Abstract
Ovarian cancer (OC) is the most deadly gynecological tumor. OC cells utilize cellular metabolic reprogramming to gain a survival advantage, particularly through aberrant lipid metabolic process. As the primary ingredient in exogenous cannabinoids, cannabidiol (CBD) has been confirmed to exhibit antitumor activity in preclinical studies. However, it is still unclear whether CBD can disrupt fatty acid metabolism and induce apoptosis in OC cells. In this study, we have demonstrated that CBD significantly inhibits the proliferation of OCs through a cannabinoid receptor type 1 (CB1R)-mediated manner. Fatty acid metabolic profiling and flow cytometry analysis revealed that CBD has the ability to decrease fatty acid levels and significantly suppress the transcription of genes involved in fatty acid uptake and synthesis in ES-2 cells. In addition, the analysis from RNA-seq and real-time RT-PCR revealed that CBD activated the endoplasmic reticulum (ER) stress pathway. Conversely, by supplementation with unsaturated fatty acid or blocking CB1R, ER stress or reactive oxygen species (ROS) signals with specific inhibitors could significantly relieve CBD induced, dose-dependent, ER stress associated apoptosis, G0-G1 phase arrest, and mitochondrial dysfunction. Taken collectively, these data indicate that CBD may disrupt lipid metabolism, and lead to ER stress-related apoptosis in OCs. Our findings may provide a theoretical mechanism for anti-ovarian cancer using CBD.
Similar content being viewed by others
Introduction
Ovarian cancer (OC) is the second most common gynecological malignancy worldwide and has the highest mortality rate with frequent relapses. Approximately 140,000 women die from OC each year worldwide1,2. Early-stage diagnostic methods are lacking, and the 5-year survival rate is approximately 50%3. OC is treated primarily by surgery followed by a combination of postoperative chemotherapy, immunotherapy, and radiotherapy4. However, these treatments have limitations, including toxicity, chemoresistance, and limited efficacy. Therefore, novel strategies are needed to reduce the risk of progression of OC and its mortality rate.
Cannabidiol (CBD), a key component of cannabis (Cannabis sativa L), has emerged as a promising candidate for neuroprotective drug development because of its antioxidant and anti-inflammatory properties and favorable pharmacological profile5. CBD has significant anticonvulsant and antiepileptogenic effects in humans and was approved by the US Food and Drug Administration as an orphan drug for the treatment of a rare form of epilepsy in children in 20186. Furthermore, a number of preclinical studies, ranging from in vitro investigations in cancer cell lines to in vivo studies in rodent models of cancer, have demonstrated that CBD has anticancer properties7,8. However, the proposed antitumor effects of CBD have yet to be confirmed by clinical trials, although there have been a few trials on the ability of CBD to reduce anxiety and pain in patients with cancer9. CBD manifests its biological activities via multiple cellular targets. Approximately 65 molecular targets for CBD have been reported, with a number of targets responsible for the various therapeutic effects of CBD10. In terms of antitumor activity, CBD has the ability to reduce expression of the Id-1 gene by regulation of activation of ERK and reactive oxygen species (ROS), consequently inhibiting the proliferation and invasion of breast cancer cells11. CBD inhibits the growth and metastasis of breast cancer cells via the epidermal growth factor receptor pathway12. In colorectal cancer cells, CBD triggers antitumor activity through a mechanism that is dependent on cannabinoid receptor 213. CBD can also induce G0–G1 phase arrest and apoptosis of OC cells by upregulating LAIR-1 and blocking the PI3K/AKT/mTOR signaling pathway14. Recently, new strategies have emerged that target key enzymes involved in lipid uptake or synthesis within cancer cells. These approaches have demonstrated significant antitumor effects and are currently being developed for use in combination with chemotherapy or immunotherapy as potential interventions for cancer15.
It remains unclear whether or not CBD can disturb fatty acid metabolism and induce apoptosis in OC cells. Fatty acids are an alternative energy source for tumor cells and are important in supporting the metabolic needs of cancer cells16. Upregulation of uptake and synthesis of fatty acids facilitates the proliferation and migration of tumor cells. At present, there is limited information regarding the relationship between CBD-regulated lipid metabolism and apoptosis. Previous studies have shown that CBD increases the production of ROS and induces endoplasmic reticulum (ER) stress-associated apoptosis13,17. ER is an important cellular organelle with a significant role in regulation of lipid metabolism and protein synthesis. ER stress can be induced by various physiological and pathological factors, including accumulation of misfolded or unfolded proteins, leading to activation of the unfolded protein response (UPR)18. The UPR is initiated and regulated by three ER sensor proteins, namely, inositol-requiring enzyme 1 (IRE1), protein kinase RNA-like ER kinase (PERK), and activating transcription factor 6 (ATF6)19. Moderate activation of the UPR assists in addressing the accumulation of misfolded proteins and restoring homeostasis in the ER20. However, chronic and unresolved ER stress results in apoptosis of cells and cell cycle arrest through various pathways, including c-Jun N-terminal kinase, C/EBP-homologous protein (CHOP), and caspase-1221. The mitochondria play a pivotal role in energy metabolism and cellular physiological processes. Multiomics studies have revealed that mitochondrial dysfunction, oxidative stress, and apoptosis signaling pathways play critical roles in the survival and development of OC cells22. Any disturbance in the ER can affect the mitochondria and vice versa, triggering a cellular response. It has been reported that mitofusion 2 is an important link between ER stress and mitochondrial metabolism23. Furthermore, when activated by ER stress, CHOP can regulate mitochondrial dysfunction and the intrinsic apoptosis pathway involving the Bcl2 family and caspase activation complex24.
Numerous studies have demonstrated that dysregulation of lipid metabolism plays a significant role in tumor progression and development of chemoresistance25 and that the ER is the primary site of lipid metabolism and includes numerous enzymes involved in this process26. Stearoyl CoA desaturase 1 (SCD1) is the enzyme responsible for the key step in conversion of saturated fatty acids (SFAs) to monounsaturated fatty acids in OC cells27. SCD1 has also been found to be expressed at high levels in OC tissue and cell lines15. Treatment with SCD1 inhibitors effectively inhibited the growth of OC stem cells in a mouse model28. Furthermore, elevated unsaturated lipid levels induced by SCD1 have been shown to protect cancer cells from ER stress and apoptosis29. These findings suggest that inhibition of SCD1 may be an effective therapeutic approach in OC.
To better understand the CBD-induced antitumor effect and the metabolic reprogramming mechanism in OC cells, we investigated the apoptosis signals and fatty acid content in these cells by transcriptomics and fatty acid metabolic profiling after treatment with CBD. Our results indicate that CBD can disrupt lipid metabolism and initiate ER stress in OC cells. Our findings may provide a theoretical mechanism for anti-ovarian cancer using CBD.
Materials and methods
Reagents
CBD was purchased from ZZStandards (Shanghai, China). Roswell park memorial institute (RMMI)-1640 and fetal bovine serum (FBS) were purchased from Thermo Fisher Scientific (Hyclone, Waltham, MA). The small-molecule inhibitors, including AM251, AM631, Capsazepine, GW9662, 4-PBA, and NAC, were purchased from Targetmol (MA, USA). OA and dimethyl sulphoxide (DMSO) were obtained from solarbio (Beijing, China). Cell cycle and apoptosis kit, as well as radio immunoprecipitation assay lysis buffer, were purchased from Beyotime (Jiangsu, China). The PE-conjugated CD36 and FITC annexin V apoptosis detection kit with propidium iodide (PI) were obtained from BioLegend (San Diego, CA, USA). BODIPY-labeled palmitate (BODIPY FLC16; Invitrogen) was utilized in conjunction with flow cytometry for the uptake experiment. The SYBR Premix Ex Taq Kit, Prime Script RT Master Kit, and the RNA-extracting reagent RNAiso Plus were procured from Takara Biotechnology (Dalian, China). Antibodies against CBR1, GRP78, ATF4, XBP1, CHOP, SCD1, Bcl2 and GAPDH were procured from ABclonal (Wuhan, China). Additionally, antibodies targeting mitofusion-2, cleaved Caspase 3, cleaved Caspase 8, cleaved Caspase 9, p65 and goat anti-rabbit IgG (conjugated to horseradish peroxidase) were obtained from Abcam.
Cell line and culture condition
Human ovarian SKOV3, ES-2, and Hey-A8 cells were obtained from the Cell Bank at the China Academy of Science (Shanghai, China) and cultured in RPMI 1640 with 10% FBS. The cells were incubated in a humidified incubator at 37 °C with 5% CO2.
Cell viability assay
The cell viability assay was performed using the cell counting kit-8 (CCK-8 Kit) in accordance with the manufacturer’s instructions (Dojindo Laboratories, Kumamoto, Japan). In brief, cells were seeded in 96-well plates at a density of 6 × 104 cells/well. After pre-treatment with small-molecule inhibitors or OA for 30 min, the cells were incubated with varying concentrations of CBD for 24 h at 37 °C and 5% CO2. DMSO was utilized as the negative control. The absorbance (Abs) was measured at 450 nm using a microplate reader. Tumor growth inhibition (%) was determined as follows: 1 - (Abs value in experimental groups / Abs value in negative control) × 100%.
RNA-sequencing and DEGs analysis
A total of 1 × 106 ovarian cancer cells were seeded in a 3.5 cm petridish in absence or presence of 40 µM CBD. The plates were incubated in a humidified atmosphere with 5% CO2 at 37 °C for 24 h. Subsequently, the cells were collected and washed with cold phosphate buffered saline (PBS). All assays were conducted in triplicate. The samples underwent transcriptome sequencing treatment by Personal Biotechnology Co. (Shanghai, China).
The edge R package (http://www.rproject.org/) was employed to identify DEGs among various treatment groups30. The gene expression levels derived from RNA-Seq data were quantified using Transcripts Per Million reads (TPM), and then converted into Log2FC values, categorizing them into three groups. Genes with expression levels of Log2FC = 2, Log2FC < 2, and Log2FC > 2 were designated as showing no change, down-regulated change, and up-regulated change, respectively. A false discovery rate (FDR) value of ≤ 0.05 and |Log2FC| > 2 was employed to screen for significant DEGs. Gene ontology (GO) enrichment was conducted using DAVID (Database for Annotation, Visualization and Integrated Discovery).
Reverse transcription quantitative PCR (RT-qPCR) analysis
After treatment with CBD and inhibitors as mentioned above, ES-2 cells were collected and total RNA was extracted from 8.4 × 105 cells using the RNA-extracting reagent RNAiso Plus. Subsequently, 0.5 µg of total RNA was reverse transcribed using a PrimeScript RT Master Kit following the manufacturer’s instructions. The resulting cDNA was utilized for RT-qPCR analysis with an SYBR Premix Ex Taq Kit and ABI Prism 7000 (Applied Biosystems, Norwalk, CT). The PCR conditions were described in a previous study31. The sequences of all primers are listed in Table 132,33,34,35. Relative transcription levels were determined employing the 2 − ΔΔCt analysis method36.
Metabolic profiling
The metabolic profiles of all samples were analyzed using the gas chromatograph/mass spectrometric (GC/MS) method, as described in previous study with minor revisions37. Briefly, 1 × 107 cell samples were ground in liquid nitrogen and ultrasonically extracted with 1 mL of chloroform-methanol (2:1, V/V). After centrifugation, the supernatant was mixed with 2 mL of 1% sulfuric acid in methanol and esterified at 80 °C for 30 min. The esterified products were extracted with 1 mL of n-hexane and centrifuged. A 100 µL aliquot of the dehydrated supernatant was diluted to 400 µL with n-hexane. Finally, 15 µL of 500 ppm methyl salicylate was added as an internal standard for GC/MS analysis (Trace 1310-ISQ 7000, USA). GC condition: Thermo TG-FAME (50 m × 0.25 mm ID × 0.20 μm); Injection volume: 1µL; Programmed heating: initial temperature 80 °C for 1 min, then increased to 160 °C at 20 °C/min for 1.5 min, followed by 196 °C at 3 °C/min for 8.5 min, and finally to 250 °C at 20 °C/min for 3 min. Carrier gas: helium at 0.63 mL/min. The MS was run in electron ionization mode (70 eV) with selected ion monitoring. The contents of each fatty acid are shown in supplementary Table S1. The measurements were carried out by Suzhou BioNovoGene Biomedical Tech Co., LTD (Jiangsu, China). Chromatographic and spectral analysis was performed using ChemStation and MassHunter (Agilent Technologies).
Flow cytometry analysis
Cell surface markers were determined by staining with fluorochrome-conjugated monoclonal antibodies (mAbs). The antibody panel included PE-conjugated CD36 for the analysis of lipoprotein receptors. Briefly, cells were incubated with inhibitors and CBD as indicated above. Subsequently, the cells (1 × 106 cells/tube) were resuspended in 50 µl of PBS containing the recommended concentrations of antibodies according to the manufacturer’s instructions, and then incubated for 30 min at 4 °C in the dark. Flow cytometry was conducted using a BD LSRFortessa, and the data was analyzed with FlowJo software.
Evaluation of cell cycle distribution and cell apoptosis by flow cytometry
Following treatment with CBD and the indicated inhibitors, PI staining was utilized for the analysis of DNA content. ES-2 cells were harvested and fixed with 70% ethanol at 4 °C overnight. Subsequently, Cell cycle and apoptosis kit was applied to assess DNA content using flow cytometry (BD Biosciences LSRFortessa, USA). The distribution of cells in the G0, G1, S, G2, and M phases was analyzed using Modfit 5.0 software.
The percentage of cells undergoing apoptosis was determined by double staining with the FITC annexin V apoptosis detection kit and PI. Treated ES-2 cells were collected and resuspended in annexin V binding buffer. A 100 µl cell suspension was transferred into a 1.5 ml tube, to which 5 µl of FITC annexin V and 10 µl of PI solution were added. After incubation for 15 min at 25 °C in the dark, 300 µl of annexin V binding buffer was added to each tube. Flow cytometry was performed using a BD Biosciences LSRFortessa, and data were analyzed with FlowJo software.
JC-1 mitochondrial membrane potential (MMP) assay
The MMP was evaluated using a mitochondria staining kit (Boyotime, Shanghai, China). The cells were seeded on 12-well plates and incubated with 5 mM NAC in 96-well plates at 37 °C and 5% CO2 for 30 min, followed by treatment with CBD at a final concentration of (30 µM, 50 µM) for 24 h. Subsequently, the cells were collected by centrifugation and resuspended in a staining solution containing 200 × JC-1 and 1 × staining buffer, then incubated at 37 °C for 20 min. Finally, the cells were washed once with JC-1 buffer. Data analysis was performed using Flowjo software. The MMP depolarization was visualized using a Leika fluorescence microscope (Leika, Wetzlar, Germany).
Determination of cellular ROS
The intracellular levels of reactive oxygen species (ROS) were analyzed using a 2′,7′-dichlorofluorescin diacetate (H2DCFDA) cellular ROS detection assay kit (KeyGen Biotech Co., Nanjing, China). After treatment with NAC and CBD as indicated above, supernatants were removed from the treated cells and replaced with 5 µM DCFH-DA solution for 30 min at 37 °C in the dark. After incubation, the cells were washed three times with PBS to remove residual particles, dead cells, and excess DCFH-DA probes. The fluorescence intensity of ROS was measured at Ex/Em 488/525 nm by a microplate reader.
Western blotting
After treatment with CBD and inhibitors as described above, a total of 4 × 106 ES-2 cells were collected. The cells were then lysed in RIPA lysis buffer supplemented with phosphatase inhibitor and protease inhibitor at 4 °C for 10 min and 12,000 rpm centrifuge for 15 min. After collecting the supernatants, the BCA assay was used to determine the protein concentrations. Samples containing equal amounts of protein (20 µg) were mixed with 5 × Laemmli buffer, boiled, and separated on 10–15% SDS-PAGE gels. Samples were transferred onto polyvinylidene fluoride membranes (Millipore, Billerica, MA). After blocking with 5% skimmed milk, the primary antibody was incubated overnight at 4 °C in an appropriate dilution. After washing, the blots were further incubated with HRP-conjugated secondary antibody for 1 h. Detection was performed using an enhanced chemiluminescence method. In our experiments, we used the following algorithm to evaluate the relative expression level of the target protein: Control group = [Control (Target protein/ Housekeeping protein) / Control (Target protein/Housekeeping protein)] = 1. Treatment group = Sample (Target protein/ Housekeeping protein) / Control (Target protein/Housekeeping protein).
Statistical analysis
All values are presented as mean ± standard deviation (SD). The data were analyzed using a two-way analysis of variance (ANOVA) method. A post-hoc correction used Benjamini-Hochberg’s false discovery rate, at a q-value of 0.05. The SPSS software 22.0 was used to perform all statistical analyses. Differences with p < 0.05 considered to be statistically significant, *p < 0.05, **p < 0.01, and ***p < 0.001.
Results
CBD inhibited proliferation of OC cells via a CBR1-mediated mechanism
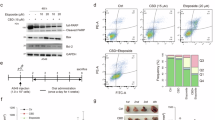
First, we performed a CCK-8 assay to investigate the impact of CBD on the viability of various types of OC cells (i.e., ES-2, SKOV3, and Hey-A8). As shown in Fig. 1A, CBD demonstrated significant antitumor activity in these cells (p < 0.01). Furthermore, at the same concentration, the antitumor efficacy of CBD was notably higher in ES-2 cells than in SKOV3 and Hey-A8 cells (p < 0.001). The biological activity of CBD is known to be regulated by several receptors, including CBR1, CBR2, the transient receptor potential vanilloid subfamily 1 (TRPV1) receptor, and peroxisome proliferator-activated receptor-gamma (PPARγ)38. Next, we examined the relevance of these receptors to CBD-induced cell death by using their selective inhibitors: AM251 (a CB1 receptor antagonist), AM631 (a CB2 receptor antagonist)39, capsazepine (a TRPV1 receptor antagonist)40, and GW9662 (a PPARγ inhibitor)38. We observed that the CBD-induced inhibition of cell viability was reversed by pre-treatment with AM251 but not by AM631. Moreover, treatment with capsazepine or GW9662 alone did not have any effect on the viability of OC cells (Fig. 1A). We also examined the expression of CBR1 in OC cells. The protein level of CBR1 was found to be significantly higher in ES-2 cells than in SKOV3 and Hey-A8 cells (Fig. 1B, C). We then subjected ES-2 cells, which showed high sensitivity to CBD, to treatment with varying concentrations of CBD for 24 and 48 h and found that CBD (20–60 µM) significantly hindered the proliferation of ES-2 cells in a manner that was dependent on both time and dosage (p < 0.05). Furthermore, an IC50 value of 32 µM was observed after 24 h of treatment (Fig. 1D). These findings indicated that CBD may restrict the growth of OC cells in a CBR1-mediated manner.
CBD repressed viability of OC cells by a CBR1-dependent mechanism. (A) OC cells were pre-incubated with the respective receptor antagonist AM251 or AM631 for 30 min at a final concentration of 100 µM and then further co-incubated with CBD (40 µM) for another 24 h. The cell inhibition rate was detected by CCK-8 assay. (B,C) Western blot analysis was conducted to detect CB1R expression in SKOV3, Hey-A8, and ES-2 cells. (D) The ES-2 cells were treated with gradient concentrations of CBD (10 µM-60 µM) for 24 and 48 h, respectively, and the cell viability was assessed by CCK-8 assay. DMSO was used as a negative control. Each value indicates the mean ± SD of results obtained from three independent experiments. *p < 0.05, **p < 0.01, *** p < 0.001.
CBD induced ER stress in OC cells
Differentially expressed genes (DEGs) were identified by RNA sequencing. As shown in Figs. 2A and 2216 genes were found to be significantly upregulated and 1908 to be significantly downregulated in ES-2 cells following treatment with 40 µM CBD. Gene Ontology (GO) analysis revealed that the upregulated DEGs were highly relevant to the ER-associated misfolded protein catabolism and PERK-mediated UPR pathways (Fig. 2B). We also observed that CBD-induced reduction in cell viability was reversed upon pre-treatment with 4-phenylbutyric acid (4-PBA), a specific and irreversible inhibitor of ER stress, in a dose-dependent manner (Fig. 2C). The transcriptome data were further validated using quantitative reverse transcription polymerase chain reaction (RT-qPCR), and the mRNA transcription levels of the genes related to ER stress were determined. The results demonstrated that the mRNA transcription levels of glucose-regulated protein 78 (GRP78, X-box binding protein 1 (XBP1), ATF6, PERK, ATF4, and CHOP were significantly higher in the treated group than in the control group (p < 0.05). The RT-qPCR results for the CBD-induced ER stress-related genes were consistent with the findings of RNA sequencing (Fig. 2D). We investigated the intrinsic mechanism of this effect by western blot analysis and found that CBD could significantly increase the mRNA transcription and protein expression of ER stress-related markers in a dose-dependent fashion (p < 0.05). Upregulation of ER stress-related markers induced by CBD was inhibited effectively by 4-PBA in a dose-dependent manner (Fig. 2E–I). These findings suggested that CBD may suppress the growth of OC cells via the ER stress pathway.
CBD could effectively induce ER stress in OC cells. (A) DEGs identified in ES-2 cells before and after being treated with 40 µM CBD. Volcano plot for the DEGs. Red, blue and gray points represent up-regulated, down-regulated, and non-regulated DEGs, respectively. (B) GO enrichment analysis of the upregulated DEGs before and after being treated with 40 µM CBD. The most significant GO terms were those with corrected p-value of < 0.05. The rich factor represents the number of DEGs that exist in this term accounting for the total number of genes of this term. (C) ES-2 cells were pre-incubated with 4-PBA, AM251 or NAC for 30 min at indicated concentrations. Cells were subsequently co-incubated with CBD at indicated concentrations. After 24 h, the cell inhibition rate was detected by CCK-8 assay. (D) ES-2 cells were pre-incubated with 4-PBA for 30 min at a final concentration of 500 µM and then further co-incubated with CBD at indicated concentrations. After 24 h, total cellular RNA was extracted and reverse transcribed. The mRNA transcription levels of ER stress-related genes GRP78, XBP1, ATF6, PERK, ATF4, and CHOP were detected by RT-qPCR as described in methods. Heatmap represents log2 values of relative mRNA transcription levels (see color scale). The log2 value of each gene in control cells was set to 0. Untreated cells served as a negative control. (E–I) The protein expression levels of the ER stress-related factors were detected by western blot analysis. Each value indicates the mean ± SD of results obtained from three independent experiments. *p < 0.05, **p < 0.01, *** p < 0.001.
CBD induced OC cells apoptosis and cell cycle arrest by regulating the ER stress signaling
The correlation between apoptosis and CBD-induced ER stress in OC cells was investigated further by flow cytometry with Annexin V/propidium iodide staining, which showed that CBD could effectively induce apoptosis in ES-2 cells, again in a dose-dependent manner (p < 0.001) (Fig. 3A, B). Furthermore, the rate of CBD-induced apoptosis was significantly reduced in the groups in which ER stress was inhibited by 4-PBA. The observed increase in apoptosis in response to CBD was consistent with greater inhibition of tumor cell growth.
CBD induced apoptosis and G0-G1 cell cycle arrest by regulating the ER stress signaling. (A,B) Flow cytometric analysis of Annexin V-FITC/PI-stained cells. ES-2 cells were pre-incubated with 4-PBA, AM251 or NAC for 30 min at indicated concentrations. Cells were subsequently co-incubated with CBD at indicated concentrations. After 24 h, cells had been harvested and stained for analysis. Dot plots of total events are shown with frequencies of cells in each quadrant. (C,D) Cell cycle distribution was determined by flow cytometry. ES-2 cells were pre-incubated with 4-PBA or AM251 for 30 min at indicated concentrations. Cells were subsequently co-incubated with CBD at indicated concentrations. After 24 h, cells were harvested and fixed with 70% ethanol at 4 °C overnight, then, stained with PI. Each value indicates the mean ± SD of results obtained from three independent experiments. *p < 0.05, **p < 0.01, *** p < 0.001.
The impact of CBD on the distribution of OC cells in various stages of the cell cycle was investigated further by flow cytometry. Exposure to CBD led to a dose-dependent increase in the percentage of cells in G0–G1 phase (Fig. 3C, D). Furthermore, addition of 4-PBA significantly increased the number of OC cells rescued from CBD-induced G0–G1 phase cycle arrest (p < 0.01). These results confirmed that CBD is able to induce apoptosis in OC cells and arrest the G0–G1 phase of the cell cycle by activation of ER stress signaling.
CBD caused disordered lipid metabolism and induced an ER stress response
Fatty acids serve as an important carbon source in the tumor microenvironment, and fatty acid-mediated lipid metabolism plays an important role in the survival of OC cells41. Elevated UFA levels have been shown to protect cancer cells against apoptosis induced by ER stress29. To investigate the impact of fatty acid metabolism on CBD-induced apoptosis in OC cells, we initially performed a GC/ MS spectrometry analysis to assess the levels of lipid metabolites in CBD-treated OC cells. As shown in Fig. 4A, levels of SFAs, including myristic acid (14 C:0), palmitic acid (16 C:0), and stearic acid (18 C:0), were significantly higher in the group treated with 40 µM CBD than in the control group (p < 0.001). At the same time, the levels of UFAs, particularly palmitoleic acid (C16:1), heptadecenoic acid (C17:1), and oleic acid (C18:1), were found to be significantly reduced in ES-2 cells following treatment with CBD (p < 0.001). Furthermore, GO analysis found a strong association of the DEGs that were downregulated with the metabolic and fatty acid biosynthetic processes in UFAs (Fig. 4B). The qPCR results demonstrated that the CBD-treated group was able to reduce the transcription levels of genes associated with fatty acid uptake, including fatty acid binding protein 5 (Fabp5), the low-density lipoprotein receptor (LDLR), and PPARγ. The proportion of CD36-positive events was reduced following treatment with CBD (Fig. 4C, D). Furthermore, the transcription levels of genes related to fatty acid biosynthesis, including acetyl-CoA carboxylase alpha (ACACA), fatty acid synthase (FASN), SCD1, and sterol regulatory element binding protein 1 (SREBP1), were decreased after treatment with CBD (Fig. 4D). We also investigated whether CBD could inhibit the uptake of fatty acids from the extracellular environment using fluorescently labeled palmitate (BODIPY FLC16)42. The findings indicated significant inhibition of uptake of palmitate in the CBD-treated group (Fig. 4E).
CBD disrupted lipid metabolism and leads to ER stress in OC cells (A) Lipid metabolism was identified in ES-2 cells before and after being treated with 40 µM CBD. Log2 values for each metabolite represent the average of triplicates. (B) GO enrichment analysis of the downregulated DEGs. The most significant GO terms were those with corrected P-value of < 0.05. The rich factor represents the number of DEGs that exist in this term accounting for the total number of genes of this term. (C) ES-2 cells were stained with PE-conjugated CD36 Abs before analysis by flow cytometry. Dot plots of total events are shown with frequencies of cells in each quadrant. (D) Total cellular RNA was extracted and reverse transcribed. The mRNA transcription levels of Lipid metabolism-related genes SCD1, FABP5, LDLR, FASN, ACACA, SREBP, and PPARγ were detected by RT-qPCR. (E) Representative plots of BODIPY FLC16 in ES-2 cells were collected and analyzed by flow cytometry after being treated with 40 µM CBD for 24 h. Untreated cells served as a negative control. (F) ES-2 cells were co-incubated with CBD at indicated concentrations for 24 h, in presence or absence of 25 µM OA. The cell inhibition rate was detected by CCK-8 assay. (G) The mRNA transcription levels of ER stress-related genes were detected by RT-qPCR. Heatmap represents log2 values of relative mRNA transcription levels (see color scale). The log2 value of each gene in control cells was set to 0. (H-N) The protein levels of GRP78, ATF4, XBP1, CHOP, SCD1, and p65 were detected by western blot analysis. Each value indicates the mean ± SD of results obtained from three independent experiments. *p < 0.05, **p < 0.01, *** p < 0.001.
Previous research has indicated that elevated levels of SFAs, as a result of inhibiting SCD1, can induce ER stress and apoptosis in several types of cancer. Furthermore, ER stress can be alleviated effectively by supplementation with UFAs29. In this study, we observed that addition of exogenous oleic acid significantly mitigated the antitumor activity of OC cells induced by administration of varying concentrations of CBD (Fig. 4F). CBD significantly inhibited the gene transcription of SCD1 and its protein expression in a dose-dependent manner. Treatment with CBD also resulted in significant dose-dependent relief of both mRNA transcription and protein expression of ER stress-related markers by oleic acid (p < 0.05, Fig. 4G–N). Overall, these data suggested that CBD interfered with lipid metabolism leading to ER stress in OC cells.
Oleic acid rescued CBD-directed ER stress-mediated apoptosis and cell cycle arrest
Levels of apoptosis in ES-2 cells were assessed by Annexin V/propidium iodide staining to determine whether UFAs protect against CBD-induced ER stress-related apoptosis. In serum-free medium, both early and late apoptosis rates were significantly higher in the CBD-treated group than in the control group (Fig. 5A, B). CBD-induced apoptosis was significantly inhibited by administration of oleic acid (p < 0.01). Furthermore, exogenous oleic acid can significantly restore the cell cycle profile of cells treated with different concentrations of CBD. (p < 0.01, Fig. 5C, D). These findings confirmed that an imbalance between UFAs and SFAs is the primary cause of CBD-induced ER stress-related apoptosis in OC cells.
OA reversed CBD induced apoptosis and G0-G1 cell cycle arrest. (A,B) Flow cytometric analysis of Annexin V-FITC/PI-stained cells. ES-2 cells were co-incubated with CBD at indicated concentrations for 24 h, in presence or absence of 25 µM OA. Dot plots of total events are shown with frequencies of cells in each quadrant. (C,D) Cell cycle distribution was determined by flow cytometry. Each value indicates the mean ± SD of results obtained from three independent experiments. *p < 0.05, **p < 0.01, *** p < 0.001.
CBD induced disordered lipid metabolism and ER stress via a CBR1-mediated mechanism
Previous research has demonstrated that several pathways involved in the growth, differentiation, and metabolism of tumor cells interact with CBR signaling43,44. To examine whether CBD-induced impairment of fatty acid metabolism and ER stress are CBR1-mediated, we used AM251, the antagonist of CBR1, to suppress CBR1 activity in ES-2 cells. CBD-induced inhibition of cell viability was reversed by pre-treatment with AM251 in a dose-dependent fashion (Fig. 2C). Furthermore, AM251 rescued a significant amount of CBD-treated cells from apoptosis and G0–G1 phase arrest (p < 0.01, Fig. 3A-D). In qPCR analyses, treatment with CBD resulted in significant dose-dependent downregulation of the transcription levels of genes associated with fatty acid metabolism (p < 0.05), which was reversed when CBR1 signaling was blocked by AM251 (Fig. 6A).
CBR1 regulated lipid metabolism and ER stress in OC cells. (A,B) ES-2 cells were pre-incubated with AM251 for 30 min at a final concentration of 100 µM and then further co-incubated with CBD at indicated concentrations. After 24 h, the mRNA transcription levels of lipid metabolism- and ER stress-related genes were detected by RT-qPCR. Heatmap represents log2 values of relative mRNA transcription levels (see color scale). The log2 value of each gene in control cells was set to 0. Untreated cells served as a negative control. (C–I) The protein levels of GRP78, ATF4, XBP1, CHOP, SCD1, and p65 were detected by western blot analysis. Each value indicates the mean ± SD of results obtained from three independent experiments. *p < 0.05, **p < 0.01, *** p < 0.001.
The balance of SFAs and UFAs is regulated by SCD1 to adjust the functions of ER stress29. Next, we investigated the levels of gene transcription and protein expression associated with the ER response after blocking CBR1. Upregulation of mRNA transcription and protein expression of ER stress-related markers was significantly alleviated by AM251 (p < 0.05, Fig. 6B–I). These findings suggested that CBD has the ability to regulate disruption of lipid metabolism in OC cells and reduce UFA synthesis through the CBR1-SCD1 signaling pathway, resulting in ER stress-induced apoptosis.
CBD induced mitochondrial dysfunction and cascade-mediated apoptotic pathways
Changes in ER stress markers are a key indicator of mitochondrial stress45. We hypothesized that CBD induces cytotoxicity by targeting the mitochondria. Our CCK-8 and Annexin V staining results indicated that the inhibitory effects of CBD on cell viability and apoptosis were significantly attenuated by pre-treatment with N-acetyl-L-cysteine (NAC), a dose-dependent scavenger of ROS (p < 0.01, Figs. 2C and 3A and B).
The mitochondrial membrane potential (MMP) and the ROS level are widely used as markers of mitochondrial function46. To investigate the effect of CBD on mitochondrial function further, we first detected MMP using the fluorescent probe JC-1. Compared with the control group, the CBD-treated group showed higher levels of JC-1 monomer green fluorescence and less aggregation of JC-1 (indicated by red fluorescence), resulting in a higher JC-1 monomer ratio. Flow cytometry showed that treatment of ES-2 cells with CBD 30 µM and 50 µM led to significant increases in the loss of MMP, which were, respectively, 8.4-fold and 14.5-fold greater than in dimethyl sulfoxide-treated control cells; intracellular levels of ROS in CBD-treated ES-2 cells also increased by up to 1.3-fold and 1.8-fold, respectively, in a dose-dependent manner. The decrease in MMP and generation of ROS could be significantly reversed by treatment with NAC (p < 0.01, Fig. 7A–C).
CBD triggers mitochondrial dysfunction and apoptosis in ES-2 cells. (A,B) Changes in MMP in ES-2 cells were determined based on JC-1 fluorescence. ES-2 cells were pre-incubated with NAC for 30 min at a final concentration of 5 mM and then further co-incubated with CBD at indicated concentrations for 24 h. The effect of CBD on the disruption of MMP was detected by JC-1 assay. Relative JC-1 red/green population is represented as a bar graph in percentage-ratio under the flow cytometry data. (C) The fluorescence intensity of ROS was measured by microplate reader. (D–I) Protein levels of mitofusion, cleaved caspase 3, 8, 9 and Bcl2 were detected by western blot analysis. Each value indicates the mean ± SD of results obtained from three independent experiments. *p < 0.05, **p < 0.01, *** p < 0.001.
In addition, western blot revealed significant downregulation of mitofusion 2 and antiapoptotic Bcl2 in the CBD-treated groups, while the expression of cleaved caspase-8, caspase-9, and caspase-3 was significantly upregulated (Fig. 7D-I). Overall, our data indicated that CBD trigger mitochondrial dysfunction and apoptosis in OC cells.
Discussion
CBD is a non-addictive compound found in the cannabis plant. Most of the previous research on CBD has focused primarily on its protective effect in the nervous system because of its ability to cross the blood–brain barrier47. However, there is now evidence demonstrating that CBD can inhibit the proliferation and metastasis of tumor cells and induce apoptosis and autophagy in various malignancies, including breast, colorectal, and gastric cancer7,13. This research has also demonstrated the role of ER and oxidative stress in induction of cell death following treatment with CBD.
Cannabinoids are a class of compounds that have been attracting considerable research interest, mainly because of their interaction with the endocannabinoid system through CB1 and CB2 receptors48. Hyperactivation of CB1 has been observed in multiple types of cancer, and extensive research has demonstrated its association with the progression of cancer and its clinical outcomes49. Importantly, high CBR1 expression have been observed in OC tissue, and the expression level of CB1R is positively correlated with the malignant potential of OC cells50. Therefore, as a specific CBR1 agonist with efficacy against OC, CBD could be used effectively in the treatment of OC. In this study, we observed high CBR1 expression in all three types of human OC cells investigated. However, no receptor-mediated mechanism contributing to the promising antitumor activity of CBD in OC cells has been identified.
Our study found a significant difference in the CBD-induced antitumor effect between the ES-2, SKOV3, and Hey-A8 cell lines. The inhibitory effects of CBD were significantly greater in ES-2 cells (45.1%) than in SKOV3 cells (25.46%) or Hey-A8 cells (21.29%). Furthermore, the western blot results indicated that the level of CBR1 protein was significantly higher in ES-2 cells than in SKOV3 and Hey-A8 cells. After inhibition of CBR1 by AM251, CBD-induced repression of cell viability was significantly reversed. These results suggest that CBD inhibits the growth of OC cells in a CBR1-mediated manner. To further elucidate the mechanism of the antitumor effect of CBD in OC cells, we initially examined the DEGs in ES-2 cells by transcriptome analysis. The results of the GO analysis suggest that the CBD-induced antitumor effect was highly relevant to the ER-associated misfolded protein catabolic process. Transcription and expression levels of genes associated with ER stress were significantly upregulated after treatment with CBD. Blocking the ER stress response with 4-PBA showed that CBD-induced apoptosis of OC cells and G0–G1 phase cycle arrest relied on activation of the XBP1 and ATF4/CHOP signaling pathways.
Fatty acids are an important energy source and serve as a key component of phospholipids in cell membranes16. Accumulation of UFAs has been shown to support growth of OC cells and to increase migration and invasion of cancer cells51. Lipid metabolism plays a complex role in multiple cell death pathways, including necroptosis and ferroptosis52. Zhao et al. recently demonstrated that controlling the balance between SFAs and UFAs can initiate ER stress in tumors29. These findings suggest that regulating lipid metabolism may be a potential therapeutic strategy in OC. Gas chromatography/mass spectrometry and RNA sequencing revealed significant downregulation of the UFA metabolic process and a decrease in transcription levels of genes associated with fatty acid uptake and synthesis in OC cells treated with CBD (Fig. 3). CD36 regulates homeostasis of cellular energy and intracellular cholesterol in OC cells53,54, and its expression was significantly decreased after treatment with CBD. We also found that CBD could convert SFAs to UFAs and palmitic and stearic acid to palmitoleic acid and oleic acid, respectively. Regulation of the conversion from SFAs to UFAs is mediated by SCD155. Meanwhile, SCD1 and its predominant product, oleic acid, have been shown to have beneficial effects in terms of restoring ER homeostasis, promoting cell cycle progression, and enhancing proliferation56, which is consistent with our finding that exogenous oleic acid significantly relieved CBD-induced antitumor activity, apoptosis, and G0–G1 phase arrest. The results of qPCR and western blot analysis indicate that CBD-induced mRNA transcription and protein expression of ER stress-related markers could be significantly mitigated in a dose-dependent manner by exogenous oleic acid, strongly suggesting that CBD may initiate ER stress-associated apoptosis by disrupting lipid metabolism.
Numerous studies published in the past decade have demonstrated that CB1 and CB2 receptor agonists can function as direct antitumor agents by activating the ERK, p38 MAPK, and JNK1/2 pathways57,58. However, a clear distinction was observed between the activity produced and the specific cancer cell line studied. Other researchers have suggested that CBD hinders the viability of cancer cells via mechanisms that bypass activation of cannabinoid receptors. Ramer et al. reported that CBD induced PPARγ-dependent toxicity in lung cancer cells59. Another study found that CBD mediated autophagy and apoptosis in endothelial cells via ROS-mediated heme oxygenase-1 but not via CBD-activated receptors (CBR1 and CBR2)31. A recent investigation found that TRPV2 is a target of cannabinoids and is involved in CBD-induced autophagic death of glioma stem-like cells60. In breast cancer cell lines, CBD induced ER stress through the TRPV1 receptor-dependent signaling pathway by increasing Ca2+ influx17. Interestingly, Brighenti et al. found that low CBD concentrations (0.01 µM to 9 µM) did not impair the viability of tumor cells but enhanced the migratory capacity of U87 glioblastoma cells. In this range, the effects of CBD are not associated with the CB1/CB2 or TRPV1 receptors; however, at higher concentrations, the effects of CBD depend on these receptors61,62. All these findings demonstrate that CBD can modulate certain pathways involved in development of cancer and exert antitumor effects. In our study, CBD had very effective antitumor activity in OC cells and significantly downregulated the transcription of genes related to fatty acid metabolism via CBR1 signaling. Notably, we also observed that CBD significantly decreased the expression of p65 protein, which is necessary for regulation of SCD1 activity28. Downregulation of CBD-induced p65 and SCD1 was also relieved by blocking CBR1. Similarly, upregulation of mRNA transcription and protein expression of ER stress-related markers was significantly alleviated by AM251. A previous study reported that increased expression of SCD1 protected OC cells from apoptosis induced by ER stress29. Overall, these results suggest that CBD may modulate ER stress-triggered apoptosis in OC cells by regulating lipid metabolism via the CBR1/SCD1 signaling pathway.
The mitochondria are essential for generating energy and play an important role in maintaining cell survival and avoiding metastasis63. Mitochondrial dysfunction, which results in depolarization of the MMP and a decrease in the ATP level, suppresses progression of OC64. Previous studies have shown that crosstalk between the mitochondria and ER is important in the regulation of cell metabolism and cell death65. CHOP-mediated apoptosis involves regulation of Bcl2 family proteins, leading to permeabilization of the mitochondrial outer membrane and caspase-dependent cell death66,67. In our study, flow cytometry demonstrated that CBD could dose-dependently increase loss of the MMP. This decrease in MMP would result in elevation of the intracellular ROS level. Figure 7C shows that the ROS level was higher in the CBD-treated group than in the control group. Full depolarization of the MMP triggers caspase-dependent apoptosis68, which occurs in the mitochondria and plays an important role in programmed cell death. The conclusions were consistent with the findings of our western blot results (Fig. 7D). We found that the expression levels of Bcl2 and mitofusion 2 were significantly downregulated and the pro-apoptotic proteins caspase-8, caspase-9, and caspase–3 were significantly upregulated after treatment with CBD. Meanwhile, CBD-induced mitochondrial dysfunction and expression of apoptotic proteins could be significantly reversed by NAC. These results indicate that CBD may trigger apoptosis of OC cells through an ER-mitochondrial pathway.
Conclusions
Preclinical studies have demonstrated that CBD, either as a monotherapy or in conjunction with other treatments, holds potential as a novel anti-tumor, anti-inflammatory, and analgesic agent. Our results demonstrated that CBD promoted OC cells apoptosis and G0-G1 phase arrest by disrupting the CBR1-mediated lipid metabolism and ER stress- and mitochondrial dysfunction-associated apoptosis signaling pathways (Fig. 8). Therefore, CBD may serve as a potential candidate for adjuvant therapy in the treatment of ovarian cancer. However, larger-scale clinical studies involving more patient samples and detailed dose-response relationship analyses are still needed to confirm the efficacy of CBD in cancer patients.
Data availability
The transcriptome sequences of ES-2 (control) and treatment with CBD groups were deposited in NCBI under the accession number PRJNA980416.
References
Bae, H., Song, G. & Lim, W. Stigmasterol causes ovarian cancer cell apoptosis by inducing endoplasmic reticulum and mitochondrial dysfunction. Pharmaceutics 12, 488 (2020).
Penny, S. M. Ovarian cancer: an overview. Radiol. Technol. 91, 561–575 (2020).
Stewart, C., Ralyea, C. & Lockwood, S. Ovarian cancer: an integrated review. Semin Oncol. Nurs. 35, 151–156 (2019).
Jin, Z., Yu, C. H. & Cheng, P. Anticancer effect of tanshinones on female breast cancer and gynecological cancer. Front. Pharmacol. 12, 824531 (2022).
Chadwick, V. L., Rohleder, C., Koethe, D. & Leweke, F. M. Cannabinoids and the endocannabinoid system in anxiety, depression, and dysregulation of emotion in humans. Curr. Opin. Psychiatry. 33, 20–42 (2020).
Cheung, K. A. K., Mitchell, M. D. & Heussler, H. S. Cannabidiol and neurodevelopmental disorders in children. Front. Psychiatry. 12, 643442 (2021).
Kis, B. et al. Cannabidiol-from plant to human body: a promising bioactive molecule with multi-target effects in cancer. Int. J. Mol. Sci. 20, 5905 (2019).
Fraguas-Sánchez, A. I. et al. Enhancing ovarian cancer conventional chemotherapy through the combination with cannabidiol loaded microparticles. Eur. J. Pharm. Biopharm. 154, 246–258 (2020).
Good, P. et al. Oral medicinal cannabinoids to relieve symptom burden in the palliative care of patients with advanced cancer: a double-blind, placebo controlled, randomised clinical trial of efficacy and safety of cannabidiol (CBD). BMC Palliat. Care. 18, 110 (2019).
Bih, C. I. et al. Molecular targets of cannabidiol in neurological disorders. Neurotherapeutics 12, 699–730 (2015).
McAllister, S. D. et al. Pathways mediating the effects of cannabidiol on the reduction of breast cancer cell proliferation, invasion, and metastasis. Breast Cancer Res. Treat. 129, 37–47 (2011).
Kisková, T., Mungenast, F., Suváková, M., Jäger, W. & Thalhammer, T. Future aspects for cannabinoids in breast cancer therapy. Int. J. Mol. Sci. 20, 1673 (2019).
Lee, H. S. et al. Cannabidiol exerts anti-proliferative activity via a cannabinoid receptor 2-mediated mechanism in human colorectal cancer cells. Int. Immunopharmacol. 108, 108865 (2022).
Ma, L. et al. A novel mechanism of cannabidiol in suppressing ovarian cancer through LAIR-1 mediated mitochondrial dysfunction and apoptosis. Environ. Toxicol. 38, 1118–1132 (2023).
Zhao, G. Y., Cardenas, H. & Matei, D. Ovarian cancer-why lipids matter. Cancers (Basel). 11, 1870 (2019).
Snaebjornsson, M. T., Janaki-Raman, S. & Schulze, A. Greasing the Wheels of the Cancer machine: the role of lipid metabolism in Cancer. Cell. Metab. 31, 62–76 (2020).
Harpe, A. D. L., Beukes, N. & Frost, C. L. CBD activation of TRPV1 induces oxidative signaling and subsequent ER stress in breast cancer cell lines. Biotechnol. Appl. Biochem. 69, 420–430 (2022).
Hetz, C. & Papa, F. R. The unfolded protein response and cell fate control. Mol. Cell. 69, 169–181 (2018).
Hetz, C., Zhang, K. Z. & Kaufman, R. J. Mechanisms, regulation and functions of the unfolded protein response. Nat. Rev. Mol. Cell. Biol. 21, 421–438 (2020).
Walter, P. & Ron, D. The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011).
Liu, D., Zhang, M. & Yin, H. C. Signaling pathways involved in endoplasmic reticulum stress-induced neuronal apoptosis. Int. J. Neurosci. 123, 155–162 (2013).
Shen, L. & Zhan, X. Q. Mitochondrial dysfunction pathway alterations offer potential biomarkers and therapeutic targets for ovarian cancer. Oxid. Med. Cell Longev. 2022, 5634724 (2022).
Doghman-Bouguerra, M. & Lalli, E. The ER-mitochondria couple: in life and death from steroidogenesis to tumorigenesis. Mol. Cell. Endocrinol. 441, 176–184 (2017).
Chen, X. Y., Shi, C. R., He, M. H., Xiong, S. Q. & Xia, X. B. Endoplasmic reticulum stress: molecular mechanism and therapeutic targets. Signal Transduct. Target Ther 8, 352 (2023).
Martinez-Outschoorn, U. E., Peiris-Pagés, M., Pestell, R. G., Sotgia, F. & Lisanti, M. P. Cancer metabolism: a therapeutic perspective. Nat. Rev. Clin. Oncol. 14, 11–31 (2017).
Moncan, M. et al. Regulation of lipid metabolism by the unfolded protein response. J. Cell. Mol. Med. 25, 1359–1370 (2021).
Tesfay, L. et al. Stearoyl-CoA desaturase 1 protects ovarian cancer cells from ferroptotic cell death. Cancer Res. 79, 5355–5366 (2019).
Li, J. J. et al. Lipid desaturation is a metabolic marker and therapeutic target of ovarian cancer stem cells. Cell. Stem Cell. 20, 303–314 (2017).
Zhao, G. Y. et al. Ovarian cancer cell fate regulation by the dynamics between saturated and unsaturated fatty acids. Proc. Natl. Acad. Sci. U.S.A 119, e2203480119 (2022).
Wang, L. K., Feng, Z. X., Wang, X. W. & Zhang, X. G. EGseq An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 26, 136–138 (2010).
Fu, X. H. et al. Staphylococcal enterotoxin C2 mutant drives T lymphocyte activation through PI3K/mTOR and NF-ĸB signaling pathways. Toxicol. Appl. Pharmacol. 333, 51–59 (2017).
Goto, K. et al. Peroxisome proliferator-activated receptor-γ in capillary endothelia promotes fatty acid uptake by heart during long-term fasting. J. Am. Heart Assoc. 2, e004861 (2013).
Wu, S. & Näär, A. M. SREBP1-dependent de novo fatty acid synthesis gene expression is elevated in malignant melanoma and represents a cellular survival trait. Sci. Rep. 9, 10369 (2019).
Liao, Z. W. et al. Exosomes from mesenchymal stem cells modulate endoplasmic reticulum stress to protect against nucleus pulposus cell death and ameliorate intervertebral disc degeneration in vivo. Theranostics 9, 4084–4100 (2019).
Wu, L. L. et al. Thapsigargin induces apoptosis in adrenocortical carcinoma by activating endoplasmic reticulum stress and the JNK signaling pathway: an in vitro and in vivo study. Drug Des. Devel Ther. 13, 2787–2798 (2019).
Fu, X. H., Xu, M. K., Zhang, H. W., Zhang, J. H. & Zhang, C. G. Enhanced interaction between Sect. 2 mutant and TCR Vβ induces MHC II-independent activation of T cells via PKCθ/NF-κB and IL-2R/STAT5 signaling pathways. J. Biol. Chem. 21, 19771–19784 (2018).
Li, H. et al. The 24-kDa subunit of mitochondrial complex I regulates growth, microsclerotia development, stress tolerance, and virulence in Verticillium dahliae. BMC Biol. 22, 289 (2024).
Shahbazi, F., Grandi, V., Banerjee, A. & Trant, J. F. Cannabinoids and cannabinoid receptors: the story so far. Iscience 23, 101301 (2020).
Böckmann, S. & Hinz, B. Cannabidiol promotes endothelial cell survival by heme oxygenase-1-mediated autophagy. Cells 9, 1703 (2020).
Terada, Y. et al. Activation and inhibition of thermosensitive TRP channels by voacangine, an alkaloid present in Voacanga Africana, an African tree. J. Nat. Prod. 77, 285–297 (2014).
Yoon, H. & Lee, S. Fatty acid metabolism in ovarian cancer: therapeutic implications. Int. J. Mol. Sci. 23, 2170 (2022).
Huang, S. C. C. et al. Cell-intrinsic lysosomal lipolysis is essential for alternative activation of macrophages. Nat. Immunol. 15, 846–855 (2014).
Wang, D. P., Kang, K., Hai, J., Lv, Q. L. & Wu, Z. B. Alleviating CB2-dependent ER stress and mitochondrial dysfunction improves chronic cerebral hypoperfusion-induced cognitive impairment. J. Neuroimmune Pharmacol. 19, 1 (2024).
Das, S., Kaul, K., Mishra, S., Charan, M. & Ganju, R. K. Cannabinoid signaling in cancer. Adv. Exp. Med. Biol. 1162, 51–61 (2019).
Li, Y. et al. GRP75 modulates endoplasmic reticulum-mitochondria coupling and accelerates Ca2+-dependent endothelial cell apoptosis in diabetic retinopathy. Biomolecules 12, 1778 (2022).
Sharma, C., Kim, S., Nam, Y., Jung, U. J. & Kim, S. R. Mitochondrial dysfunction as a driver of cognitive impairment in Alzheimer’s disease. Int. J. Mol. Sci. 22, 4850 (2021).
Bhunia, S., Kolishetti, N., Arias, A. Y., Vashist, A. & Nair, M. Cannabidiol for neurodegenerative disorders: A comprehensive review. Front Pharmacol 13, 989717 (2022).
Castillo-Arellano, J., Canseco-Alba, A., Cutler, S. J. & León, F. The polypharmacological effects of cannabidiol. Molecules 28, 3271 (2023).
Stasiulewicz, A., Znajdek, K., Grudzień, M., Pawiński, T. & Sulkowska A.J.I. A guide to targeting the endocannabinoid system in drug design. Int. J. Mol. Sci. 21, 2778 (2020).
Messalli, E. M., Grauso, F., Luise, R., Angelini, A. & Rossiello, R. Cannabinoid receptor type 1 immunoreactivity and disease severity in human epithelial ovarian tumors. Am. J. Obstet. Gynecol. 211, 234e1–234e6 (2014).
Chaudhry, S. & Thomas, S. N. Targeting lipid metabolism in the treatment of ovarian cancer. Oncotarget 13, 768–783 (2022).
Magtanong, L., Ko, P. J. & Dixon, S. J. Emerging roles for lipids in non-apoptotic cell death. Cell. Death Differ. 23, 1099–1109 (2016).
Su, X. & Abumrad, N. A. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol. Metab. 20, 72–77 (2009).
Ladanyi, A. et al. Adipocyte-induced CD36 expression drives ovarian cancer progression and metastasis. Oncogene 37, 2285–2301 (2018).
Iwai, T. et al. Stearoyl-coa desaturase-1 protects cells against lipotoxicity-mediated apoptosis in proximal tubular cells. Int. J. Mol. Sci. 17, 1868 (2016).
Zhao, X. Y., Wang, M., Liu, J. J. & Su, X. Stearoyl coA desaturase 1 and inositol-requiring protein 1 α determine the efficiency of oleic acid in alleviating silica nanoparticle-induced insulin resistance. J. Biomed. Nanotechnol. 17, 1349–1363 (2021).
Galve-Roperh, I. et al. Anti-tumoral action of cannabinoids: involvement of sustained ceramide accumulation and extracellular signal-regulated kinase activation. Nat. Med. 6, 313–319 (2000).
Sarfaraz, S., Afaq, F., Adhami, V. M., Malik, A. & Mukhtar, H. Cannabinoid receptor agonist-induced apoptosis of human prostate cancer cells LNCaP proceeds through sustained activation of ERK1/2 leading to G1 cell cycle arrest. J. Biol. Chem. 281, 39480–39491 (2006).
Ramer, R. et al. COX-2 and PPAR-γ confer cannabidiol-induced apoptosis of human lung cancer cells. Mol. Cancer Ther. 12, 69–82 (2013).
Nabissi, M. et al. Cannabidiol stimulates Aml-1a-dependent glial differentiation and inhibits glioma stem-like cells proliferation by inducing autophagy in a TRPV2-dependent manner. Int. J. Cancer. 137, 1855–1869 (2015).
Bisogno, T. et al. Molecular targets for cannabidiol and its synthetic analogues: effect on vanilloid VR1 receptors and on the cellular uptake and enzymatic hydrolysis of anandamide. Br. J. Pharmacol. 134, 845–852 (2001).
Vaccani, A., Massi, P., Colombo, A., Rubino, T. & Parolaro, D. Cannabidiol inhibits human glioma cell migration through a cannabinoid receptor-independent mechanism. Br. J. Pharmacol. 144, 1032–1036 (2005).
Zong, W. X., Rabinowitz, J. D. & White, E. Mitochondria and cancer. Mol. Cell. 61, 667–676 (2016).
Ashraf, R. & Kumar, S. Mfn2-mediated mitochondrial fusion promotes autophagy and suppresses ovarian cancer progression by reducing ROS through AMPK/mTOR/ERK signaling. Cell Mol. Life Sci. 79, 573. (2022).
Gutiérrez, T. & Simmen, T. Endoplasmic reticulum chaperones tweak the mitochondrial calcium rheostat to control metabolism and cell death. Cell. Calcium. 70, 64–75 (2018).
Ghribi, O., DeWitt, D. A., Forbes, M. S., Herman, M. M. & Savory, J. Co-involvement of mitochondria and endoplasmic reticulum in regulation of apoptosis: changes in cytochrome c, Bcl-2 and Bax in the hippocampus of aluminum-treated rabbits. Brain Res. 903, 66–73 (2011).
Pan, M. Y. et al. Prodigiosin activates endoplasmic reticulum stress cell death pathway in human breast carcinoma cell lines. Toxicol. Appl. Pharmacol. 265, 325–334 (2012).
Gharbaran, R., Shi, C., Onwumere, O. & Redenti, S. Plumbagin induces cytotoxicity via loss of mitochondrial membrane potential and caspase activation in metastatic retinoblastoma. Anticancer Res. 41, 4725–4732 (2021).
Acknowledgements
This research was suppoted by grant from the Liaoning Province Science and Technology plan joint plan (Natural Science Foundation-General Program) (2024-MSLH-457), Natural Science Foundation of Liaoning Province of China (2022-MS-409), Science and Technology Innovation Project of Shenyang (RC210215). We thank Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization, X.F., and G.L.; Formal analysis, X.F., Z.Y., Y.B., W.Z., B.Y., Y.S., Z.J., and X.T.; Funding acquisition, G.L.; Investigation, X.F., Z.Y., X.T., Z.J., and B.Y.; Methodology, X.F., G.L. and Z.Y.; Project administration, G.L.; Software, X.F., F.F., Z.Y., B.Y. Y.S. and X.T.; Supervision, G.L.; Validation, G.L.; Writing – original draft, X.F.; Writing – review & editing, G.L.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Fu, X., Yu, Z., Fang, F. et al. Cannabidiol attenuates lipid metabolism and induces CB1 receptor-mediated ER stress associated apoptosis in ovarian cancer cells. Sci Rep 15, 4307 (2025). https://doi.org/10.1038/s41598-025-88917-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-88917-1