Abstract
The crosstalk between cancers and the immune microenvironment plays a critical role in malignant progression. FMS-like tyrosine kinase 3 (FLT3) is a frequently mutated gene in acute myeloid leukemia (AML). However, its role in solid cancers remains poorly understood. We analyzed the frequency of FLT3 alterations, its mRNA expression levels, and its prognostic implications across multiple cancer types. Additionally, we explored genes co-expressed with FLT3 and performed gene ontology analysis to identify associated biological processes. We also examined the relationship between FLT3 expression and markers of various immune cells, tertiary lymphoid structures (TLSs), and epithelial-mesenchymal transition. Furthermore, we validated these findings in our own cohort of hepatocellular carcinoma (HCC) patients. We found that FLT3 alteration and expression were both significantly upregulated in AML and were associated with poor prognosis, which is opposite to its role in solid cancers. The genes co-expressed with FLT3 in solid cancers were correlated with the regulation of the immune microenvironment. FLT3 was positively correlated with the formation of TLSs in only solid cancers, which was especially relevant to central memory T cells. We also found that FLT3 was positively correlated with the infiltration of NK cells, B cells, and DCs. It also positively correlated with the occurrence of apoptosis in solid cancers, but exhibited opposite roles in AML. The structural factors of the TLSs were positively correlated with FLT3 in solid cancers, but exhibited a negative correlation in AML. Meanwhile, we further validated the above conclusions in our own HCC cohort and demonstrated that FLT3 could serve as a predictive indicator of PD-1 treatment efficacy in HCC. In summary, the role of FLT3 is different in AML and solid cancers. FLT3 is associated with dendritic cell infiltration, tertiary lymphoid structure construction, and predict response to checkpoint inhibitors immunotherapy in HCC.
Similar content being viewed by others
Introduction
Cancer is the leading cause of death and a major public health issue. Despite the availability of various treatment strategies, the overall prognosis for cancer patients remains poor due to the high frequency of recurrence, metastasis, and low therapeutic response1. The tumor microenvironment (TME) plays a critical role in influencing the clinical outcomes of malignant cancers, as it houses cancer cells that interact with surrounding cells via the circulatory and lymphatic systems, thereby impacting the development and progression of cancer2. The recruitment, activation, and reprogramming of immune cells are outcomes of reciprocal interactions between cancer cells and the inflammatory microenvironment, key processes that contribute to TME remodeling3. The TME is subject to specific gene regulation, which varies across different cancer types, and its impact on various forms of malignant progression remains unclear.
FMS-like tyrosine kinase 3 (FLT3), which encodes a tyrosine kinase that activates pathways involved in proliferation and differentiation, is a commonly mutated gene in acute myeloid leukemia (AML)4. FLT3 mutations occur in approximately 30% of AML patients and are associated with poorer overall survival (OS) and the recurrent development of resistance5. Several small-molecule FLT3 tyrosine kinase inhibitors, including sorafenib, lestaurtinib, quizartinib, crenolanib, gilteritinib, and midostaurin, are currently being evaluated and have shown significant improvements in patient outcomes6. However, FLT3 inhibition, whether alone or in combination with other targets, is unable to fully block the malignant progression of solid cancers, despite showing antitumor effects in vitro and in vivo7. The roles of FLT3 gene alterations in solid cancers remain largely undefined, and the sensitivity to FLT3 inhibition in these cancers is unknown. Therefore, a deeper understanding of FLT3 genetic alterations in solid cancers, particularly from the perspective of the tumor inflammatory microenvironment, is crucial for developing better treatment strategies for patients with solid cancers.
In the present study, we analyzed the frequency of FLT3 alterations and its mRNA expression levels, exploring their correlation with prognosis across multiple cancer types. We also examined the relationship between FLT3 expression and the formation of tertiary lymphoid structures (TLSs), immune cell infiltration (including lymphocytes, monocytes, and dendritic cells (DCs)), and markers of epithelial-mesenchymal transition (EMT). Additionally, we assessed the correlation between FLT3 expression and lymphotoxins, chemokines, and adhesion molecules involved in TLS formation. Notably, FLT3 exhibited opposing roles in solid cancers compared to its role in AML. Using hepatocellular carcinoma (HCC) as a representative solid cancer, we further confirmed this conclusion.
Materials and methods
FLT3 alteration frequency, mRNA expression level, and survival analysis
The cBioPortal (http://www.cbioportal.org/) for cancer genomics provides information regarding the integrative analysis of complex cancer genomics and clinical profiles from 105 cancer studies in The Cancer Genome Atlas (TCGA) pipeline8,9. FLT3 expression and the frequency of FLT3 alterations (including mutation, fusion, amplification, deep deletion, and multiple alterations), as well as the prognostic value of FLT3 mutations in different cancer types, were assessed using the TCGA Pan-Cancer Atlas (comprising 10,967 samples from 32 studies) following the online instructions of cBioPortal.
GEPIA2 (http://gepia2.cancer-pku.cn/#index), an updated version of GEPIA, analyzes RNA sequencing expression data from 9,736 cancer samples and 8,587 normal samples from the TCGA and GTEx projects, using a standardized processing pipeline10. FLT3 cancer/normal differential expression and the correlation between FLT3 expression levels and survival outcomes across various cancer types were analyzed using GEPIA2. Additionally, GEPIA2 was used to explore the correlation between FLT3 expression and various immune cells, TLSs, EMT markers, as well as pro-apoptotic and anti-apoptotic markers.
The correlation between FLT3 expression and survival across various cancer types was further analyzed using the PrognoScan database (http://www.abren.net/PrognoScan/)11. PrognoScan identifies relationships between gene expression and patient prognosis, such as OS and disease-free survival (DFS), across a large collection of publicly available cancer microarray datasets. The threshold for significance was set at a Cox p-value < 0.05.
Genes co-expressed with FLT3
Genes co-expressed with FLT3 in various cancer types were analyzed using the LinkedOmics database (http://www.linkedomics.org), a publicly available portal that provides multi-omics data from all 33 TCGA cancer types12. The top 100 genes with significant positive correlations with FLT3 were identified in acute myeloid leukemia (LAML), liver hepatocellular carcinoma (HCC, known as LIHC in the database), head and neck squamous cell carcinoma (HNSC), breast invasive carcinoma (BRCA), and skin cutaneous melanoma (SKCM) based on this platform.
Functional enrichment analysis
Metascape (http://metascape.org) is a free, well-maintained, and user-friendly gene-list tool for gene annotation and analysis13. It serves as an automated meta-analysis platform to identify common and unique pathways across various orthogonal target-discovery studies. In this study, Metascape was used to perform pathway and process enrichment analysis for the top 100 genes positively correlated with FLT3 across multiple cancer types. Gene Ontology (GO) terms for biological processes, cellular components, and molecular functions, as well as relevant pathways, were enriched using the Metascape tool.
FLT3 expression and immune cell infiltration in various cancer types
TIMER 2.0 (http://timer.cistrome.org/) is a comprehensive resource for the systematic analysis of immune infiltrates across diverse cancer types14. Expression levels of FLT3 in cancer versus adjacent normal tissues across different cancer types were analyzed using the ‘Gene_DE’ module in TIMER 2.0, based on data from all TCGA cancers. Relative immune cell fractions were estimated using the CIBERSORT algorithm, which relies on a reference expression signature (LM22) containing 547 genes to distinguish 22 immune cell subtypes15. Data for immune cell infiltration estimates in LAML, LIHC, HNSC, and SKCM were obtained from the ‘Estimation’ module in TIMER 2.0.
Tissue samples, immunohistochemistry (IHC) and real-time quantitative polymerase chain reaction (RT-qPCR)
This study was approved by the Clinical Research Ethics Committee of Qilu Hospital, Shandong University, and informed consent was obtained from all participants. Tissues and matched adjacent normal tissue samples were surgically resected at the hospital, with all postoperative pathology reports confirming the diagnosis of hepatocellular carcinoma. The samples were stored in liquid nitrogen, with a portion fixed in 10% formalin and embedded in paraffin for long-term preservation. These tissue blocks were sectioned into 4 μm thick slices for immunohistochemical (IHC) staining. IHC procedures followed standard protocols, including dewaxing, antigen retrieval, antibody incubation (Servicebio, 1:100), and DAB development (Dako REAL). The double staining of immunohistochemistry was performed under the guidance of the instructions in the kit (ZSJB-BIO). RNA extraction and reverse transcription were performed according to the manufacturer’s instructions (Vazyme), followed by RT-qPCR analysis.
Statistical analysis
Data statistics, analysis, and graphing were performed using SPSS (version 17.0), R (version 4.2.1), ImageJ (version 1.8.0), and GraphPad Prism (version 8.0). Venn diagrams and bar plots were generated using ImageGP. Survival analysis was conducted using Kaplan-Meier survival curves, with statistical significance assessed by the log-rank test. Gene expression correlations were evaluated using Spearman’s correlation. A p-value < 0.05 was considered statistically significant.
Results
Genetic alteration frequency and mRNA level of FLT3 in AML is higher than in solid cancers, and FLT3 alteration is associated with a poor prognosis only in AML
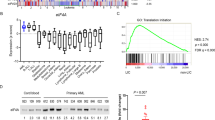
The data obtained from cBioPortal encompassed a combined study of 74,247 cancer samples. Pan-cancer analysis revealed FLT3 alterations (mutations, deletions, and amplifications) across multiple cancer types. Four solid cancers—kidney chromophobe (KICH), mesothelioma (MESO), thymoma (THYM), and uveal melanoma (UVM)—showed no FLT3 alterations, indicated by a tail without alteration. Conversely, six cancer types—AML, SKCM, uterine corpus endometrial carcinoma (UCEC), colon adenocarcinoma (COAD), esophageal carcinoma (ESCA), and stomach adenocarcinoma (STAD)—exhibited a “head” with frequent FLT3 alterations (more than 5%). Among these, AML displayed the highest frequency of FLT3 alterations, accounting for over 30% (Fig. 1A).
Genetic alteration frequency and mRNA levels of FLT3 in pan cancer and its association with prognosis. (A) FLT3 alterations (including mutations, deletions, and amplifications) were observed in pan cancer using the cBioPortal database. (B) The role of an FLT3 alteration in OS was evaluated using the cBioPortal database. The FLT3 alteration group exhibited a shorter OS than the FLT3 unaltered group in pan-cancers. (C) FLT3 expressions were observed in pan cancer using the cBioPortal database and the FLT3 mRNA level was significantly higher in AML than in any other solid cancer. (D) The role of the FLT3 mRNA level in OS and RFS were evaluated using the GEPIA2 database. A high level of FLT3 mRNA was significantly correlated with a poor prognosis only in AML.
Next, we explored whether FLT3 alterations were associated with cancer patient prognosis. Using the cBioPortal database, we evaluated the impact of FLT3 alterations on survival and found that patients with FLT3 alterations had a significantly shorter overall survival (OS) compared to those without FLT3 alterations across pan-cancer types (p = 0.0311, Fig. 1B). Kaplan-Meier survival plots were then generated for specific cancers (AML, SKCM, UCEC, COAD, ESCA, and STAD) to further investigate the relationship between FLT3 alterations and survival. This analysis confirmed that FLT3 alterations were associated with poor prognosis only in AML, while in STAD, FLT3 alterations were associated with better prognosis compared to the unaltered group (Supplementary Fig. 1). These findings suggest that FLT3 alterations occur more frequently in AML than in solid cancers, and are associated with poor prognosis specifically in AML.
To further investigate the role of FLT3 in cancer, we assessed FLT3 mRNA expression across various cancer types using cBioPortal. FLT3 expression was found to be highly variable across cancers, with significantly higher mRNA levels in AML compared to any other solid cancers (Fig. 1C). We then examined whether FLT3 expression correlated with patient prognosis in different cancers using the GEPIA2 database, which includes data from 33 human cancer types. In ten cancer types (BRCA, HNSC, LIHC, SKCM, adrenocortical carcinoma (ACC), cervical and endocervical cancers (CESC), COAD, kidney renal clear cell carcinoma (KIRC), sarcoma (SARC), and uterine carcinosarcoma (UCS)), high FLT3 mRNA levels were associated with better prognosis, showing significant differences in OS and/or relapse-free survival (RFS). In another seven cancers, high FLT3 expression was linked to a longer OS and/or RFS, although without significant differences. In AML, however, high FLT3 expression correlated with a significantly poorer prognosis (Fig. 1D). These trends were confirmed in human cancer datasets from GEO (Table 1). Collectively, these findings suggest that FLT3 mRNA levels are higher in AML compared to solid cancers, and that high FLT3 expression is associated with poor prognosis only in AML, contrary to its potential protective role in many solid cancers.
Functions of genes co-expressed with FLT3 in solid cancers were correlated with the regulation of lymphocyte activation and the adaptive immune response
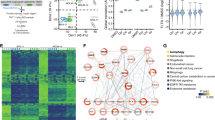
Based on survival analysis, high levels of FLT3 mRNA in BRCA, LIHC, HNSC, and SKCM were significantly associated with better prognosis for both OS and RFS. Consequently, these four cancer types were included in a systematic analysis using the LinkedOmics database, which compiles co-expression data, to identify genes correlated with FLT3 expression (Fig. 2A). We further explored the top 100 genes co-expressed with FLT3 in these cancers using Metascape. Accumulative hypergeometric p-values were calculated and presented as a heatmap for the top 20 enriched GO/KEGG term clusters. Genes co-expressed with FLT3 were significantly enriched in lymphocyte activation and the adaptive immune response (BRCA data missing; Fig. 2B). We then performed a similar analysis using the LinkedOmics database to identify genes correlated with FLT3 expression in AML. The top 100 genes co-expressed with FLT3 in AML were also analyzed using Metascape (Fig. 2C). The results showed genes that co-expressed with FLT3 were significantly enriched in RNA degradation, which was different from the enriched gene function in selected solid cancers (Fig. 2D). These results suggest that genes co-expressed with FLT3 are closely related to the TME in most solid cancers, which contrasts with its role in AML.
Functions of genes that co-expressed with FLT3 in solid cancers were associated with the tumor microenvironment. (A) The top 100 genes correlated with FLT3 were analyzed using the LinkedOmics database in BRCA, LIHC, HNSC, and SKCM. (B) The top 20 clusters with enriched GO/KEGG terms were performed using Metascape, and the genes that co-expressed with FLT3 were significantly enriched in lymphocyte activation and the adaptive immune response. (C) The top 100 genes correlated with FLT3 were analyzed using the LinkedOmics database in AML. (D) Genes that co-expressed with FLT3 were significantly enriched in RNA degradation, according to Metascape.
FLT3 was positively correlated with the formation of TLSs in solid cancers, but exhibited a negative correlation with TLSs in AML
Numerous experimental studies have established that TLSs in TME are independent predictors of better prognosis, primarily through the recruitment of functional T cells to the cancer site and their role in mediating an effective antitumor immune response16,17. Based on relevant literature, we selected eight biomarkers that represent TLSs in cancer tissues, including CD4, CD8, CD163, FOXP3, MS4A1, CD3, FCER2, and PECAM1 18,19. The mRNA levels of these biomarkers, indicative of TLSs, were assessed across different human cancer types using the GEPIA2 database. No significant differences were observed among the solid cancers, nor between solid cancers and AML (Fig. 3A). Next, we compared immune cell infiltration characteristics in AML and three selected solid cancers (HNSC, LIHC, and SKCM). Notable differences in immune cell infiltration were identified, with AML showing a higher proportion of monocytes, macrophages, and T cells compared to solid cancers (Fig. 3B). To further investigate the relationship between FLT3 expression and the TLS biomarkers, we examined 29 cancer types. FLT3 expression was positively correlated with these biomarkers in 27 out of 28 solid cancer types, particularly in HNSC, SKCM, COAD, ESCA, READ, BLCA, STAD, and UVM. However, MESO showed weak or no correlation with the biomarkers. In contrast, AML exhibited negative or no correlations between FLT3 expression and the TLS biomarkers. Specifically, CD3 and CD8 (p = 0.0025) showed negative correlations with FLT3 expression in AML, while the mRNA levels of CD4, CD163, FOXP3, FCER2, and PECAM1 showed no correlation with FLT3 expression in AML (Fig. 3C). These results suggest that FLT3 is positively correlated with TLSs formation in solid cancers but exhibits a negative correlation in AML.
FLT3 correlation with the formation of tertiary lymphoid structures (TLSs) in solid cancers. (A) Eight biomarkers that represent the TLSs exhibited no significant differences among a variety of cancers using the GEPIA2 database. (B) The characteristics of immune cell infiltration in AML. Three selected solid cancers were compared and the differences in immune cell infiltration were found using the TIMER database. (C) Eight biomarkers that represent the TLSs exhibited a positive correlation with the expression of FLT3 in most solid cancers using the GEPIA2 database.
FLT3 mainly correlated with TH1 immune cells, especially central memory T cells in solid cancers, but exhibited a negative correlation with TH1 in AML
T cells are classified into several subtypes based on their functions and surface markers, each playing distinct roles in the initiation and progression of malignant cancers. TH1 effector cells produce cytokines such as interleukin (IL)-2, interferon (IFN)-γ, and tumor necrosis factor (TNF)-α, which primarily mediate antitumor immunity20. In this study, FLT3 expression was positively correlated with the infiltration of TH1 cells in 27 out of 28 solid cancer types, particularly in HNSC, SKCM, CESC, COAD, cholangiocarcinoma (CHOL), ESCA, LUAD, READ, STAD, TGCT, and THCA. However, MESO showed weak or no correlation with FLT3. In contrast, in AML, FLT3 mRNA levels exhibited a negative correlation with TH1 cell infiltration (Fig. 4A).
FLT3 correlation with the TH1 immune system in solid cancers. (A) Based on surface markers, T cells are categorized into TH1 and TH2 cells. FLT3 expression was positively correlated with the levels of infiltrating TH1 cells in 27/28 types of solid cancer and exhibited a negative correlation with TH1 cells in AML. (B) FLT3 expression exhibited a strong positive correlation with the levels of infiltrating effector T, effector memory T, and central memory T cells in 27/28 types of solid cancer. (C) CCR7, SELL, and IL7R represented central memory T cells and exhibited a strong positive correlation with the expression of FLT3.
Additionally, FLT3 expression strongly correlated with the infiltration of effector T cells, effector memory T cells, and central memory T cells in 27 out of 28 solid cancer types, with the most notable correlation seen with central memory T cells (Fig. 4B). We further validated the positive correlation between FLT3 expression and the surface markers of central memory T cells, including CCR7, SELL, and IL7R, in the selected 28 cancer types. In AML, however, FLT3 expression showed a negative or no correlation with these surface markers, which was in stark contrast to the trend observed in solid cancers (Fig. 4C). These findings suggest that FLT3 is predominantly associated with the TH1 immune response, especially central memory T cells, in solid cancers, while exhibiting a negative correlation with TH1 cells in AML.
FLT3 was positively correlated with the infiltration of NK cells, B cells, and DCs, and with the occurrence of apoptosis in solid cancers, but exhibited opposite roles in AML
B cell-, NK cell-, and macrophage-mediated immune responses are crucial components of the immune system, particularly in TLSs, and play significant roles in antitumor immunity21. NK cells, part of the innate immune system, do not require antigen recognition in the context of MHC class I. Their activity is modulated by a balance between inhibitory and activating receptors. In this study, we observed that FLT3 expression was positively correlated with the markers CD11b, CCR7, CXCR3, and NKG2D in 26 out of 28 solid cancer types, except for MESO and BRCA, which showed a weak positive correlation with FLT3 expression. In AML, FLT3 mRNA levels exhibited no or negative correlations with NK cell-related markers. Additionally, FLT3 expression showed a positive correlation with the surface markers of B cells, such as CD11C and CD20, in 26 out of 28 cancers. A positive correlation was also observed between FLT3 expression and dendritic cell (DC) biomarkers, including CD83 and CD1a, in 19 out of 28 cancers (Fig. 5A).
FLT3 correlation with the infiltration of NK cells, B cells, monocytes, and DCs in solid cancers. (A) FLT3 was partially positively correlated with the infiltration of NK cells, B cells, and DCs in solid cancers, but exhibited no or negative correlations with these markers in AML. (B) Monocytes exhibited a positive relationship with the expression of FLT3 in most of the solid cancers, but M1 cells were not correlated with the expression of FLT3 in AML. (C) Lymphotoxins, chemokines, and adhesion molecules involved in the formation of TLSs correlated with the level of FLT3 in solid cancers.
To further explore the relationship between FLT3 expression and macrophage subtypes, we focused on markers for monocytes and M1/M2 macrophages using GEPIA2. A positive correlation was found between FLT3 expression and the surface markers CD86 and CD115 in monocytes across 25 out of 28 selected cancers. Moreover, FLT3 expression was positively correlated with infiltrating M2 macrophages in 24 out of 28 cancers, particularly in SKCM, COAD, CHOL, KICH, BLCA, THCA and UVM. The correlations between FLT3 expression and M1 macrophages were weak or absent. In contrast, in AML, FLT3 expression showed negative or no correlation with the markers of B cells, NK cells, monocytes and macrophages (Fig. 5B).
EMT is a critical process in metastasis22. To evaluate the correlation between FLT3 expression and mesenchymal markers, we focused on epithelial and mesenchymal phenotype markers in 28 cancer types using GEPIA2. No significant differences were observed between solid cancers and AML (Supplementary Fig. 2A). Furthermore, we assessed the correlation between FLT3 expression and apoptosis markers. We found that BLD, a pro-apoptosis gene, was positively correlated with FLT3 mRNA levels in 26 out of 28 solid cancers. In AML, there was only a weak correlation between FLT3 expression and BLK. We also observed that BCLW, an anti-apoptosis gene, showed weakly positive or negative correlations with FLT3 expression in 26 out of 28 solid cancers but had a significantly positive correlation in AML (Supplementary Fig. 2B).
These findings indicate that FLT3 is partially positively correlated with the infiltration of NK cells, B cells, and DCs, as well as with the occurrence of apoptosis in solid cancers. However, the role of FLT3 in AML is opposite to its role in solid cancers.
Lymphotoxins, chemokines, and adhesion molecules involved in the formation of TLSs were positively correlated with the level of FLT3 in solid cancers, but exhibited a negative correlation in AML
The genesis of TLSs is driven by a set of chemotactic cytokines that regulate cell positioning and interactions23,24. In this study, FLT3 expression was positively correlated with 22 biomarkers related to TLS formation, including lymphotoxins, chemokines, cytokines, and adhesion molecules. In AML, the mRNA levels of these biomarkers showed negative or no correlation with FLT3 expression. In contrast, these biomarkers exhibited significantly positive correlations with FLT3 expression in solid cancers, such as LTA (25/28), LTB (26/28), CCL2 (25/28), CCL19 (27/28), CXCL9 (27/28), VCAM1 (27/28), TIGIT (28/28) and CSF1 (26/28). Notably, significant positive correlations were observed in several solid cancers, including HNSC, SKCM, COAD and THCA. In contrast, FLT3 mRNA levels in AML were negatively correlated with these biomarkers (Fig. 5C). These findings suggest that FLT3 expression in solid cancers is positively correlated with the formation of TLSs, whereas in AML, the correlation is negative.
FLT3 could serve as a predictive indicator of PD-1 treatment efficacy
To further support the above conclusions, we conducted validation in HCC samples (Fig. 6A; Table 2). The results demonstrated a significant correlation between FLT3 expression levels and DC infiltration in HCC. Specifically, high FLT3 expression was associated with increased levels of CD1a + DC, while low FLT3 expression corresponded to lower levels of CD1a + DC (Fig. 6B, Supplementary Fig. 3A). However, this correlation was not observed in para-carcinoma tissue (Fig. 6B). Spearman analysis further confirmed a significant positive correlation between FLT3 expression and CD1a + DC infiltration in HCC, which was absent in peritumoral tissues (Fig. 6C). Interestingly, the expression of FLT3 in cirrhotic tissue was significantly higher compared to tissue from patients without cirrhosis (Fig. 6D; Table 2), potentially reflecting liver cell regeneration during cirrhosis. Of course, we also examined the correlation between FLT3 expression levels and infiltration of other immune cells in HCC. The results showed that there was a correlation between the expression level of FLT3 and the infiltration of CD8 + T cells, NK cells, and B cells in HCC (Supplementary Fig. 3B). When FLT3 was highly expressed, the infiltration of these cells also increased significantly. However, the correlation between these cells and FLT3 expression levels was not as strong as that of CD1a + DC cells (Supplementary Fig. 3B-C).
The expression of FLT3 was associated with CD1a+DC immune infiltration in HCC. (A) The information of HCC cohort. (B-C) FLT3 expression was positively correlated with CD1a+DC immune infiltration in HCC. (B): IHC results, (C): correlation analysis. (D) The expression levels of FLT3 was significantly correlated with liver cirrhosis. ***: p<0.001.
Additionally, using the median FLT3 expression as a cutoff, we found that high FLT3 expression tended to correlate with better OS and RFS, although the difference was not statistically significant (Fig. 7A-B). Furthermore, we examined FLT3 expression in samples from patients treated with PD-1 inhibitors (including Tirelizumab, Sintilimab, Toripalimab, and Camrelizumab) (Table 3). The samples were categorized into PD-1 effective (24 complete remission, 21 partial remission) and PD-1 ineffective (24 ineffective) groups (Fig. 7C). The results showed that FLT3 expression was significantly higher in patients who responded to PD-1 treatment (Fig. 7D). These findings suggest that FLT3 expression could serve as a potential predictive marker for the efficacy of PD-1-based therapies.
FLT3 could serve as a predictive indicator of PD-1 treatment efficacy in HCC. (A-B) High expression of FLT3 could indicate better OS and RFS, but there was no statistically significant difference. (C-D) The expression level of FLT3 was significantly increased in patients with PD-1 treatment effectiveness. ***: p<0.001.
Discussion
Solid cancers that appear in sites such as the head and neck, lung, breast, liver, stomach, colon, prostate, and skin are constructed by different proportions and types of malignant cells, endowing diverse features of cancer malignancy and drug responses25. And there are significant differences between these solid cancers and AML in various aspects. Therefore, understanding these differences is crucial for identifying why the same drug can have varying effects on different types of cancers. Receptor tyrosine kinases (RTKs) are key mediators of signaling pathways involved in the development of stem and progenitor cells, thereby contributing to cancer development26. FLT3 is an RTK predominantly expressed in hematopoietic stem and progenitor cell populations, and it has garnered significant attention in pharmacotherapy. Mutations in FLT3 are found in 25–35% of patients with AML. Recently, RNA sequencing revealed that FLT3 mRNA is not only expressed in AML but also in several human organs, including the appendix, lymph nodes, and spleen27. In solid cancers, the gene alterations and roles of FLT3 remain unclear, and the sensitivity of solid cancers to FLT3 inhibition is unknown. In this study, we identified six types of cancers—AML, SKCM, UCEC, COAD, ESCA, and STAD—that exhibited a relatively higher frequency of FLT3 gene alterations, with alteration frequencies of more than 5%. In contrast to solid cancers, AML has the highest frequency of FLT3 alterations, accounting for more than 30%. FLT3 alteration is also associated with a poor prognosis in AML, with a shorter OS compared to those in the FLT3-unaltered group. The mRNA levels of FLT3 were higher in AML than in solid cancers, and high FLT3 expression was associated with a poor prognosis only in AML. Multi-kinase inhibitors, such as sunitinib and sorafenib—first-generation FLT3 inhibitors—could improve outcomes in AML28. However, according to published reports, the therapeutic effect of these inhibitors in solid cancers has been unsatisfactory. A better understanding of the genetic alterations of FLT3 in solid cancers is crucial for evaluating the reasons why FLT3 inhibitors show poor curative effects and for defining suitable therapeutic strategies.
Cancers support a complex microenvironment characterized by the presence of various immune cell populations, reflecting the capacity of the immune system to sense cancer cells. Cancer development is closely associated with the physiological state of the TME, which orchestrate permissive niches for cancer progression29. In human cancer, a series of genetic and phenotypic changes, along with corresponding cytokines, chemokines, and metabolites derived from cancer cells, have a significant impact on the TME30. The genetics of AML also inform the composition of the immune microenvironment and contribute to leukemia cell growth31. A series of studies suggest that these soluble factors may be hijacked by cancer cells, promoting disease progression and recurrence. Recently, structural factors of the bone marrow (BM) niche were found to contribute to treatment resistance and relapse in AML, highlighting opportunities to manipulate, target, and evade the anti-inflammatory leukemic microenvironment32. Taking this as a reference, targeting the altered TME of cancer cells is a promising therapeutic approach to prevent cancer progression and relapse33. However, the differences between the BM microenvironment of AML and the TME of solid cancers remain vague. The development of effective anti-cancer therapies is challenged by the TME. So, it is necessary to explore more suitable treatment options for solid cancers. In this study, we explored the correlation between FLT3 and immune infiltration in solid tumors. We also evaluated lymphotoxins, chemokines, and adhesion molecules involved in the formation of TLSs, showing a positive correlation with FLT3 levels in solid cancers.
The prognostic value of TLSs has been evaluated in many types of cancers34. In general, TLSs in cancers are influenced by the number, location, and frequency of cellular components, and are correlated with active immune responses and a favorable prognosis16. In this study, we showed varying kinds of immune cell infiltration in different types of cancers. However, no significant differences or observable trends were found in TLSs across various cancer types, including AML. We also found that FLT3 was positively correlated with the formation of TLSs and the TH1 immune system, especially with central memory T cells, in solid cancers. Interestingly, FLT3 exhibited a negative correlation with TH1 in AML. Therefore, we concluded that FLT3 inhibitors could improve the survival of AML patients but may not be suitable for treating solid cancers, which is consistent with published reports7. Furthermore, the in-depth mechanisms were further analyzed. At present, it is generally believed that the structure of CD20 + B cells surrounded by T cells is the main feature of TLSs. Therefore, based on the results of our HCC cohort, we speculated that there was a significant correlation between the expression level of FLT3 and TLSs in HCC. However, due to the uniqueness of TLSs, they do not conform to the classical definition of organs and require numerous markers for display. There is still a lack of direct evidence in our HCC cohort.
DCs are professional antigen-presenting cells (APCs), whose primary function is to process and present antigens to B and T lymphocytes, thereby mediating adaptive immunity35. FLT3 is highly expressed in DCs, a specific mature hematopoietic lineage, and activation of FLT3 signaling dramatically increases the number of DCs in humans36. DCs with high FLT3 expression represent a unique population of APCs capable of sensitizing T cells to both new and recalled antigens37. AML cells exploit stromal-dependent pro-survival signals and shape the BM microenvironment to create a permissive niche favorable for the maintenance and progression of AML. In this study, we demonstrated a positive relationship between the mRNA level of FLT3 and the biomarkers of DCs, CD83 and CD1a, in most solid cancers, with a negative relationship observed only in AML. In solid cancers, the positive correlation between FLT3 expression and the TME prompted the hypothesis that poor drug efficacy could drive the formation of a negative TME following FLT3 inhibition. These conclusions were further validated in our HCC cohort, where FLT3 was shown to serve as a predictive indicator of PD-1 treatment efficacy. However, its role in other solid cancers warrants further investigation.
We showed that although FLT3 acts as an oncogene in AML, it functions as a cancer suppressor gene in most solid cancers. In AML, FLT3 inhibition, combined with the regulation of the immune microenvironment using the cytokines mentioned above, could enhance its anti-cancer efficacy. In solid cancers, FLT3 inhibitors or multi-kinase agents targeting FLT3 are not recommended unless supplemented with the aforementioned immune factors. In summary, the role of FLT3 differs between AML and solid cancers. FLT3 is associated with dendritic cell infiltration, tertiary lymphoid structure formation, and can predict response to checkpoint inhibitor immunotherapy in HCC. Combining immune-modulating agents may be key to effective control in most solid cancers. It is critical to adopt a comprehensive therapeutic approach.
Data availability
Our own dataset used and/or analyzed during the current study are available from the corresponding authors on reasonable request. The data are not publicly available due to privacy. Some data that supports the findings of this study are available from the repository of TCGA (https://portal.gdc.cancer.gov).
Abbreviations
- ACC:
-
Adrenocortical carcinoma
- AML:
-
Acute myeloid leukemia
- BLCA:
-
Bladder urothelial carcinoma
- BRCA:
-
Breast invasive carcinoma
- ccRCC:
-
Clear cell renal carcinoma
- CESC:
-
Cervical and endocervical cancers
- CHOL:
-
Cholangiocarcinoma
- COAD:
-
Colon adenocarcinoma
- DLBC:
-
Lymphoid neoplasm diffuse large B-cell lymphoma
- ESCA:
-
Esophageal carcinoma
- GBM:
-
Glioblastoma multiforme
- HNSC:
-
Head and neck squamous cell carcinoma
- KICH:
-
Kidney chromophobe
- KIRC:
-
Kidney renal clear cell carcinoma
- KIRP:
-
Kidney renal papillary cell carcinoma
- LAML:
-
Acute Myeloid Leukemia
- LGG:
-
Brain lower grade glioma
- LIHC:
-
Liver hepatocellular carcinoma
- LUAD:
-
Lung adenocarcinoma
- MESO:
-
Mesothelioma
- OV:
-
Ovarian serous cystadenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PCPG:
-
Pheochromocytoma and paraganglioma
- PRAD:
-
Prostate adenocarcinoma
- pRCC:
-
Papillary renal cell carcinoma
- SARC:
-
Sarcoma
- SKCM:
-
Skin cutaneous melanoma
- STAD:
-
Stomach adenocarcinoma
- TGCT:
-
Testicular germ cell tumors
- THCA:
-
Thyroid carcinoma
- THYM:
-
Thymoma
- UCEC:
-
Uterine corpus endometrial carcinoma
- UCS:
-
Uterine carcinosarcoma
- UVM:
-
Uveal melanoma
- APCs:
-
Antigen-presenting cells
- BM:
-
Bone marrow
- DCs:
-
Dendritic cells
- EMT:
-
Epithelial-mesenchymal transition
- FLT3:
-
FMS-like tyrosine kinase 3
- GO:
-
Gene ontologies
- DMFS:
-
Distant metastasis free survival
- DSS:
-
Disease specific survival
- OS:
-
Overall survival
- RFS:
-
Recurrent free survival
- TCGA:
-
The cancer genome atlas
- TME:
-
Tumor microenvironment
References
Boumahdi, S. & de Sauvage, F. J. The great escape: tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov. 19, 39–56. https://doi.org/10.1038/s41573-019-0044-1 (2020).
Hirata, E. & Sahai, E. Tumor Microenvironment and Differential responses to Therapy. Cold Spring Harb Perspect. Med. 7 https://doi.org/10.1101/cshperspect.a026781 (2017).
Fu, Y., Liu, S., Zeng, S. & Shen, H. From bench to bed: the tumor immune microenvironment and current immunotherapeutic strategies for hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 38, 396. https://doi.org/10.1186/s13046-019-1396-4 (2019).
Kiyoi, H., Kawashima, N. & Ishikawa, Y. FLT3 mutations in acute myeloid leukemia: therapeutic paradigm beyond inhibitor development. Cancer Sci. 111, 312–322. https://doi.org/10.1111/cas.14274 (2020).
Daver, N., Schlenk, R. F., Russell, N. H. & Levis, M. J. Targeting FLT3 mutations in AML: review of current knowledge and evidence. Leukemia 33, 299–312. https://doi.org/10.1038/s41375-018-0357-9 (2019).
Wu, M., Li, C. & Zhu, X. FLT3 inhibitors in acute myeloid leukemia. J. Hematol. Oncol. 11, 133. https://doi.org/10.1186/s13045-018-0675-4 (2018).
Klaeger, S. et al. The target landscape of clinical kinase drugs. Science 358 https://doi.org/10.1126/science.aan4368 (2017).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1. https://doi.org/10.1126/scisignal.2004088 (2013).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404. https://doi.org/10.1158/2159-8290.CD-12-0095 (2012).
Tang, Z., Kang, B., Li, C., Chen, T. & Zhang, Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560. https://doi.org/10.1093/nar/gkz430 (2019).
Mizuno, H., Kitada, K., Nakai, K. & Sarai, A. PrognoScan: a new database for meta-analysis of the prognostic value of genes. BMC Med. Genomics. 2, 18. https://doi.org/10.1186/1755-8794-2-18 (2009).
Vasaikar, S. V., Straub, P., Wang, J. & Zhang, B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 46, D956–D963 (2017).
Zhou, Y. et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523. https://doi.org/10.1038/s41467-019-09234-6 (2019).
Li, T. et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 48, W509–W514. https://doi.org/10.1093/nar/gkaa407 (2020).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 12, 453–457. https://doi.org/10.1038/nmeth.3337 (2015).
Sautes-Fridman, C., Petitprez, F., Calderaro, J. & Fridman, W. H. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer. 19, 307–325. https://doi.org/10.1038/s41568-019-0144-6 (2019).
Colbeck, E. J., Ager, A., Gallimore, A. & Jones, G. W. Tertiary lymphoid structures in Cancer: drivers of Antitumor Immunity, Immunosuppression, or Bystander sentinels in Disease? Front. Immunol. 8, 1830. https://doi.org/10.3389/fimmu.2017.01830 (2017).
Sofopoulos, M. et al. The prognostic significance of peritumoral tertiary lymphoid structures in breast cancer. Cancer Immunol. Immunother. 68, 1733–1745. https://doi.org/10.1007/s00262-019-02407-8 (2019).
Lin, L., Hu, X., Zhang, H. & Hu, H. Tertiary lymphoid organs in Cancer Immunology: mechanisms and the New Strategy for Immunotherapy. Front. Immunol. 10, 1398. https://doi.org/10.3389/fimmu.2019.01398 (2019).
Chraa, D., Naim, A., Olive, D. & Badou, A. T lymphocyte subsets in cancer immunity: friends or foes. J. Leukoc. Biol. 105, 243–255. https://doi.org/10.1002/JLB.MR0318-097R (2019).
Germain, C., Gnjatic, S. & Dieu-Nosjean, M. C. Tertiary lymphoid structure-Associated B cells are key players in Anti-tumor Immunity. Front. Immunol. 6, 67. https://doi.org/10.3389/fimmu.2015.00067 (2015).
Aiello, N. M. et al. Upholding a role for EMT in pancreatic cancer metastasis. Nature 547, E7–E8. https://doi.org/10.1038/nature22963 (2017).
Tang, H., Zhu, M., Qiao, J. & Fu, Y. X. Lymphotoxin signalling in tertiary lymphoid structures and immunotherapy. Cell. Mol. Immunol. 14, 809–818. https://doi.org/10.1038/cmi.2017.13 (2017).
Hiraoka, N., Ino, Y. & Yamazaki-Itoh, R. Tertiary lymphoid organs in Cancer tissues. Front. Immunol. 7, 244. https://doi.org/10.3389/fimmu.2016.00244 (2016).
Quail, D. F. & Joyce, J. A. Microenvironmental regulation of tumor progression and metastasis. Nat. Med. 19, 1423–1437. https://doi.org/10.1038/nm.3394 (2013).
Du, Z. & Lovly, C. M. Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer. 17 https://doi.org/10.1186/s12943-018-0782-4 (2018).
Fagerberg, L. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol. Cell. Proteom. 13, 397–406. https://doi.org/10.1074/mcp.M113.035600 (2014).
Antar, A. I., Otrock, Z. K., Jabbour, E., Mohty, M. & Bazarbachi, A. FLT3 inhibitors in acute myeloid leukemia: ten frequently asked questions. Leukemia 34, 682–696. https://doi.org/10.1038/s41375-019-0694-3 (2020).
Sullivan, M. R. et al. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. Elife 8 https://doi.org/10.7554/eLife.44235 (2019).
Pelizzo, G. et al. Microenvironment in neuroblastoma: isolation and characterization of tumor-derived mesenchymal stromal cells. BMC Cancer. 18, 1176. https://doi.org/10.1186/s12885-018-5082-2 (2018).
Kumar, B. et al. Acute myeloid leukemia transforms the bone marrow niche into a leukemia-permissive microenvironment through exosome secretion. Leukemia 32, 575–587. https://doi.org/10.1038/leu.2017.259 (2018).
Epperly, R., Gottschalk, S. & Velasquez, M. P. A bump in the Road: how the hostile AML Microenvironment affects CAR T cell therapy. Front. Oncol. 10, 262. https://doi.org/10.3389/fonc.2020.00262 (2020).
Roma-Rodrigues, C., Mendes, R., Baptista, P. V. & Fernandes, A. R. Targeting tumor microenvironment for cancer therapy. Int. J. Mol. Sci. 20 https://doi.org/10.3390/ijms20040840 (2019).
Kourea, H. & Kotoula, V. Towards tumor immunodiagnostics. Ann. Transl Med. 4, 263. https://doi.org/10.21037/atm.2016.07.07 (2016).
Patente, T. A. et al. Human dendritic cells: their heterogeneity and clinical application potential in Cancer Immunotherapy. Front. Immunol. 9, 3176. https://doi.org/10.3389/fimmu.2018.03176 (2018).
Liu, K. & Nussenzweig, M. C. Origin and development of dendritic cells. Immunol. Rev. 234, 45–54. https://doi.org/10.1111/j.0105-2896.2009.00879.x (2010).
Lamberti, M. J. et al. Dendritic cells and immunogenic cancer cell death: A combination for improving Antitumor immunity. Pharmaceutics 12, (2020). https://doi.org/10.3390/pharmaceutics12030256
Funding
This work was supported by the Youth Fund of Natural Science Foundation of Shandong Province (No. ZR2023QH535); the Wujieping Medical Foundation (No. 3206750); the Natural Science Foundation of Shandong Province (No. ZR2020MH253).
Author information
Authors and Affiliations
Contributions
Y.T. and Q.Z. were involved in the conception and design of the study. Y.T., H.W., J.Z., C.Y., F.X. and Q.Z. did the statistical analyses and wrote the manuscript. Y.S. and T.L. collected the data and perform the follow-up. Y.T. and Q.Z. provided resources and administrative support. Q.Z. critically reviewed or revised the manuscript for important intellectual content. All authors reviewed the final version of the manuscript, and agree with its content and submission.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study protocol received approval from the Ethics Committee of Qilu Hospital, Shandong University.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tang, Y., Wang, H., Zhang, J. et al. FLT3 is associated with dendritic cell infiltration, tertiary lymphoid structure construction, and predict response to checkpoint inhibitors immunotherapy in solid cancers. Sci Rep 15, 2477 (2025). https://doi.org/10.1038/s41598-025-86185-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-86185-7
Keywords
This article is cited by
-
Accurate prediction of anti-cancer drug responses using grey wolf optimization and multidimensional molecular data
Network Modeling Analysis in Health Informatics and Bioinformatics (2025)