance using several immune deconvolution methods, allowing users to dynamically generate high-quality figures that explore immunological, clinical, and genomic features. TIMER was utilized to analyze the correlation between CDKN2A expression and immune infiltrates, focusing on the clinical relevance of CDKN2A based on TCGA datasets. Specifically, the relationship between CDKN2A and immune cell types, including CD8+ T cells, CD4+ T cells, B cells, neutrophils, dendritic cells, and others, was examined.
Somatic copy number alteration analysis
LinkedOmics (https://www.linkedomics.org/login.php) is a publicly accessible portal that encompasses multi-omics data from all 32 TCGA cancer types and 10 CPTAC cancer cohorts45.
Integrated analysis using LinkedOmics was carried out to gain a deeper understanding of CDKN2A expression patterns and its association with clinical outcomes in UCEC. Initially, analytical tools provided by LinkedOmics were used to perform a joint analysis of the CDKN2A-SCNA and RNA-seq datasets related to 548 UCEC samples from TCGA database (accessible at https://pdc.cancer.gov/pdc/) in order to explore the relationship between CDKN2A CNVs and its mRNA expression levels. The analysis focused on statistically significant gene expression (adjusted p-value < 0.05) and the magnitude of CNVs (log2 fold change > 1). This comprehensive approach was aimed at identifying whether CDKN2A gene CNVs influence its expression in UCEC and further assessing the impact of these variations on patient prognosis. All analyses were conducted following standardized bioinformatics protocols and were aided by the advanced visualization tools offered by LinkedOmics to facilitate the interpretation of the results.
Drug sensitivity analysis
Gene Set Cancer Analysis (GSCA) (GSCA: Gene Set Cancer Analysis (wchscu.cn)) is a database that enables the searching, investigating, and exploring of gene-set cancer analyses pertaining to mRNA expression, mutations, immune infiltration, and drug resistance. GSCA integrates over 10,000 multidimensional genomic data points for 33 distinct cancer types from TCGA and more than 750 small molecule drugs from GDSC (Genomics of Drug Sensitivity in Cancer) and CTRP (The Cancer Therapeutics Response Portal)46.
The GSCA platform was used to examine CDKN2A drug sensitivity in a range of cell lines, thereby offering insights that can inform the integration of clinical data and small molecule drugs to identify potential therapeutic agents.
Data availability
The datasets generated and analyzed during the current study are available in the GTEx(https://gtexportal.org/home/), TCGA (https://portal.gdc.cancer.gov/) and CPTAC(https://gdc.cancer.gov/) repository.
References
Zheng, R. S. et al. Cancer incidence and mortality in China, 2022. Zhonghua Zhong Liu Za Zhi. 46, 221–231. https://doi.org/10.3760/cma.j.cn112152-20240119-00035 (2024).
Crosbie, E. et al. Endometrial cancer 399, 1412–1428, doi:https://doi.org/10.1016/s0140-6736(22)00323-3 (2022).
Getz, G. et al. Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73. https://doi.org/10.1038/nature12113 (2013).
McCluggage, W. G., Singh, N. & Gilks, C. B. Key changes to the World Health Organization (WHO) classification of female genital tumours introduced in the 5th edition Histopathology 80, 762–778, (2020). https://doi.org/10.1111/his.14609 (2022).
Concin, N. et al. ESGO/ESTRO/ESP guidelines for the management of patients with endometrial carcinoma. Int. J. Gynecologic Cancer. 31, 12–39. https://doi.org/10.1136/ijgc-2020-002230 (2021).
Imboden, S. et al. Implementation of the 2021 molecular ESGO/ESTRO/ESP risk groups in endometrial cancer. Gynecol. Oncol. 162, 394–400. https://doi.org/10.1016/j.ygyno.2021.05.026 (2021).
Kim, W. Y. & Sharpless, N. E. The regulation of INK4/ARF in cancer and aging. Cell 127, 265–275. https://doi.org/10.1016/j.cell.2006.10.003 (2006).
Zhao, L., Zhou, X., Li, H., Yin, T. & Jiang, Y. Prognosis of immunotherapy for non-small cell lung cancer with CDKN2A loss of function. J. Thorac. Disease. 16, 507–515. https://doi.org/10.21037/jtd-23-1017 (2024).
Luo, J., Wang, J. & Huang, J. -h. CDKN2A is a prognostic biomarker and correlated with immune infiltrates in hepatocellular carcinoma. Bioscience Rep. 41 https://doi.org/10.1042/bsr20211103 (2021).
Xu, W. et al. Genomic alteration of MTAP/CDKN2A predicts sarcomatoid differentiation and poor prognosis and modulates response to immune checkpoint blockade in renal cell carcinoma. Front. Immunol. 13 https://doi.org/10.3389/fimmu.2022.953721 (2022).
Cheng, T. et al. CDKN2A-mediated molecular subtypes characterize the hallmarks of tumor microenvironment and guide precision medicine in triple-negative breast cancer. Front. Immunol. 13 https://doi.org/10.3389/fimmu.2022.970950 (2022).
Liu, Q., Yu, M. & Zhang, T. Construction of oxidative stress-related genes risk Model predicts the prognosis of Uterine Corpus Endometrial Cancer patients. Cancers (Basel) 14. https://doi.org/10.3390/cancers14225572 (2022).
Pineau, P., Marchio, A., Cordina, E., Tiollais, P. & Dejean, A. Homozygous deletions scanning in tumor cell lines detects previously unsuspected loci. Int. J. Cancer. 106, 216–223. https://doi.org/10.1002/ijc.11214 (2003).
Wang, J. Z. et al. Increased mRNA expression of CDKN2A is a transcriptomic marker of clinically aggressive meningiomas. Acta Neuropathol. 146, 145–162. https://doi.org/10.1007/s00401-023-02571-3 (2023).
Stott, F. et al. The alternative product from the human CDKN2A locus, p14(ARF), participates in a regulatory feedback loop with p53 and MDM2. 17, 5001–5014, (1998). https://doi.org/10.1093/emboj/17.17.5001
Pant, T., Uche, N., Juric, M., Zielonka, J. & Bai, X. Regulation of immunomodulatory networks by Nrf2-activation in immune cells: Redox control and therapeutic potential in inflammatory diseases. Redox Biol. 70, 103077. https://doi.org/10.1016/j.redox.2024.103077 (2024).
Reina-Campos, M., Scharping, N. E. & Goldrath, A. W. CD8 + T cell metabolism in infection and cancer. Nat. Rev. Immunol. 21, 718–738. https://doi.org/10.1038/s41577-021-00537-8 (2021).
Hassin, O. & Oren, M. Drugging p53 in cancer: one protein, many targets. Nat. Rev. Drug Discovery. 22, 127–144. https://doi.org/10.1038/s41573-022-00571-8 (2022).
Dostert, C., Grusdat, M., Letellier, E. & Brenner, D. The TNF family of ligands and receptors: communication modules in the Immune System and Beyond. Physiol. Rev. 99, 115–160. https://doi.org/10.1152/physrev.00045.2017 (2019).
Briukhovetska, D. et al. Interleukins in cancer: from biology to therapy. Nat. Rev. Cancer. 21, 481–499. https://doi.org/10.1038/s41568-021-00363-z (2021).
Dranoff, G. Cytokines in cancer pathogenesis and cancer therapy. Nat. Rev. Cancer. 4, 11–22. https://doi.org/10.1038/nrc1252 (2004).
Steele, C. D. et al. Signatures of copy number alterations in human cancer. Nature 606, 984–991. https://doi.org/10.1038/s41586-022-04738-6 (2022).
Bogani, G. et al. Uterine serous carcinoma. Gynecol. Oncol. 162, 226–234. https://doi.org/10.1016/j.ygyno.2021.04.029 (2021).
Kurnit, K. C. et al. CTNNB1 (beta-catenin) mutation identifies low grade, early stage endometrial cancer patients at increased risk of recurrence. Mod. Pathol. 30, 1032–1041. https://doi.org/10.1038/modpathol.2017.15 (2017).
Urick, M. E. & Bell, D. W. Clinical actionability of molecular targets in endometrial cancer. Nat. Rev. Cancer. 19, 510–521. https://doi.org/10.1038/s41568-019-0177-x (2019).
Gonzalez-Bosquet, J. et al. PP2A and E3 ubiquitin ligase deficiencies: seminal biological drivers in endometrial cancer. Gynecol. Oncol. 162, 182–189. https://doi.org/10.1016/j.ygyno.2021.04.008 (2021).
Hussein, Y. R. & Soslow, R. A. Molecular insights into the classification of high-grade endometrial carcinoma. Pathology 50, 151–161. https://doi.org/10.1016/j.pathol.2017.09.010 (2018).
Santos, M. et al. Novel copy-number variations in pharmacogenes contribute to interindividual differences in drug pharmacokinetics. Genet. Med. 20, 622–629. https://doi.org/10.1038/gim.2017.156 (2018).
Wang, C. et al. The prediction of drug sensitivity by multi-omics fusion reveals the heterogeneity of drug response in pan-cancer. Comput. Biol. Med. 163 https://doi.org/10.1016/j.compbiomed.2023.107220 (2023).
Chen, W. et al. Applications and mechanisms of the cyclin-dependent kinase 4/6 inhibitor, PD-0332991, in solid tumors. Cell. Oncol. 45, 1053–1071. https://doi.org/10.1007/s13402-022-00714-4 (2022).
Zajkowicz, A. et al. Nutlin-3a, an MDM2 antagonist and p53 activator, helps to preserve the replicative potential of cancer cells treated with a genotoxic dose of resveratrol. Mol. Biol. Rep. 40, 5013–5026. https://doi.org/10.1007/s11033-013-2602-7 (2013).
Zatloukalová, P., Galoczová, M. & Vojtěšek, B. Prima-1 and APR-246 in Cancer Therapy. Klinicka Onkologie. 31 https://doi.org/10.14735/amko20182S71 (2018).
Tang, Z. F., Kang, B. X., Li, C. W., Chen, T. X. & Zhang, Z. M. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560. https://doi.org/10.1093/nar/gkz430 (2019).
Consortium, G. T. The genotype-tissue expression (GTEx) project. Nat. Genet. 45, 580–585. https://doi.org/10.1038/ng.2653 (2013).
Huang, D. W. et al. The DAVID Gene Functional Classification Tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol 8 (2007).
Ashburner, M. et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. https://doi.org/10.1038/75556 (2000).
Kanehisa, M., Goto, S., Kawashima, S. & Nakaya, A. The KEGG databases at GenomeNet. Nucleic Acids Res. 30, 42–46. https://doi.org/10.1093/nar/30.1.42 (2002).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592. https://doi.org/10.1093/nar/gkac963 (2023).
Chandrashekar, D. S. et al. An update to the integrated cancer data analysis platform. Neoplasia 25. UALCAN, 18–27. https://doi.org/10.1016/j.neo.2022.01.001 (2022).
Cancer Genome Atlas Research. The Cancer Genome Atlas Pan-cancer analysis project. Nat. Genet. 45, 1113–1120. https://doi.org/10.1038/ng.2764 (2013).
Li, Y. et al. Proteogenomic data and resources for pan-cancer analysis. Cancer Cell. 41, 1397–1406. https://doi.org/10.1016/j.ccell.2023.06.009 (2023).
Mertins, P. et al. Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc. 13, 1632–1661. https://doi.org/10.1038/s41596-018-0006-9 (2018).
Li, T. W. et al. A web server for Comprehensive Analysis of Tumor-infiltrating Immune cells. Cancer Res. 77, E108–E110. https://doi.org/10.1158/0008-5472.Can-17-0307 (2017).
Vasaikar, S. V., Straub, P., Wang, J. & Zhang, B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963. https://doi.org/10.1093/nar/gkx1090 (2018).
Liu, C. J. et al. GSCALite: a web server for gene set cancer analysis. Bioinformatics 34, 3771–3772. https://doi.org/10.1093/bioinformatics/bty411 (2018).
Acknowledgements
Not applicable.
Author information
Authors and Affiliations
Contributions
Conceptualization, Jingxian Wu and Yanfei Gao; methodology, Jingxian Wu; software, Liang Ma; validation, Liang Ma, Yuling Li; formal analysis, Liang Ma; investigation, Yuling Li; resources, Yuling Li; writing—original draft preparation, Liang Ma, Yuling Li; writing—review and editing, Jingxian Wu, Yanfei Gao; visualization, Liang Ma; supervision, Jingxian Wu; project administration, Jingxian Wu. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ma, L., Li, Y., Wu, J. et al. Bioinformatics approaches to multi-omics analysis of the potential of CDKN2A as a biomarker and therapeutic target for uterine corpus endometrial carcinoma. Sci Rep 15, 895 (2025). https://doi.org/10.1038/s41598-025-85364-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-85364-w