Abstract
Neuromuscular diseases usually manifest as abnormalities involving motor neurons, neuromuscular junctions, and skeletal muscle (SkM) in postnatal stage. Present in vitro models of neuromuscular interactions require a long time and lack neuroglia involvement. Our study aimed to construct rodent bioengineered spinal cord neural network-skeletal muscle (NN-SkM) assembloids to elucidate the interactions between spinal cord neural stem cells (SC-NSCs) and SkM cells and their biological effects on the development and maturation of postnatal spinal cord motor neural circuits. After coculture with SkM cells, SC-NSCs developed into neural networks (NNs) and exhibited a high proportion of glutamatergic and cholinergic neurons, low proportion of neuroglia and gamma-aminobutyric acidergic neurons, and increased expression of synaptic markers. In NN-SkM assembloids, the acetylcholine receptors of SkM cells were upregulated, generating neuromuscular junction-like structures with NNs. The amplitude and frequency of SkM cell contraction in NN-SkM assembloids were increased by optogenetic and glutamate stimulation and blocked by tetrodotoxin and dizocilpine, respectively, confirming the existence of multisynaptic motor NNs. The coculture process involves the secretion of neurotrophin-3 and insulin growth factor-1 by SkM cells, which activate the related ERK-MAPK and PI3K-AKT signaling pathways in NNs. Inhibition of the ERK-MAPK and PI3K-AKT pathways significantly reduces neuronal differentiation and synaptic maturation of neural cells in NN-SkM assembloids, while also decreasing acetylcholine receptor formation on SkM cells. In brief, NN-SkM assembloids simulate the composition of spinal cord motor NNs and respond to motor regulatory signals, providing an in vitro model for studying postnatal development and maturation of spinal cord motor NNs.
Similar content being viewed by others
Introduction
Elucidation of the interaction mechanisms between spinal cord-derived neural stem cells (SC-NSCs) and skeletal muscle (SkM) cells is important for understanding the functional formation and maturation of motor neural networks (NNs) during development in mammals1,2. This knowledge will also help clarify the mechanisms underlying the occurrence and progression of neuromuscular diseases3,4,5.
The development of organoid technology, particularly the generation of self-organizing neuromuscular organoids derived from human induced pluripotent stem cells (iPSCs), provides an interactions system of spinal cord neurons and SkM cells for modeling neuromuscular diseases successfully based on individual human genetic backgrounds and capable of forming mature, and functional NMJs6,7,8,9, . However, iPSCs-derived neuromuscular organoids replicate the neuromuscular interactions and provide opportunities to study functional neuromuscular networks emerge during development, while most neuromuscular diseases occur in postnatal stage10,11,12. Currently, there is a lack of in vitro models that accurately represent the postnatal regulation effect of SkM cells on the differentiation, development and function maturation of SC-NSCs13,14. Also, the self-organizing neuromuscular organoids typically require 30 days or longer for the development of physiologically functional neuromuscular junctions (NMJs)15,16,17. This extended time period not only increases cost but also introduces additional uncertainties such as contamination into the process18,19. Thus, developing a spinal cord neural network-skeletal muscle (NN-SkM) interaction system that responds to motor regulatory signals in a short period would promote the experimental analysis of neuromuscular diseases and result in more effective elucidation of the mechanisms underlying the formation and regulation of spinal cord motor NNs20.
Our study aimed to construct a dynamic coculture system by seeding rodent SC-NSCs into 3-dimensional (3D) sponge scaffolds and subsequently coculturing them with 2-dimensional (2D) SkM cells, thereby generating NN-SkM assembloids21. Our objective was to investigate the postnatal biological effects and underlying mechanisms of SkM cells on the differentiation of SC-NSCs and the formation of spinal cord motor NNs.
Results
The generation of NN-SkM assembloids
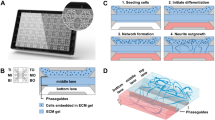
To generate NN-SkM assembloids, SC-NSCs and SkM cells were obtained as seed cells. Prior to seeding neurospheres into the collagen sponge, we confirmed the immunoreactivity of Nestin, Sox2 and HoxD922–24 in the SC-NSCs (Fig. 1a-c) and HoxD9 in the NSCs from hippocampus (Fig. 1d). Similarly, the immunoreactivity of myogenic determination protein 1 (MyoD) and myogenin (MyoG) confirmed the differentiation of the SkM cells (Fig. 1e and f). Subsequently, we cocultured SC-NSC-derived NNs with SkM cells for 7 days to form NN-SkM assembloids (Fig. 1g).
Construction of NN-SkM assembloids. (a–c) SC-NSCs expressing Nestin, Sox2 and HoxD9. (d) HoxD9 expression in NSCs from the hippocampus. (e) and (f) SkM cells exhibited MyoD-positive and MyoG-positive immunoreactivity. (g) A schematic diagram summarizing the assembly process of the NN-SkM assembloids. The cell nuclei were counterstained with Hoechst 33,342 (Hoe). Scale bars: 10 μm in (a), (c), and (d); 20 μm in (b), (e) and (f).
The formation of an excitatory NN
To investigate the biological effects of SkM cells on the differentiation of SC-NSCs, NN-SkM assembloids were utilized as the experimental group and separately cultured NNs as the control group. Immunofluorescence staining revealed that the SC-NSCs in both the NN-SkM group and the NN group exhibited neuronal, astrocytic, and oligodendrocytic differentiation potential (Fig. 2a-f). Statistical analysis revealed that the NN-SkM group had a greater percentage of microtubule-associated protein 2 (Map2)-positive neurons than did the NN group (P < 0.01, Fig. 2a, b and g). Furthermore, in the NN-SkM group, 17.1% of the cells expressed oligodendrocyte transcription factor 2 (Olig2), while 2.1% expressed the astrocyte marker glial fibrillary acidic protein (GFAP); these values were notably lower than those in the NN group (Fig. 2g). Additionally, we compared the transcriptomic profiles of the SCNSCs, NN, and NN-SkM groups and explored the expression of genes related to neural differentiation. Clustering heatmap analysis of mRNAs related to trilineage differentiation (Fig. 2h) and quantitative RNA expression analysis revealed significant upregulation of mRNAs related to neuron differentiation (Sod2, Vefc, and Cdnf; Fig. 2i) in the NN-SkM group. Conversely, mRNAs associated with oligodendrocyte differentiation (Cntnp, Gjc3, and Ak4, Fig. 2j) and astrocyte differentiation (Gpr3711, Csf1r, and Agt, Fig. 2k) were downregulated in the NN-SkM assembloids compared to those in the NN groups. These findings collectively demonstrate that cocultivation with SkM cells facilitates the differentiation into neurons.
The differentiation of SC-NSCs in the NN and NN-SkM assembloids. After 7 days of cultivation, the SC-NSCs differentiated into Map2-positive neurons (arrows in a and b), Olig2-positive oligodendrocytes (arrows in c and d) and GFAP-positive astrocytes (arrows in e and f) in the NN and NN-SkM assembloids. (g) Bar chart showing that the NN-SkM group had more Map2-positive cells and fewer GFAP-positive and Olig2-positive cells than the NN group (n = 6; **P < 0.01). (h) Heatmap representation of genes related to neural differentiation that were significantly differentially expressed (P < 0.05) between the NN-SkM, NN and SC-NSCs groups. Z-scoring label-free quantification (LFQ) intensities are depicted; red and blue represent increased and decreased values, respectively. Expression of genes associated with neuron differentiation (i), oligodendrocyte differentiation (j) and astrocyte differentiation (k). The results were significant across every group, with data input as values to the base 10 to allow log adjustment (n = 3, ***P < 0.001, **P < 0.01, *P < 0.05). The data are presented as means ± standard deviations (SD) in (g,i–k). Scale bars = 20 μm in (a–f).
The expression of neurotransmitters in the NN-SkM and NN groups was compared. Immunofluorescence analysis revealed choline acetyltransferase (ChAT)-positive, vesicular glutamate transporter 1 (Vglut1)-positive, and glutamate decarboxylase 67 (GAD67)-positive neurons in both the NN-SkM and NN groups (Fig. 3a-f). The NN-SkM group had an increase in the proportion of the cholinergic neuron marker ChAT, which accounts for 36.2% of the cell population, and the glutamatergic neuron marker Vglut1, which constitutes 33.7% of the cells, compared to than that in the NN group (Fig. 3h). Moreover, a decrease in the proportion of the gamma-aminobutyric acidergic (GABAergic) neuron marker GAD67 were reported in the NN-SkM group, accounting for 12.5% of the population, which was significantly lower than that in the NN group (Fig. 3h). Clustering heatmap analysis of neurotransmitter-related mRNA expression (Fig. 3g) and quantitative mRNA expression analysis demonstrated significant upregulation of cholinergic neuron-related mRNAs (e.g., Prkca, Cacna1c, and Rbp1, Fig. 3i) and glutamatergic neuron-related mRNAs (e.g., Jak2, Sparc, and Vgf, Fig. 3j) in the NN-SkM group compared to the NN and SC-NSCs groups. Moreover, mRNAs associated with GABAergic neuron differentiation (e.g., Gabrr1, Efna5, and Cacna1s; Fig. 3k) were significantly downregulated in the NN-SkM group compared with NN group. These findings suggest that cocultivation with SkM cells promotes the differentiation of SC-NSCs into excitatory neurons, contributing to the formation of an excitatory NN.
Neurotransmitter expression in NN and NN-SkM assembloids after 7 days of culture. (a–c) The expression of ChAT, Vglut1 and GAD67 in the NN. (d–f) The expression of ChAT, Vglut1 and GAD67 in the NN-SkM assembloids. (h) Bar chart showing that the NN-SkM group had more ChAT-positive and Vglut1-positive neurons and fewer GAD67-positive cells than the NN group (n = 6, ***P < 0.001, **P < 0.01). (g) Heatmap representation of genes related to neurotransmitter expression that were significantly different (P < 0.05) between the NN-SkM, NN and SC-NSCs groups. Z-scored LFQ intensities are depicted; red and blue represent increased and decreased values, respectively. Expression of genes associated with cholinergic (i), glutamatergic (j), and GABAergic synapses (k). The results were significant across every group, with data with data log-transformed to base 10 (n = 3, **P < 0.01). The data are presented as the means ± SDs in (h–k). Scale bars: 20 μm in (a–f).
Gene expression related to neuronal function in the NN-SkM assembloids
We conducted RNA-seq analysis of the NN-SkM, NN, and SC-NSC groups. Among 15,479 genes common to all three groups, the highly expressed unique genes associated with neuronal function were examined. Hierarchical clustering heatmap analysis (Fig. 4a) revealed that compared to those in the NN and SC-NSC groups, the neural cells in the NN-SkM group exhibited significantly increased expression of genes related to axon function, neuron projection, and synapse formation. Quantitative mRNA analysis revealed notable upregulation of genes associated with axon function (Timp2, Hspb1, and Tubb3, Fig. 4b), neuron projection (Epha3, Ephb4, and Wfs1, Fig. 4c), and synapse formation (JAK2, P2rx5, and Epha3, Fig. 4d) in the NN-SkM group compared with the NN and SC-NSC groups. Gene Ontology (GO) enrichment analysis revealed significant differences in the expression of genes related to nerve cell components between the NN-SkM and NN groups (Fig. 4e). The scatter plot shows increased expression of genes related to neuronal function, including neuronal activation and neurotrophic factor-related signaling, in both the NN-SkM and NN groups (Fig. 4f).
The expression of neuronal function-related genes in the NN-SkM assembloids. (a) Heatmap constructed from 109 upregulated (red) and downregulated (blue) genes with significant differences related to axon function, neuron projection and synapse formation by clustering analysis. Genes associated with axon function (b), neuron projection (c) and synapse formation (d). The results were significant across every group, with data input as values to the base 10 to allow log adjustment (n = 3, ***P < 0.001, **P < 0.01, *P < 0.05). (e) GO term enrichment analysis of the most highly expressed genes in the NN-SkM assembloids. (f) The scatter plot illustrates the expression levels of genes associated with neuronal activation and growth factor-related signaling across the NN and NN-SkM groups. Each point in the scatter plot corresponds to a gene, with the x-axis denoting the gene expression levels in the NN group and the y-axis representing the gene expression levels in the NN-SkM group. Genes exhibiting significant differences are depicted by red and orange dots, respectively, while genes with no significant differences are depicted in gray. The data are presented as the means ± SDs (n = 3) in (b–d). BP = biological process; CC = cellular component; MF = molecular function.
Formation of synapses and NMJs of the NN-SkM assembloid
To investigate the impact of SkM cells on synaptic formation in NNs, we conducted immunofluorescence assays and Western blotting to assess the expression of synaptic markers, including postsynaptic density protein 95 (PSD95) and synaptophysin (SYP), in both the NN-SkM and NN groups (Fig. 5a-c). A significant increase in SYP expression was observed in the NN-SkM group (Fig. 5c and d) compared to the NN group, indicating the maturity and synaptic formation of the NN in NN-SkM assembloids.
Synapse and NMJ formation in the NN-SkM assembloids. After 7 days of cultivation, neuronal cells in the NN (a) and NN-SkM assembloids (b) expressed postsynaptic density protein 95 (PSD95, green), a postsynaptic marker, and synaptophysin (SYP, white), a presynaptic marker, after 7 days of culture. (c) and (d) Western blot showing differential expression between the NN and NN-SkM groups (*P < 0.05). After 2D cultivation (e) and coculture with NN (f), SkM cells exhibited clusters with BTX. (g) Quantification of BTX clusters per SkM cell in the SkM and NN-SkM groups (**P < 0.01). (h) NF-positive neurons juxtaposed with BTX-positive SkM cells in the NN-SkM assembloids. Scale bars: 20 μm in (a–b, e–f); 5 μm in (h). Blots shown in Supplementary Fig. 2. The data are presented as the means ± SDs in (d) and (g). Arrows in Fig. 5e and f indicate the BTX clusters.
Furthermore, to explore the biological effects of NN on NMJ formation, SkM cells from NN-SkM assembloids were served as the experimental group and 2D SkM cells as the control group (SkM group). Acetylcholine receptors on SkM cells were labeled and quantified using α-bungarotoxin (BTX). Immunofluorescence assays demonstrated a substantial increase in the amount of BTX on SkM cells in the NN-SkM group compared to the SkM group (Fig. 5e-g). Immunofluorescence revealed the colocalization of neurofilament (NF) from the NN with BTX in SkM cells, indicating the establishment of a structure resembling NMJs (Fig. 5h).
Functional assessment of the NN-SkM assembloids
To investigate the function of NN-SkM assembloids in motor regulation, we subjected the neurons in the NN-SkM assembloids after 7days of cultivation to optogenetic stimulation and the voltage-gated sodium channel blocker TTX and observed muscle contraction before stimulation, after stimulation, and following TTX intervention (Fig. 6a-c, Video S1).
Under resting conditions, SkM cells exhibited regular contraction with a frequency of approximately 30 contractions per 20 s and a normalized displacement of approximately 7.5%. After optogenetic stimulation, the frequency and amplitude of SkM cell contraction in the NN-SkM assembloids were significantly greater than those before stimulation (Fig. 6d and e). The subsequent use of TTX inhibited SkM cell contraction in the NN-SkM assembloids (Fig. 6d). These findings suggest that muscle contraction in NN-SkM assembloids is initiated and modulated by action potentials in the NN.
To determine whether this response was specifically mediated by glutamatergic transmission, glutamate and the NMDA receptor antagonist MK-801 were added to the culture medium of NN-SkM assembloids (Fig. 6f, Video S2). After glutamate stimulation, the amplitude and frequency of SkM cell contraction in the NN-SkM assembloids were notably greater than those before stimulation (Fig. 6g and h). However, upon treatment with MK-801, SkM cell contraction in the NN-SkM assembloids was inhibited (Fig. 6g). The immunofluorescence results in the NN-SkM and NN groups demonstrated mutual contact between Vglut1-positive neurons and ChAT-positive neurons, suggesting the presence of synaptic connections between glutamatergic and cholinergic neurons in the NN-SkM assembloids (Fig. 6i and j). This finding suggested that SkM contraction in NN-SkM assembloids is regulated by glutamatergic neurons in the NN, which transmit excitatory neural impulses and thereby modulate the motor output of cholinergic neurons (Fig. 6k).
Furthermore, after coculture for 21 days, the contraction of the skeletal muscle mediated by glutamate in the NN-SkM assembloids were observed (see Video S3, showing SkM cell contraction before glutamate stimulation, after glutamate stimulation, and after the application of MK-801), indicating that the excitatory NN-SkM assembloids allow a long time window for functional observation.
NN control of muscle activity. (a) Schematics showing the NN-SkM assembloid setup. (b) Representative image showing an NN-SkM assembloid. In the NN, the expression of Thy1-ChR2/EYFP resulted in green fluorescence. (c) Representative traces of SkM cell contraction after normalization to the prestimulation baseline, after optogenetic stimulation and after TTX application in the NN-SkM assembloids. (d) Quantification of SkM cell contraction before and after optogenetic stimulation and after TTX application showing the median number of events per subfield within a field over a 20-second interval (**P < 0.01; ***P < 0.001). (e) Quantification of displacement normalized to the baseline levels before and after optogenetic stimulation. (f) Representative traces of SkM cell contraction normalized to the prestimulation baseline, after glutamate stimulation, and after the application of MK-801 to the NN-SkM assembloids. (g) Quantification of SkM cell contraction before glutamate stimulation, after glutamate stimulation and after MK-801 application showing the median number of events in subfields per field in 20 s (**P < 0.01; ***P < 0.001). (h) Quantification of displacement normalized to the baseline before and after glutamate stimulation. (i) and (j) Neurons in the NN and NN-SkM groups were triple-stained for ChAT, Vglut1, and SYP, revealing the synaptic connections between glutamatergic and cholinergic neurons. (k) Schematic diagram of the multisynaptic motor regulation system in the NN-SkM assembloids. Scale bars: 10 μm in (i–j).
The interaction mechanism between NN and SkM cells
To explore the interaction mechanism between NN and SkM cells, we first investigated differential gene expression between the NN-SkM group and the NN group. NN-SkM assembloids showed upregulation of genes that positively regulated neuron differentiation, axon regeneration, neurotransmitter release, growth factor-related signaling and signaling pathway transduction through a gene interaction regulatory network (Fig. 7a). Based on the genes related to the neural functional sets, a network diagram was generated showing the association of genes enriched in the following categories: neuron differentiation, axongenesis, growth factor-related signaling, the PI3K-AKT and MAPK signaling pathways, regulation of the insulin growth factor 1 receptor (IGF1-R) signaling pathway, and regulation of the ERK1 and ERK2 pathways (Fig. 7b).
The interaction mechanism between NN and SkM cells. (a) The network diagram represents the expression levels of five functional gene sets in the NN-SkM group compared with those in the NN group. Individual nodes between functional sets represent individual genes, and the color represents the log2FC value. (b) Network diagram showing the associations of genes enriched in different functional sets in the NN-SkM group compared with those in the NN group. The node size of the functional set represents the total number of candidate genes according to GO. (c) The expression profiles of SkM cells in the SkM and NN-SkM groups. (d) The expression profiles of IGF1-R, TrkC, p-AKT, AKT, p-PI3K, PI3K, p-ERK and ERK in the NN and NN-SkM groups are shown. (e) and (f) show the expression of TrkC in the NN and NN-SkM groups. (g) and (h) show the expression of IGF1-R in the NN and NN-SkM groups. (i) Relative expression levels of NT-3 and IGF-1 in SkM cells in the SkM and NN-SkM groups. (j) Bar chart showing the quantification of protein expression in the NN and NN-SkM groups based on Western blotting (*P < 0.05, ***P < 0.001). (k) Schematic diagram showing that neurotrophic factors secreted by SkM cells activate the PI3K-AKT and MAPK-ERK pathways within NNs. The data are presented as the means ± SDs (n = 3) in (i and j). Scale bars: 10 μm in (e–h). Blots shown in Supplementary Fig. 2.
The expression of neurotrophin-3 (NT-3) and insulin growth factor 1 (IGF-1) in SkM cells in NN-SkM assembloids was quantitatively evaluated via Western blotting, with single cultured SkM cells serving as the control group. Both the SkMs in the NN-SkM group and the individually cultured SkM cells expressed NT-3 and IGF-1 (Fig. 7c). Notably, the NN-SkM group exhibited significant increases in the expression of NT-3 and IGF-1 (Fig. 7i). This finding suggested that the cocultivation of NN and SkM cells facilitates the secretion of neurotrophic factors, including IGF-1 and NT-3, from SkM cells.
Subsequently, the expression of neurotrophic factor receptors in NNs in NN-SkM assembloids were quantitatively evaluated, with single-cultured NNs serving as the control group. Increased expression of IGF1-R and the receptor tyrosine protein kinase C (TrkC) was observed in the NN-SkM group (Fig. 7d and j). Immunofluorescence staining revealed a greater population of cells positive for IGF1-R and TrkC in the NN-SkM group than in the NN group (Fig. 7e-h). Furthermore, compared to those in the NN group, the expression levels of p-PI3K, p-ERK, and p-AKT were notably increased in the NN of the NN-SkM assembloids (Fig. 7d and j). This finding suggested that after the cocultivation of NN with SkM cells, the expression of the NT-3 receptors TrkC and IGF1-R increased in NNs, subsequently activating downstream signaling pathways such as the PI3K-AKT and MAPK-ERK pathways (Fig. 7k).
To investigate the role of the PI3K/AKT and MAPK/ERK signaling pathways in NN-SkM assembloids, we examined neural differentiation potential, synaptic maturation of the NNs, and acetylcholine receptor formation after treatment with Linsitinib (IGF1-R inhibitor), LY294002 (PI3K inhibitor), MK-2206 (AKT inhibitor), TrkC-Fc (TrkC blocker), SB203580 (MAPK inhibitor), and FR180204 (ERK inhibitor). The NN-SkM group served as control. Immunofluorescence staining and statistical analysis revealed that, in the Linsitinib, LY294002, MK-2206, TrkC-Fc, SB203580, and FR180204 groups, the percentage of Map2-positive neurons decreased, while Olig2-positive oligodendrocytes increased in the NN-SkM assembloids compared to the NN-SkM group (Figure S4a-4 h). The expression of PSD95 and SYP in the NN-SkM assembloids was reduced (Figure S4i and Figure S5a-5n) compared to the NN-SkM group. The expression of acetylcholine receptors in SkM cells within NN-SkM assembloids was also reduced compared to the SkM cells in the NN-SkM group (Figure S4j and Figure S5o-5u). Under resting conditions, SkM cells in the NN-SkM group exhibited regular contraction. In contrast, in the Linsitinib, LY294002, MK-2206, TrkC-Fc, SB203580, and FR180204 groups, the frequency of SkM cell contraction in the NN-SkM assembloids was significantly lower than in the NN-SkM group (Figure S3j and Video S4).
Discussion
The interaction between SC-NSCs and SkM cells plays a crucial role during the development of spinal cord motor NNs and neuromuscular diseases25,26. Utilizing tissue engineering technology, we construct NN-SkM assembloids in a short assembly time which simulate the composition of spinal cord motor NNs including neurons and neuroglia, and are capable of responding to signals transmitted by glutamate. This approach revealed the biological effects of SkM cells on the postnatal differentiation of SC-NSCs and the maturation of spinal cord motor NNs, elucidating the multisynaptic motor regulation of spinal cord excitatory NNs (Fig. S1).
Construction of excitatory NN-SkM assembloids responsive to motor regulatory signals
Establishing NMJs between motor neurons and SkM cells is a crucial prerequisite for in vitro modeling of neuromuscular diseases27,28,29,30,31. In NN-SkM assembloids, motor neurons extend their axons to SkM cells, promoting the expression of acetylcholine receptors in SkM cells and the formation of NMJ-like structures4,5,32. In contrast to previous in vitro culture methods that merely establish functional connections between motor neurons and SkM cells7,33,34, NN-SkM assembloids contain not only predominantly cholinergic and glutamatergic excitatory neurons but also a lower proportion of GABAergic inhibitory neurons, oligodendrocytes, and astrocytes within the NN. Therefore, we speculate that in this interaction system, the regulation of SkM cells by motor neurons involves various types of neurons and glial cells, resembling the motor regulation of the spinal cord in vivo35,36. Moreover, a significant increase in both the frequency and amplitude of SkM cell contractions was observed following stimulation with blue light or glutamate upon NN treatment. Conversely, the administration of TTX or the glutamate receptor blocker MK-801 significantly reduced SkM cell activity. TTX was used to investigate the regulation of muscle function by the NNs as a whole, rather than focusing solely on cholinergic neurons and the neuromuscular junction. Additionally, the application of MK801 suggests that the control of muscle contraction by cholinergic neurons may be influenced by NMDA-mediated Ca2+ influx, which is regulated by glutamatergic neurons within the spinal cord network. These results indicate that in NN-SkM assembloids, motor neuron-innervated SkM cells can respond to excitatory regulation by glutamatergic neurons, similar to spinal motor neurons in vivo19,37,38.
Compared to self-organized neuromuscular organoid systems, which lack involvement of neuroglia, and require a longer time (more than 30 days) to achieve neuronal control of muscles32,39,40, the NN control of SkM cells occurred as early as the 7th day after 3D NNs cocultured with 2D SkM cells. Furthermore, this neuronal control persisted for up to 21 days. In other words, excitatory NN-SkM assembloids excitatory NN-SkM assembloids robustly reproduce the postnatal interaction between spinal cord NNs and SkM cells, and offer a long functional observation window for studying the development of neuromuscular diseases and drug screening in vitro. Additionally, self-organized organoids exhibit volume enlargement, increased cell apoptosis, and phenotypic differences after prolonged culture10,39, while engineered excitatory NN-SkM assembloids can be standardized in construction, with a controllable size and the ability to achieve neuronal-muscle control in a short time frame. Moreover, this model can be adapted to meet the demands of high-throughput testing.
The functional maturation mechanism of NN-SkM assembloids
Current studies on neuromuscular organoids have focused on the neuronal innervation of SkM cells17,26,41,42, with less attention given to the biological effects of SkM cells on the differentiation of SC-NSCs and the functional maturation of motor NNs. However, the immunofluorescence and RNA-seq results in our study demonstrated that SkM cells can induce a greater proportion of SC-NSCs in NN-SkM assembloids to differentiate into neurons than can SC-NSCs in 3D cultivation alone. Moreover, a larger proportion of neurons in the NN-SkM assembloids exhibited excitatory neurotransmitter activity, differentiated into glutamatergic and cholinergic neurons, and expressed more synaptic-related proteins, such as PSD95 and SYP, while neurons in NN group showed relatively lower expression of mRNA and proteins related to synaptic formation, axon growth and neuron projection (Fig. 4a-d). These results confirm that the interaction with SkM cells plays a crucial role in promoting the functional maturation of the motor nervous system during postnatal development and suggest that the established excitatory NN-SkM assembloids can be further utilized for studies focusing on spinal cord motor NN development or drug screening for neuromuscular diseases.
We speculate that SkM cells primarily promote SC-NSC differentiation and neuronal functional maturation by activating the membrane receptors of NSCs or neurons through secreted bioactive factors. Western blot analysis demonstrated the upregulation of NT-3 and IGF-1 expression in SkM cells, while the expression of membrane receptors for neurotrophic factors, such as IGF1-R and TrkC, was significantly upregulated in the NNs, suggesting that NT-3 and IGF-1 secreted by SkM cells may be important bioactive factors43,44,45,46,47. Furthermore, RNA-seq revealed upregulated genes related to the IGF-1 signaling pathway, PI3K-AKT pathway, and MAPK-ERK pathway (Fig. 7b). Additionally, inhibition of the downstream PI3K-AKT or MAPK-ERK signaling pathways led to a decrease in neuronal differentiation, impaired synaptic maturation in the NNs, and reduced acetylcholine receptor formation in SkM cells (Figure S4 and S5). Thus, after NT-3 and IGF-1 secreted by SkM cells activate membrane receptors in NNs, the downstream PI3K-AKT pathway and MAPK-ERK pathway collectively promote the establishment of NMJs and regulate the functional maturation of NNs (Fig. 7k)40,48,49,50.
The advantages and limitations of the NN-SkM assembloids
Coculturing SkM cells may be an important factor accelerating the functional maturation process of NNs47,51,52, and the selection of seed cells for NNs could be another factor. In this study, we chose rodent SC-NSCs obtained from neonatal rats for use as seed cells rather than iPSC-derived NSCs. Theoretically, SC-NSCs exhibit directional caudal development during central nervous system development, reducing the induction time and the use of a series of specific induction factors53,54,55. Additionally, although the excitatory NN in NN-SkM assembloids mainly consists of excitatory neurons, it contains a low proportion of inhibitory neurons, oligodendrocytes, and astrocytes, the composition of which more closely resembles the NN within gray matter, showing spinal network complexity compared with those human in vitro model56,57. Compared with reported 3D neuromuscular organoids or 2D neuromuscular coculture systems5,37, the NN-SkM assembloids constructed in our study may better simulate the postnatal regulation of SkM by the spinal cord under physiological conditions and reflect the authentic response of the spinal cord during disease modeling or drug screening58,59. However, neuromuscular organoids derived from iPSC-derived NSCs can mimic the development of human organs19,39,60, showing notable advantages in studying possible developmental origins of neuromuscular diseases37. Moreover, the application of rodent-based in vitro model allows for faster, standardized, and multi-channel drug screening and gene manipulation to simulate postnatal neuromuscular diseases, with the potential to significantly reduce the number of animals sacrificed in in vivo animal experiments.
Because the cocultivation of SkM cells can promote the formation of excitatory NNs, we speculate that a coculture system of NN-SkM assembloids can be designed to construct excitatory NNs for transplantation to repair the injured spinal cord, serving as a substitute for damaged spinal cord tissue. Compared with NNs derived from forebrain NSCs, NNs derived from SC-NSCs exhibit a greater propensity for establishing synaptic connections with descending nerve fibers such as those in the corticospinal tract61,62,63. Thus, the transplantation of SC-NSC-derived NNs induced by SkM cells to injury sites would facilitate synaptic connections with brain-derived nerve fibers to relay excitatory neural information from the brain and integrate with motor neurons below the injured spinal cord segment, thereby relaying neural information from the motor cortex to control SkM contraction64,65,66. However, our study did not conduct extensive in vitro genetic manipulation, drug screening, or in vivo transplantation validation, which could serve as focal points for future research9.
Conclusion
In this study, we constructed excitatory NN-SkM assembloids that respond to motor regulation signals using tissue engineering techniques. This approach not only mimics the control of SkM by spinal cord NNs but also elucidates the biological mechanism of SkM cells in the postnatal differentiation of SC-NSCs and the maturation of spinal cord motor NNs. The development of NN-SkM assembloids achieved rapid and standardized construction, making them suitable for further applications in studying postnatal spinal cord neural development and neuromuscular disease while also meeting the demands of high-throughput testing and motor NN transplantation to repair spinal cord injuries.
Methods
Ethics statement
All animal experiments were approved by the Institutional Animal Care and Use Committee of Sun Yat-sen University (Approval Number: No. 2021PS704K). All methods were carried out in accordance with relevant regulations and ARRIVE guidelines.
Cultivation of SC-NSCs and SkM cells
SC-NSCs were obtained from three-day-old wild-type or green fluorescent protein (GFP)-expressing Sprague‒Dawley (SD) rats or three-day-old transgenic mice [B6. Cg-Tg (Thy1-ChR2/EYFP) 18Gfng/J; Jackson Laboratories, Bar Harbor, ME, USA] using established methods (Wang et al., 2007). Briefly, the rats and mice were anesthetized by CO2 inhalation in a controlled chamber until respiration ceased for 2 min. Then the entire spinal cord was carefully dissected in D-Hank’s solution. After removal of the epineurium, the spinal cord tissue was sectioned into small pieces in a Petri dish (Corning, USA) and digested at 37 °C with 4 mL of collagenase II to obtain a single-cell suspension. Subsequently, the cells were resuspended in SC-NSC medium consisting of Dulbecco’s modified Eagle’s medium (DMEM)/F12 (1:1, Life Technologies, USA) supplemented with 1% B27 and 20 ng/mL bFGF. The cells grew as neurospheres in suspension and were subjected to mechanical dissociation every day. The neurosphere progenitor content was assessed via nestin immunostaining.
SkM cells were derived from seven-day-old wild-type or GFP-expressing SD rats or seven-day-old transgenic mice [B6. Cg-Tg (Thy1-ChR2/EYFP) 18Gfng/J]. SkM bundles were carefully isolated from the thigh, and the nerves, fascia, and blood vessels were removed in D-Hank’s solution. After being minced into 2 × 2 mm pieces, the SkM bundles were digested for 15 min at 37 °C with 4 mL of collagenase II. The isolated cells were centrifuged to remove residual enzyme, resuspended in D-Hank’s solution, and then preplated for 30 min to reduce the fibroblast fraction. Subsequently, the cells were plated onto Matrigel-coated (1:50; Corning, 356230) 6-well plates. The cells were cultured in 40% DMEM (high glucose, Gibco) and 40% Ham’s F12 medium supplemented with 20% fetal bovine serum (FBS, Gemini), 100 U/mL penicillin (Gibco), and 100 U/mL streptomycin (Gibco) at 37 °C in 5% CO2. Once they reached 90% confluence, the cells were passaged at a 1:4 ratio using 0.25% trypsin (Gibco) and then cultured in differentiation medium (high glucose [DMEM] supplemented with 2% horse serum).
Generation of NNs from SC-NSCs
The collagen sponge (BIOT Biology, Wuxi, China) scaffold was initially tailored into a square column (5 × 5 × 1 mm3). A total of 5 × 105 SC-NSCs per 10 µL of culture medium were seeded in each collagen sponge scaffold to generate 3D NNs. To promote cell adhesion, the scaffolds seeded with SC-NSCs were incubated in a 12-well plate containing 50 µL of medium at 37 °C for 30 min. Then, the scaffolds were further incubated for 1 day in 500 µL of culture medium (DMEM/F12 supplemented with 1% B27 and 1% FBS) to form the SC-NSC-derived NNs.
Construction of NN-SkM assembloids
For the generation of NN-SkM assembloids, P2-P3 SkM cells were cultured in Matrigel-coated 6-well plates with differentiation medium (DMEM/F12 supplemented with 1% FBS) to generate 2D SkM cells. Next, the SC-NSC-derived NNs were placed above the 2D SkM cells so that they were in contact with one another and were allowed to interact. The coculture system was maintained in 1000 µL of culture medium for 7 days, with medium changing every day. SC-NSC-derived NNs that were not cocultured with SkM cells served as controls to assess neuronal differentiation and neurotransmitter expression. To determine the role of the PI3K/AKT and MAPK/ERK signaling pathways in NN-SkM assembloids, Linsitinib (IGF1-R inhibitor, 1 µM), LY294002 (PI3K inhibitor, 10 µM), MK-2206 (AKT inhibitor, 10 µM), TrkC-Fc (TrkC blocker, 1 µg/mL), SB203580 (MAPK inhibitor, 1 µM), and FR180204 (ERK inhibitor, 1 µM) were added to the culture medium respectively and incubated for 7 days. NN-SkM assembloids without inhibitors served as the control group.
Immunofluorescence
NN, SkM cells and NN-SkM assembloids were fixed in 4% paraformaldehyde for 20 min at room temperature. After three washes with phosphate-buffered saline (PBS), the samples were permeabilized with PBS containing 0.2% Triton X-100 and 10% goat serum for 30 min. The samples were incubated with primary antibodies (Table S1) overnight at 4 °C. After three rinses with PBS, the samples were incubated with secondary antibodies for 1 h at 37 °C. Nuclear counterstaining was conducted with Hoechst 33,342. Images were taken with a confocal microscope (Dragonfly, CR-DFLY-202 2540; Andor Technology, Belfast, UK).
Western blotting
SC-NSC-derived NNs and SkM cells were used for protein extraction. Equal amounts of protein were separated by electrophoresis, followed by transfer onto a polyvinylidene fluoride membrane. The membrane was incubated with primary antibodies at 4 °C overnight, followed by incubation with horseradish peroxidase-conjugated secondary antibodies. The bands were detected with an enhanced chemiluminescence substrate kit.
Optogenetic and glutamate stimulation and blockade
For optogenetic and glutamate stimulation of NN-SkM assembloids, we imaged NN-SkM assembloids in six-well plates using a 10x or 20x objective in an automated microscope (BioTek-Lionheart FX) under environmentally controlled conditions (37 °C, 5% CO2). For optogenetic stimulation, the contraction of SkM cells in NN-SkM assembloids without stimulation were recorded as control. Then, optogenetic stimulation consisting of 66 pulses of blue light (488 nm, 911 ms in duration each) was delivered using an optical fiber-coupled LED that was directed toward the NN. After optogenetic stimulation, 0.5 µM tetrodotoxin (TTX) was used to block the voltage-gated sodium channels of the NN. Each period of contraction was recorded for 60 s at a frame rate of 10 frames/second.
For activation of the glutamatergic neurons in the NN, the contraction of muscle fibers in NN-SkM assembloids without stimulation was recorded as a control, and then glutamate was added to the culture medium of the NN-SkM assembloids at a final concentration of 1 mM. Next, the NMDA receptors were blocked by dizocilpine (MK-801) at a final concentration of 50 µM. Each period of contraction was recorded for 60 s at a frame rate of 10 frames/second.
Muscle contraction was quantified using the automated, open-source ImageJ plugin MUSCLEMOTION (https://github.com/l-sala/MUSCLEMOTION)26,39. MUSCLEMOTION quantifies movement by subtracting the absolute changes in pixels between a reference frame and the frame of interest. One to two fields were imaged per assembloid. 2–3 regions of interest (ROIs) were selected in each imaging field, and the analysis was individually performed for each of the ROIs. For the analysis of contractions, events over 3 median absolute deviations were considered contraction events. Displacement over time was calculated by normalizing all values to the 50 frames preceding stimulation.
mRNA sequencing
After coculture for 7 days, the NNs in the NN-SkM assembloids were mechanically separated from the SkM cells for mRNA sequencing (RNA-seq), and the NNs cultured alone and SC-NSCs were used as controls. RNA was extracted via standard methods. The RNA integrity was determined by agarose gel electrophoresis (28 S:18 S ≥ 1.5). The RNA purity was assessed using a NanoDrop spectrophotometer (optical density [OD] 260/280: 1.8–2.2), and the RNA concentration was accurately quantified using a Qubit fluorometer (≥ 500 ng/mL). RNA-Seq analysis of the samples was performed by Genergy Bio-Technology Co., Ltd. (Shanghai, China). Data generated by a high-throughput sequencer were then converted into sequenced reads by CASAVA base calling. The original sequence counts of known genes for all samples were determined by StringTie, and the expression levels of known genes were calculated as fragments per kilobase of transcript per million fragments mapped. DESeq2 software (version number: 1.44.0; https://github.com/thelovelab/DESeq2) was used to identify differentially expressed genes between groups with |log2(fold change [FC]) | values ≥ 1 and P values < 0.05.
Statistical analysis
The data are expressed as the means ± standard deviations (SDs). The data were analyzed using GraphPad Prism software (version number: 9.5.0; GraphPad Software, San Diego, CA, USA; https://www.graphpad.com). Two-sample t tests were used for comparisons of two groups, and one-way analysis of variance (ANOVA) with a Tukey post hoc correction was used for comparisons involving more than two groups. For all experiments, values of P < 0.05 were considered to indicate statistical significance.
Data availability
The datasets generated and analyzed in this study are available at the China National Center for bioinformation (GSA: CRA017096) repository, accessible at https://ngdc.cncb.ac.cn/gsa/browse/CRA017096.
References
Harley, P. et al. 3D Compartmentalised Human pluripotent stem cell-derived neuromuscular co-cultures. Bio-Protoc 13, e4624 (2023).
Leng, Y. et al. Advances in in vitro models of neuromuscular junction: Focusing on organ-on-a-chip, organoids, and biohybrid robotics. Adv. Mater. Deerfield Beach Fla. 35, e2211059 (2023).
Klim, J. R. et al. ALS-implicated protein TDP-43 sustains levels of STMN2, a mediator of motor neuron growth and repair. Nat. Neurosci. 22, 167–179 (2019).
Zhang, K., Bai, L., Xu, W. & Shen, C. Human neuromuscular junction three-dimensional organoid models and the insight in motor disorders. J. Mol. Cell. Biol. 13, 767–773 (2022).
Kim, H., Kim, G. S., Hyun, S. H. & Kim, E. Advancements in 2D and 3D in Vitro models for studying neuromuscular diseases. Int. J. Mol. Sci. 24, 17006 (2023).
Melamed, Z. et al. Premature polyadenylation-mediated loss of stathmin-2 is a hallmark of TDP-43-dependent neurodegeneration. Nat. Neurosci. 22, 180–190 (2019).
Pereira, J. D. et al. Human sensorimotor organoids derived from healthy and amyotrophic lateral sclerosis stem cells form neuromuscular junctions. Nat. Commun. 12, 4744 (2021).
Shimizu, K. et al. Alignment of skeletal muscle cells facilitates acetylcholine receptor clustering and neuromuscular junction formation with co-cultured human iPSC-derived motor neurons. Cells 11, 3760 (2022).
Urzi, A. et al. Efficient generation of a self-organizing neuromuscular junction model from human pluripotent stem cells. Nat. Commun. 14, 8043 (2023).
Bucchia, M., Merwin, S. J., Re, D. B. & Kariya, S. Limitations and challenges in modeling diseases involving spinal motor neuron degeneration in vitro. Front. Cell. Neurosci. 12, 61 (2018).
Kabiljo, R., Iacoangeli, A., Al-Chalabi, A. & Rosenzweig, I. Amyotrophic lateral sclerosis and cerebellum. Sci. Rep. 12, 12586 (2022).
Berdyński, M. et al. SOD1 mutations associated with amyotrophic lateral sclerosis analysis of variant severity. Sci. Rep. 12, 103 (2022).
Borciani, G., Montalbano, G., Baldini, N., Vitale-Brovarone, C. & Ciapetti, G. Protocol of co-culture of human osteoblasts and osteoclasts to test biomaterials for bone tissue Engineering. Methods Protoc. 5, 8 (2022).
Zschüntzsch, J. et al. The evolution of complex muscle cell in vitro models to study pathomechanisms and drug development of neuromuscular disease. Cells 11, 1233 (2022).
Dimos, J. T. et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science 321, 1218–1221 (2008).
Xu, C. et al. A zebrafish embryo culture system defines factors that promote vertebrate myogenesis across species. Cell 155, 909–921 (2013).
Guo, X. et al. A human-based functional NMJ system for personalized ALS modeling and drug testing. Adv. Ther. 3, 2000133 (2020).
Takasato, M. et al. Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568 (2015).
Osaki, T., Uzel, S. G. M. & Kamm, R. D. Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci. Adv. 4, eaat5847 (2018).
Buchner, F., Dokuzluoglu, Z. & Grass, T. Rodriguez-Muela, N. Spinal cord organoids to study motor neuron development and disease. Life Basel Switz. 13, 1254 (2023).
Olmsted, Z. T. et al. Fabrication of homotypic neural ribbons as a multiplex platform optimized for spinal cord delivery. Sci. Rep. 10, 12939 (2020).
Miller, A. & Dasen, J. S. Establishing and maintaining hox profiles during spinal cord development. Semin Cell. Dev. Biol. 152–153, 44–57 (2024).
Imai, F., Adam, M., Potter, S. S. & Yoshida, Y. HoxD transcription factors define monosynaptic sensory-motor specificity in the developing spinal cord. Dev. Camb. Engl. 148, dev191122 (2021).
Philippidou, P. & Dasen, J. S. Hox genes: Choreographers in neural development, architects of circuit organization. Neuron 80, 12–34 (2013).
Quadrato, G. et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature 545, 48–53 (2017).
Gazzola, M. & Martinat, C. Unlocking the complexity of neuromuscular diseases: insights from human pluripotent stem cell-derived neuromuscular junctions. Int. J. Mol. Sci. 24, 15291 (2023).
Fischer, L. R. et al. Amyotrophic lateral sclerosis is a distal axonopathy: Evidence in mice and man. Exp. Neurol. 185, 232–240 (2004).
McCampbell, A. et al. Antisense oligonucleotides extend survival and reverse decrement in muscle response in ALS models. J. Clin. Invest. 128, 3558–3567 (2018).
Moakley, D. et al. Pharmacological profiling of purified human stem cell-derived and primary mouse motor neurons. Sci. Rep. 9, 10835 (2019).
Cy, L. et al. iPSC-derived functional human neuromuscular junctions model the pathophysiology of neuromuscular diseases. JCI Insight 4, (2019).
M, A. B. et al. A 3D culture model of innervated human skeletal muscle enables studies of the adult neuromuscular junction. eLife 8, (2019).
Baldassari, S. et al. Brain organoids as model systems for genetic neurodevelopmental disorders. Front. Cell. Dev. Biol. 8, 590119 (2020).
Mitchell, J. C. et al. Wild type human TDP-43 potentiates ALS-linked mutant TDP-43 driven progressive motor and cortical neuron degeneration with pathological features of ALS. Acta Neuropathol. Commun. 3, 36 (2015).
Sharma, A. et al. ALS-associated mutant FUS induces selective motor neuron degeneration through toxic gain of function. Nat. Commun. 7, 10465 (2016).
Chen, J., Leong, S. Y. & Schachner, M. Differential expression of cell fate determinants in neurons and glial cells of adult mouse spinal cord after compression injury. Eur. J. Neurosci. 22, 1895–1906 (2005).
O’Shea, T. M., Burda, J. E. & Sofroniew, M. V. Cell biology of spinal cord injury and repair. J. Clin. Invest. 127, 3259–3270 (2017).
Faustino Martins, J. M. et al. Self-Organizing 3D human trunk neuromuscular organoids. Cell. Stem Cell. 27, 498 (2020).
Lynch, E. et al. Current progress in the creation, characterization, and application of human stem cell-derived in vitro neuromuscular junction models. Stem Cell. Rev. Rep. 18, 768–780 (2022).
Andersen, J. et al. Generation of functional human 3D cortico-motor assembloids. Cell 183, 1913–1929e26 (2020).
de Vieira, R., Cañizares Luna, M. & Pasterkamp, R. J. Advances in central nervous system organoids: A focus on organoid-based models for motor neuron disease. Tissue Eng. Part. C Methods 27, 213–224 (2021).
Shin, M. K. et al. Generation of skeletal muscle organoids from human pluripotent stem cells to model myogenesis and muscle regeneration. Int. J. Mol. Sci. 23, 5108 (2022).
Gao, C. et al. Neuromuscular organoids model spinal neuromuscular pathologies in C9orf72 amyotrophic lateral sclerosis. Cell. Rep. 43, 113892 (2024).
Gómez-Pinilla, F., Ying, Z., Opazo, P., Roy, R. R. & Edgerton, V. R. Differential regulation by exercise of BDNF and NT-3 in rat spinal cord and skeletal muscle. Eur. J. Neurosci. 13, 1078–1084 (2001).
Omura, T. et al. Different expressions of BDNF, NT3, and NT4 in muscle and nerve after various types of peripheral nerve injuries. J. Peripher Nerv. Syst. JPNS 10, 293–300 (2005).
Dutt, V., Gupta, S., Dabur, R., Injeti, E. & Mittal, A. Skeletal muscle atrophy: Potential therapeutic agents and their mechanisms of action. Pharmacol. Res. 99, 86–100 (2015).
Oliveira Fernandes, M. & Tourtellotte, W. G. Egr3-dependent muscle spindle stretch receptor intrafusal muscle fiber differentiation and fusimotor innervation homeostasis. J. Neurosci. Off J. Soc. Neurosci. 35, 5566–5578 (2015).
Syverud, B. C., VanDusen, K. W. & Larkin, L. M. Growth factors for skeletal muscle tissue engineering. Cells Tissues Organs. 202, 169–179 (2016).
Ah, D. & D, T. C, V., A, S. The role of insulin-like Growth factor 1 (IGF-1) in brain development, maturation and neuroplasticity. Neuroscience 325, (2016).
Suo, N., Guo, Y. E., He, B., Gu, H. & Xie, X. Inhibition of MAPK/ERK pathway promotes oligodendrocytes generation and recovery of demyelinating diseases. Glia 67, 1320–1332 (2019).
Shan, X. et al. Cyanidin-related antidepressant-like efficacy requires PI3K/AKT/FoxG1/FGF-2 pathway modulated enhancement of neuronal differentiation and dendritic maturation. Phytomedicine Int. J. Phytother Phytopharm. 76, 153269 (2020).
Smith, A. S. T. et al. Creating interactions between tissue-engineered skeletal muscle and the peripheral nervous system. Cells Tissues Organs. 202, 143–158 (2016).
Qiao, Y., Cong, M., Li, J., Li, H. & Li, Z. The effects of neuregulin-1β on intrafusal muscle fiber formation in neuromuscular coculture of dorsal root ganglion explants and skeletal muscle cells. Skelet. Muscle. 8, 29 (2018).
Liu, X. et al. 3D bioprinted neural tissue constructs for spinal cord injury repair. Biomaterials 272, 120771 (2021).
Xu, B. et al. Transplantation of neural stem progenitor cells from different sources for severe spinal cord injury repair in rat. Bioact Mater. 23, 300–313 (2023).
You, Z. et al. Microvascular endothelial cells derived from spinal cord promote spinal cord injury repair. Bioact Mater. 29, 36–49 (2023).
Al-Khater, K. M. & Todd, A. J. Collateral projections of neurons in laminae I, III, and IV of rat spinal cord to thalamus, periaqueductal gray matter, and lateral parabrachial area. J. Comp. Neurol. 515, 629–646 (2009).
Donkels, C. et al. Oligodendrocyte lineage and myelination are compromised in the gray matter of focal cortical dysplasia type IIa. Epilepsia 61, 171–184 (2020).
Ziganshin, A. U., Khairullin, A. E., Hoyle, C. H. V. & Grishin, S. N. Modulatory roles of ATP and adenosine in cholinergic neuromuscular transmission. Int. J. Mol. Sci. 21, 6423 (2020).
Davis, L. A., Fogarty, M. J., Brown, A. & Sieck, G. C. Structure and function of the mammalian neuromuscular junction. Compr. Physiol. 12, 3731–3766 (2022).
Chambers, S. M. et al. Combined small-molecule inhibition accelerates developmental timing and converts human pluripotent stem cells into nociceptors. Nat. Biotechnol. 30, 715–720 (2012).
Horiguchi, S. et al. Neural precursor cells derived from human embryonic brain retain regional specificity. J. Neurosci. Res. 75, 817–824 (2004).
Watanabe, K. et al. Comparison between fetal spinal-cord- and forebrain-derived neural stem/progenitor cells as a source of transplantation for spinal cord injury. Dev. Neurosci. 26, 275–287 (2004).
Kadoya, K. et al. Spinal cord reconstitution with homologous neural grafts enables robust corticospinal regeneration. Nat. Med. 22, 479–487 (2016).
Lai, B. Q. et al. Transplantation of tissue engineering neural network and formation of neuronal relay into the transected rat spinal cord. Biomaterials 109, 40–54 (2016).
Lai, B. Q. et al. Stem cell-derived neuronal relay strategies and functional electrical stimulation for treatment of spinal cord injury. Biomaterials 279, 121211 (2021).
Yu, H. et al. Activating endogenous neurogenesis for spinal cord injury repair: Recent advances and future prospects. Neurospine 20, 164–180 (2023).
Acknowledgements
We would like to express our gratitude to Biorender (www.biorender.com) for providing the tools to create the graphical abstract in this paper.
Funding
This research was supported by grants from the National Key R&D Program of China, No. 2022YFA1105900 (to B-QL); the Chinese National Natural Science Foundation (Grant Nos. 81891003 to Y-SZ and 82371400 to B-QL); the Natural Science Foundation of Guangdong Province, China, (No. 2021B1515020045 to B-QL and No. 2024A1515013164; 2024A1515012413 to Q-JZ); and the Science and Technology Planning Project of Guangdong Province (No. 2023B1212060018 to B-QL).
Author information
Authors and Affiliations
Contributions
Conceptualization, B.L., Y.Z., Q.Z., and H.Y.; methodology, B.L., Y.Z., Q.Z., and H.Y.; software, H.Y. and S.Y.; validation, H.Y., S.Y.and B.L.; formal analysis, H.Y., S.Y.and J.X; investigation, B.L., H.Y, and S.Y.; resources, B.L., Y.Z., Y.C.,and Q.Z.; data curation, B.L., H.Y., S.Y., J.X., C.W., R.W., Y.G., and Y.Y.; writing—original draft preparation, B.L., H.Y., and S.Y.; writing—review and editing, B.L., H.Y., S.Y., Y.C., C.W., J.X., Y.Y., R.W., Y.G., Z.C., Y.D., X.Z., G.L., Y.M., Q.Z., and Y.Z.; visualization, H.Y., S.Y.and B.L; supervision, Y.C., Y.D., X.Z., G.L. and Y.M.; project administration, B.L., Y.Z., Y.C., and Q.Z.; funding acquisition, B.L., Y.Z., and Q.Z. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Material 5
Supplementary Material 6
Supplementary Material 7
Supplementary Material 8
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yu, H., Yang, S., Chen, Y. et al. Construction of a rodent neural network-skeletal muscle assembloid that simulate the postnatal development of spinal cord motor neuronal network. Sci Rep 15, 3635 (2025). https://doi.org/10.1038/s41598-025-88292-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-88292-x