Abstract
Liquid–liquid phase separation (LLPS) associates with immune infiltration in multiple diseases. Nonetheless, the role of LLPS-related genes (LLPS-RGs) in immune infiltration of ulcerative colitis (UC) is still elusive. We identified the hub LLPS-RGs (DE-LLPS-RGs) (HSPB3, SLC16A1, TRIM22, SRI, PLEKHG6, GBP1, PADI2) by machine learning algorithms. Hub genes were screened that displayed high prediction accuracy of UC patients. Both the microarray and scRNA-seq datasets showed a strong correlation with immune cell infiltration and cytokines, especially GBP1, TRIM22, SRI. And qRT-PCR analysis showed that GBP1 play a pro-inflammatory role in UC. Two distinct clusters were identified, in which cluster A displayed higher immune infiltration level compared with the cluster B. The top targeted biological pathways of two clusters were distinct, glutamate receptor antagonist ranked top for cluster A while HDAC inhibitor ranked top in cluster B. External cohort and UC cell model validation indicated the similar immune infiltration levels, gene expression and cytokine expression patterns. We determined the seven high accuracy diagnostic genes of UC patients and provide a new perspective on immunoregulation in UC pathogenesis. And suggest patient stratification and candidate targets for precision treatment based on hub genes screened.
Similar content being viewed by others
Introduction
Ulcerative colitis (UC) is one of the inflammatory bowel diseases (IBD), characterized by a chronic inflammatory process involving the proximal colon or rectum1. To date, the etiology of UC remains unclear, but evidence suggests that genetic factors, luminal factors, gut microbiota, and immune dysfunction, including dis-regulated innate or adaptive immunity, participate in the development of UC1,2,3. UC is known to be driven by type II helper T cells and multiple cytokines produced by T cells or macrophages, such as IL-13, TNF, TGF-β, IL-9, and IL-23, in the lumina propria4,5, and patients with UC may benefit from the anti-cytokine therapy such as ustekinumab, which targets IL23p40 and shows an anti-inflammatory effect in IBD patients6. In addition, neutrophilic immune responses also play a key role in UC immune dysfunction. Studies have shown that enzymes and proteins that make up neutrophil extranuclear traps (NETs), including neutrophil elastase and myeloperoxidase, are significantly increased in the mucosa of patients with UC, even when they are in remission7. However, a detailed understanding of the immune response in UC remains elusive, and whether liquid–liquid phase separation (LLPS), specifically, regulates immune dysfunction in patients with UC remains unclear.
Various biological reactions occur in specific intracellular spaces, particularly in membranous organelles such as the endoplasmic reticulum and mitochondria8. Proteins, nucleic acids, and other vital molecules are limited to a compartment in these organelles unless a disease state triggers cell apoptosis and an innate immune response, causing leakage of cytochrome C or nucleic acids into the cytoplasm, for example9,10. However, the question remains as to how organelles without membranes or compartment-free areas of a cell limit molecular reactions. LLPS, which was first described by Edmund Beecher Wilson in 1899, could explain this phenomenon11. LLPS is associated with crucial biological functions, including gene expression, autophagy, and neuronal synaptic signaling12,13. Aberrant LLPS has been implicated in many diseases, including neurodegenerative disease, cancers, SARS-CoV-2 infection, and cardiovascular disease12,14,15. Notably, LLPS also plays a crucial role in immune responses16. Receptors and their ligands on the surfaces of immune cells form microclusters. A typical example is TCR signaling through T cell microclusters, which involves multivalent interactions of three key proteins, LAT, SOS1, and GRB2, at high density16,17. In contrast, B cell receptor (BCR) signaling begins in the cytoplasm before integration into the membrane where the BCR signaling pathway is activated18. Similar to T cell microclusters, innate immune receptors and ligands also form microclusters on cell surfaces, including the FcεRI receptor on mast cells19. And the role of LLPS in UC immune response is still a mystery.
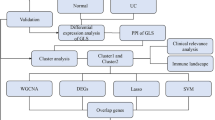
To investigate whether LLPS is associated with or regulates immune responses in patients with UC, we first acquired gene expression profiles of UC and normal tissue samples from the GEO database. Differential expressed-LLPS-related genes (DE-LLPS-RGs) were extracted and used to construct multiple machine learning algorithms to identify hub LLPS-RGs in UC patients. In addition, the potential biological functions of these genes and immune landscapes were investigated and verified in Single cell RNA sequence (scRNA-seq) data, external dataset or experiment. Moreover, 268 patients with UC were separated into two subgroups based on expression patterns of hub genes. The underlying biological differences and candidate drugs of two distinct cluster were investigated. Our study provides insights into the immunoregulation of LLPS in UC and suggests potential treatment targets for patients with this disease.
Method
Data collection and preprocessing
GSE87473 (21 normal samples and 106 UC samples) and GSE92415 (21 normal samples and 162 UC samples) were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/) and transformed to log2 form. Subsequently, the two datasets were merged and the batch effect was eliminated by the R package “sva”20. DrLLPS data resource (http://llps.biocuckoo.cn/index.php) version 1.0 incorporates 987 regulators, 150 scaffolds, and 8,148 potential clients to obtain DE-LLPS-RGs. Due to species limitations, 3,633 LLPS-RGs were extracted for subsequent analyses (supplementary Table S1)21.
Identification of differential expression of LLPS genes in UC samples
The differentially expressed genes (DEGs) of GSE87473 and GSE92415 were obtained under the threshold of adjust.p<0.05 and |logFC| ≥ 1 using the R package “limma”22 and displayed in a heatmap. DEGs from the merge dataset were integrated by the R package “RRA”.
Biological functions analysis
Ontology (GO) enrichment, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis23, Gene Set Enrichment Analysis (GSEA) and single sample GSEA (ssGSEA) were adopted to determine potential biological functions or immune cell infiltration by using R package “clusterProfiler”24. p < 0.05 and FDR < 0.25 were considered statistically significant.
Machine learning approach
The shared hub genes were filtered for subsequent analysis using three machine learning algorithms: least absolute shrinkage and selection operator (LASSO), support vector machine (SVM), and random forest (RF). The LASSO regression, SVM, and RF analyses were carried out using the R packages “glmnet”25, “kernlab”26, and “randomForest, ” whereas the “circlize” R package27 was used to illustrate interactions between the hub genes. A receiver operating characteristic (ROC) analysis was performed using the “pROC” R package to further evaluate gene prediction capacity and the ability to discriminate UC from non-UC samples28.
Immune landscape analysis
We used the CIBERSORT deconvolution algorithm to identify differences in infiltrating abundances of 22 immune cells29. The outcomes were visualized in a violin graph produced using the “corrplot” and “ggplot2” packages in R. The relationships between hub genes and immune cells were estimated using Spearman’s correlation coefficients. The relationships between hub genes and key cytokines in UC pathology (obtained from the literature) was estimated using Spearman’s correlation coefficients.
scRNA-seq analysis the hub genes in UC patients
The dataset GSE162335 which contained UC patients scRNA-seq was downloaded from GEO database. Before quality control, there were a total of 55,931 cells, of which 18,375 were inflamed UC cells. Following quality control procedures, cells and genes exhibiting low expression levels were excluded to maintain a mitochondrial percentage per cell below 5%, while ensuring gene features ranged between 200 and 2500. After filtering, a total of 15,012 cells were identified. Single-cell data was processed using the “Seurat 5.1” package30, whereby all cells were segregated into distinct clusters. The “Harmony” package was adopted to remove batch effects. Specificlly, the “LogNormalize” method was adopted to normalized and the “vst” method was performed and identified 2000 highly variable genes (HVGs). According to the expression of HVGs, PCA was performed and 20 principal components were selected for t-SNE analysis with a resolution of “0.1” to identify distinct clusters. The “FindAllMarkers” function with a logfc.threshold of 0.25 was employed to identify the DEGs in each cluster, and marker genes for each cluster were identified using avg_log2FC > 1 and p.val.adj < 0.05. To annotate each cluster, the “BlueprintEncodeData” dataset from the R package “celldex” served as a reference, and “SingleR” was utilized.
Subgroups identification based on hub gene expression patterns
Firstly, we performed an unsupervised hierarchical clustering analysis to divide UC patients into two clusters based on expression patterns of hub genes using the R package “ConsensusClusterPlus”31 (k = 2). Next, DEGs and potential functions of genes in the two clusters were determined31, and the immune landscape of the two clusters were assessed.
Potential drugs analysis for UC clusters
We performed the Connectivity Map (cMAP) analysis (https://clue.io/) to identify the candidate drugs for two subgroups of UC patients. cMAP analysis, a novel biomedical research tool, was constructed to connect the genes, drugs and diseases based on the shared gene-expression signatures whereby to identify candidate targeting drugs for disease32,33. Specifically, DEGs between two subgroups were identified using R packages “limma”, and the top 150 down-regulated genes and all the 119 up-regulated genes were adopted to cMAP analysis with the query parameters: “Gene expression (L1000), Touchstone, Individual query, 1.0”.
Cell culture, UC cell model construction and transfection
CRC cell line HT29 was purchased from ATCC and cultured at 37℃ incubator with 5% CO2 concentration supplemented with total RPMI1640 medium (with 10% fetal bovine serum and 1% penicillin streptomycin)34. 100 ng/ml TNF-α was added to treat group for 24 h to construct UC cell model. GBP1 shRNA plasmids (MiaoLing biology, China) were transfected into HT29 to construct stable knockdown cell following the standard protocol.
Tissue collection
Human colon biopsies including 4 healthy individuals and 6 UC were obtained from the Department of Gastroenterology, Zhongnan Hospital of Wuhan University. All tissues were flash-preserved at − 80 °C. All human biopsies were collected with informed permission from all subjects, and the Ethics Committee at Zhongnan Hospital of Wuhan University [No. 2024062 K]. All procedures performed with human participants were in accordance with the 1964 Declaration of Helsinki and its later amendments or comparable ethical standards.
Real time quantitative PCR (RT-qPCR) analysis
Briefly, total RNA was extracted by Trizol Reagent (Invitrogen) and 1 µg RNA was reversed to cDNA follow up the instruction of cDNA Synthesis Kit (Vazyme). RT-qPCR assays were performed follow up the instruction of CFX Connect™ PCR System (Bio-Rad)35. All the primers were displayed in supplementary Table S2.
Western-blot
The whole protein from the UC cell model was extracted using RIPA (Biosharp, China). Following 8–12% SDS-PAGE separation of the protein, the protein was transferred to PVDF (polyvinylidene difluoride) membranes. Next, the membranes were incubated with primary antibody GBP1 (15303-1-AP, Proteintech, China), TRIM22 (13744-1-AP, Proteintech, China) for an overnight period at 4 °C after being blocked with quick-block fluid. Following one hour of room temperature incubation with secondary antibodies, the bands were quantified using an ECL substrate.
Statistical analysis
All the data were analyzed by R software (version 4.2.1). Differences analysis was using Wilcoxon rank-sum test and one-way ANOVA test or Student’s t test. Correlations analysis was determined by the Spearman correlation analysis. The threshold of statistical significance was p < 0.05. * p < 0.05, ** p < 0.01, *** p < 0.001.
Results
Screening the DE-LLPS-RGs in UC samples
UC and normal colonic tissue RNA-seqs were downloaded from GEO database, 3633 human LLPS-RGs were obtained from DrLLPS database (version 1.0). Firstly, we adapted R package “limma” to identify the DEGs in GSE87473 and GSE92415 sets, respectively (Fig. 1A–B), and performed RRA method to rank the shared DEGs between two sets. Next, we merged GSE87473 and GSE92415 sets and identified batch DEGs (Fig. 1C), batch effect was eliminated by R packages “combat” (Supplementary Fig. 1A–B). Figure 1D displayed the 40 DE-LLPS-RGs in UC samples and all the DE-LLPS-RGs harbored strong interactions (Supplementary Fig. 1C).
GO and KEEG enrichments analysis of DE-LLPS-RGs
To further investigated the underlying biological function of DE-LLPS-RGs, we conducted the GO and KEEG pathways enrichment analysis. The results showed that DE-LLPS-RGs participated several molecular functions, cellular components and biological processes, including maintenance of location, positive regulation of inflammatory response, secretory granule lumen, cytoplasmic vesicle lumen, actin binding, Toll-like receptor binding, respectively (Fig. 2A). Besides, the DE-LLPS-RGs were enriched in IL-17 signaling pathway, nitrogen metabolism pathway and B cell receptor signaling pathway (Fig. 2B).
Identification of the hub LLPS-signatures via machine learning
In order to identified the hub DE-LLPS-RGs in UC patients, we performed three methods: LASSO, SVM-RFE and RF algorithms. For lasso regression, 15 hub genes: HSPB3, SLC16A1, ANXA5, TRIM22, S100A8, SRI, PLEKHG6, GBP1, C3, PADI2, BASP1, FOSL1, CCL21, LRRK2, FOS were screened (Fig. 3A–B). For SVM-RFE, the minimum error was reached while feature numbers were 16, indicating 16 hub DE-LLPS-RGs: SLC16A1, TRIM22, LRRK2, CCL21, SRI, PADI2, SLC4A4, FOSL1, BASP1, S100A8, PLEKHG6, C3, AIFM3, GBP1, CPT1A, HSPB3 were identified (Fig. 3C). RF algorithm screened 19 hub gens: SLC16A1, HSPB3, CPT1A, SRI, S100A11, FOS, PLEKHG6, SCIN, TWIST1, GBP1, AQP11, ANXA5, AKR1B10, ZBP1, TRIM22, CDC25B, SLC4A4, PLAUR, PADI2 (Fig. 3D). Then, seven shared hub genes: HSPB3, SLC16A1, TRIM22, SRI, PLEKHG6, GBP1, PADI2 of three algorithms were extracted through Veen diagram (Fig. 3E), all the hub genes were downregulated in UC samples except the GBP1 and TRIM22. To evaluate the prediction accuracy, ROC curve was conducted and all genes showed excellent prediction accuracy, and those genes displayed strong interactions (Fig. 3F and Supplementary Fig. 1C).
Functional and pathways evaluation of hub genes through GSVA
The functional enrichment of 7 hub genes were investigated through GSVA analysis. Fig. 4A1–A7 showed the results of GO enrichments. For the hub up-regulated LLPS-RGs in UC, GBP1 and TRIM22, both enriched in adaptive immune response, adaptive immune response based on somatic recombination of immune response built form immunoglobulin superfamily domains, cellular response to molecule of bacterial origin. For hub down-regulated LLPSRGs in UC, most genes enriched in adaptive immune response, adaptive immune response based on somatic recombination of immune response built form immunoglobulin superfamily domains, lymphocyte mediated immunity, B cell activation, except HSPB3. HSPB3 was enriched in catalytic activity on a nucleic acid, DNA dependent DNA replication, G-protein coupled receptor activity. For KEEG pathways enrichment, GBP1 and TRIM22 enriched in cytokine cytokine receptor interaction, chemokine signaling pathway, hematopoietic cell lineage, toll like receptor signaling pathway. For hub down-regulated LLPS-RGs in UC, cytokine cytokine receptor interaction, hematopoietic cell lineage, chemokine signaling pathways were observed enrichment while HSPB3 were mismatch repair, DNA replication, nucleotide excision repair (Fig. 4B1–B7).
Comprehensive immune landscape evaluation of hub genes in UC patients
According to the GSEA analysis in merged datasets (the batch effect was eliminated by the R package “sva”), we observed that majority of hub genes were related to immune function or pathways, therefore, we comprehensive investigated the immune landscape of 7 hub genes.
Firstly, we evaluate the immune infiltration between normal and UC samples, the level of T cells CD4 memory activated, T cell follicular helper, T cell gramma delta, neutrophils, mast cells activated, dendritic cells activated, macrophages M0/M1/M2, monocytes in UC samples were significant higher than those in normal tissues while T cells CD8 ,T cells regulatory (Tregs), dendritic cells resting, mast cells resting were lower compared with those in normal samples (p<0.05) (Supplementary Fig. 1D). Next, we accessed the immune infiltration of hub genes, respectively. Our results demonstrated that GBP1 and TRIM22 were strong positive correlation with macrophages M1, neutrophils, T cells CD4 memory activated while negative correlation with macrophages M2, Tregs, T cell CD8. For hub down-regulated LLPSRGs in UC, macrophages M0/M1, T cells CD4 memory activated displayed negative correlation while macrophages M2, eosinophils, dendritic cell resting showed positive correlation (Fig. 5A). We then investigated the relationship between the immune regulators (include immunosuppressive and immunostimulatory factors) and hub genes (Fig. 5B–C). Previous study demonstrated that both cytokines disorder and immune response disorder were observed in UC tissues and contributed to the development of UC3. To further investigate the detailed immunoregulation pattern in UC patients, we analyzed the correlation between the key cytokines in UC immune response regulation (extraction from previous study3) with hub genes screened (Fig. 5D).
ScRNA-seq analysis of hub gene expression patterns in immune cells of UC patients
To investigate the expression pattern of hub genes in immune cells of UC patients, we obtained the GSE162335 dataset which contained UC scRNA-seq from the GEO dataset. Figure 6A showed the expression characteristics of the 11 samples. After filtering the data, we obtained 15,012 cells from the lamina propria of the colon in individuals with inflammatory UC. We then normalized the data and used the “VST” method to identify 1,500 highly variable genes (HVGs), with the top 10 shown in Fig. 6B and Supplementary Fig. 2A–C. Using the t-distributed stochastic neighbor embedding method (t-SNE), all immune cells were divided into 7 clusters and immune cells were annotated into seven cell types (Fig. 6C). Similarly, hub genes may correlate with immune cells, specifically the monocytes, (with GBP1, SRI, TRIM22), CD4+T cells (with GBP1, SRI, SCL16A1, TRIM22), and B cells (with SRI, TRIM22) (Fig. 6D–J).
ScRNA-seq analysis of hub gene expression patterns in immune cells of UC patients. (A) Immune cells of GSE162335 following quality control. (B) The gene scatter plot displays the top 10 highly variable genes. In this plot, red dots represent the highly variable genes, while black dots indicate the other genes. (C) tSNE algorithm divided all immune cells into clusters that were annotated as 7 kinds of cells. (D–J) The expression of GBP1, SRI, PAID2, SLC16A1, TRIM22, PLEKHG6 in immune cells.
External dataset and RT-qPCR validate the expression levels of hub genes and their correlation with immune landscape
GSE87466 that downloaded from GEO database contained 21normal and 87 UC samples was utilized to validate the expression levels of hub signatures. Boxplot demonstrated that the expression level of the 7 hub signatures screened were consistent with the differential expression analysis in Fig. 1A–C (Fig. 7A). Moreover, UC cell model of HT29 cell line also showed the similarity expression pattern of public database except gene SLC16A1 (Fig. 7B–C). Meanwhile, Fig. 7D showed that GBP1 and TRIM22 were highly expressed in UC tissues. We further assessed the prediction accuracy of signatures screened in validation set and ROC curve displayed a great accuracy of hub genes in UC patients (Fig. 7E). Immune landscape was also tested in validation set and was coherent. GBP1 and TRIM22 were strong positive correlation with macrophages M1, neutrophils, T cells CD4 memory activated while negative correlation with macrophages M2, monocytes, T cell CD8, which were accordance with our results. For hub down-regulated signatures, macrophages M0/M1, or neutrophils displayed negative correlation while macrophages M2, eosinophils or dendritic cell resting showed positive correlation (Fig. 7F). The correlation between immune regulators (include immunosuppressive and immunostimulatory factors) and hub genes were validated and was coherent (Fig. 7G–H). Moreover, we further tested the correlation between the key cytokines in UC immune response regulation (extraction from previous study) with hub genes screened in the independent cohort GSE87466 (Fig. 7I). In summary, gene expression pattern, prediction accuracy and immune landscape that were tested in validation set or cell line demonstrated the similarity with our research.
External validation. (A) Boxplot of DEGs, by “limma” packages of R. (B–C) Gene expression validation in vitro. (D) The expression of GBP1 and TRIM22 in normal and UC tissues. (E) ROC curve of hub gene signatures. (F) Immune infiltration in validation set. (G–H) Boxplot of the correlation between immunosuppressive, immunostimulatory factors and hub genes. (I) Boxplot of the relationship of hub genes and key cytokines. *p<0.05, **p<0.01, ***p<0.001.
GBP1 positive correlation with cytokines in UC
GBP1 and TRIM22 were upregulated in UC patients and correlated strongly with immune cells. Previous study demonstrated that TRIM22 was upregulated in UC cells and mouse models and correlated positively with IL-6 and TNF-α; moreover, TRIM22 promoted the development of UC via the NF-κB signaling pathway36. However, to our knowledge, the role of GPB1 in UC has not been reported. Thus, we aimed to elucidate the role of GBP1in UC. We constructed a UC cell model with GBP1 knock down (Fig. 8A–B). In this model, we found that the expression of multiple cytokines, including IL-6 (Fig. 8C), TNF (Fig. 8D), IL-23 (Fig. 8E), and IL-12B (Fig. 8F) were downregulated. These findings indicate that GBP1 may play a pro-inflammatory role in in UC.
GBP1 positive correlation with cytokines in UC. (A) RT-qPCR showed the knockdown of GBP1 mRNA expression. (B) Western-blot showed the knockdown of GBP1 protein expression. (C–F) The expression of cytokines IL-6, TNF, IL-23, IL-12B were down-regulated accompanied GBP1 knockdown in UC cell model. *p<0.05, **p<0.01, ***p<0.001.
Subgroups identification based on hub LLPS-RGs expression
R package “ConsensusClusterPlus” was utilized to perform consensus clustering analysis. Based on the expression patterns of 7 hub DE-LLPS-RGs screened, 268 UC patient samples of GSE87473 and GSE92415 that obtained from GEO database were divided into two subgroups (k = 2) and 180 samples divided into cluster A while 88 into cluster B (Fig. 9A–B). The PCA diagram demonstrated that the expression level of two clusters were distinct (Fig. 9C). We then accessed the expression level of hub DE-LLPS-RGs between two clusters, GBP1 and TRIM22 were up-regulated in cluster A while the reminders were down-regulated (Fig. 9D).
Functional and immune infiltration landscape evaluation of subgroups
GSEA and ssGSEA were conducted to investigate the underlying biological function and immune infiltration difference between cluster A and B. For cluster A, GO enrichment showed that adaptive immune response, adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains, B cell activated, lymphocyte mediated immunity and positive regulation of immune response were enriched (Fig. 10A–B). KEEG enrichment displayed cytokine signaling pathway, cytokine cytokine receptor interaction, leishmania infection and hematopoietic cell lineage were enriched in cluster A (Fig. 10C–D). The result of ssGSEA showed that the immune infiltration level of cluster A were comprehensive higher than cluster B, especially in MDSC, neutrophil, activated B cell, Treg, type 2 T helper cell (Fig. 10E).
Candidate drugs identification for two subgroups of UC patients by cMAP analysis
Recently, personalized medical management of patients with UC has gained considerable attention. We used cMAP (https://clue.io/) analysis to identify candidate drugs among two clusters of patients with UC. According to the cMAP instructions, all the prediction drugs were ranked according to connectivity scores, in which the top or bottom ranked drugs were most strongly associated with query genes33. DEGs between two clusters included: top 150 downregulated genes and 119 upregulated genes. To obtain the reliable results, we applied a score of less than − 95 for further analysis. For patients in cluster A, we identified 93 molecular pathways targeted by 119 compounds (Fig. 11A). Among the targeted biological pathways, glutamate receptor antagonist, acetylcholine receptor agonist, serotonin receptor antagonist, adrenergic receptor antagonist, and MEK inhibitor exhibited the largest number of targeted compounds. For patients in cluster B, we identified 100 molecular pathways targeted by 139 compounds. Among the targeted biological pathways, HDAC inhibitor, topoisomerase inhibitor, CDK inhibitor, MEK inhibitor, and JNK inhibitor pathways exhibited the largest number of targeted compounds (Fig. 11B).
Discussion
Accumulating evidence indicates that immune response disorders, including those affecting both innate and adaptive immunity, are crucial for the pathogenesis of UC3. This especially involves an atypical Th2 response and cytokines produced by T cells or macrophages, such as IL-13, IL-5, TNF, TGF-β, IL-9, and IL-23, in the lumina propria4,37. Moreover, multiple therapeutic advances, especially targeted drugs based on an understanding of immune disorders associated with UC, have shifted the traditional treatments and provided novel treatment choices. Therefore, it is pivotal to assess the underlying immune response mechanism to explore new therapeutic targets. Recently, one biological process, namely LLPS, was found to result in the formation of relatively enclosed compartments that facilitate chemical reactions, and it plays a vital role in the pathogenesis of multiple diseases and is associated with immune responses12,16. However, the association between LLPS and the immune response in UC remains unknown, and investigations of LLPS immunoregulation in patients with UC could provide new insights into disease pathogenesis.
In the present study, 40 DE-LLPS-RGs were identified between patients with UC and healthy controls based on transcription profiles and LLPS-RGs from the GEO database and the DrLLPS data resource, respectively. Potential biological functions were predicted using GO and KEGG pathway analyses. Our results indicated that these DE-LLPS-RGs were enriched in and positively regulated inflammatory responses, Toll-like receptor binding, the IL-17 signaling pathway, and the B cell receptor signaling pathway in patients with UC, indicating that the LLPS process is involved in the UC-associated immune response. IL-17 comprises a family of cytokines that includes IL-17 A–IL-17 F. These cytokines play crucial roles in pro-inflammatory and immune disorders38. IL-17-producing cells were significantly increased in the healthy colonic epithelium. In contrast, UC tissues showed increased IL-17 levels, indicative of dysregulation of the IL-17 signaling pathway in patients with UC. Multiple studies have indicated that IL-17 and Th17 cells that produce these cytokines play key roles in the pathogenesis of immune-mediated UC39,40. For example, anti-IL-17 treatment was determined to minimize disease progression in a mouse model of UC41, and the expression level of IL-17 was found to be significantly increased in patients with active UC42,43. Similarly, Toll-like receptors also play a dual role in intestinal immunity by maintaining tolerance to the intestinal microbiota and inducing inflammation44. In patients with UC, the expression of Toll-like receptor 4 was found to be upregulated, and Toll-like receptor 2, which has anti- and pro-inflammatory properties, contributes to the differentiation of Th17 cells45,46. Moreover, the B cell receptor (BCR) signaling pathway was enriched in DE-LLPS-RGs, indicating that LLPS might participate in the regulation of adaptive immune responses in patients with UC. Evidence has shown that B cells are involved in UC pathogenesis. B cells, with markers such as CD24high and CD38high, as well as Bregs, which can secrete IL-10 to trigger inflammation in UC tissues, were found to be decreased in patients with UC compared with numbers in healthy controls47. However, LLPS regulates the BCR signaling pathway via microclusters of BCRs, which are found in the cytoplasm and at the cell surfaces18. Therefore, our results suggested that LLPS might be involved in immunoregulation in patients with UC.
To identify the key LLPS-RGs, machine learning algorithms (LSAAO, SVM, and RF) were used, and seven shared hub genes, specifically HSPB3, SLC16A1, TRIM22, SRI, PLEKHG6, GBP1, and PADI2, with potential involvement in immunoregulation in UC were finally identified. A previous study demonstrated that SLC16A1 expression is downregulated in UC tissues compared with that in paired normal controls, and its expression was found to be regulated by TNF-α48. TNFα is elevated in patients with UC, and this results in weakening of the mucosal barrier3. Moreover, our result showed that the expression of SLC16A1 was negatively related to TNFα (Fig. 5D), which supports the potential role of SLC16A1 in cytokine or immune regulation. TRIM22 expression was reported to be both upregulated in UC cell and mouse models and positively associated with IL-6 and TNF-α, which is consistent with the findings of our study (Fig. 5D); moreover, it might promote the development of UC via the NF-κB signaling pathway36. Specially, GBP1, a pyroptosis-related gene, is part of a potential signature of UC that is composed of a pyroptosis score, which is positively related to the inflammatory microenvironment (such as IL6–JAK–STAT3) and immune cell infiltration (such as Tregs and neutrophils)49. However, this result was slightly different from our results (GBP1 was strongly positively related to neutrophils but negatively related to Tregs (Fig. 5A)). One possible explanation for this difference is that the pyroptosis score in the previous study included four additional signatures. In addition, GBP1 was found to be overexpressed in UC and associated with poor outcomes after anti-TNF drug therapy, indicating the potential correlation between GBP1 and TNF50, which was consistent with our results (Fig. 5D). Besides, scRNA-seq analysis showed that GBP1 harbored a strong correlation with macrophages and CD4 + T cells (Fig. 6D). Macrophages play a significant role in active phage of UC, especially M1 macrophages, could destroy tight junction proteins and the epithelial barrier51. However, the roles of HSPB3, SRI, PLEKHG6, and PADI2 in UC have not been reported. To verify the expression pattern of the hub genes screened, we constructed a UC cell model using HT29 cells treated with TNF-α, and RT-qPCR showed that the expression pattern of all hub genes, except SLC16A1, was consistent with the bioinformatics analysis. SLC16A1 expression is downregulated in UC tissues compared with that in healthy controls48, and the minor differences noted could have been caused by different cell culture conditions or the selection of cell lines52.
We further investigated the biological function of each hub gene. Evidently, the hub genes were mainly related to adaptive immune responses or cytokine pathways. The immune landscape of patients with UC was also assessed and investigated by scRNA-seq. An adaptive immune response, which mainly involves two types of immune cells, CD8 + and CD4 + T cells, is essential for UC pathogenesis. According to our data on immune infiltration, the expression level of activated CD8 T cells, TFH cells, was higher in patients with UC than that in controls. CD8 + T cells in patients with UC contribute to the formation of ulcers and damage the epithelium through the production of diverse cytokines53. TFH cells are also associated with active UC and are significantly increased in UC tissues and positively associated with IL-21 and C-reactive protein levels, as well as Mayo scores, according to one study involving 31 patients with UC and 29 controls54. In contrast, we observed that Treg numbers were lower in patients with UC than those in the control group and negatively correlated with the expression of hub genes, which might play a protective role in healthy controls. This suggests that an adaptive immune response disorder underlies UC pathogenesis. The immune response disorder and cytokine production are inseparable in UC pathogenesis, and our results showed that levels of IL-5, IL-6, IL-12, IL-23 A, and TNF were strongly correlated with the expression of all hub genes screened (positive correlation with GBP1 and TRIM22, and negative correlation with genes for which expression was downregulated). The cytokine levels are increased in patients with UC and that these cells play a proinflammatory role in UC pathogenesis55,56. For example, IL-6 from macrophages and DCs acts as a pro-inflammatory cytokine and helps facilitate the differentiation of CD4+ T cells into Th17 cells57. Collectively, these findings suggest that LLPS-RGs participate in the regulation of immune disorders in UC and that they could be new targets for UC treatment.
Therapeutic advances in recent years, especially precise treatments, have shifted the traditional therapy for UC. Targeted drugs greatly alleviate the progression of UC while avoiding the adverse effects of traditional drugs. To explore the potential biological differences between and candidate drugs for UC subtypes, we grouped patients with UC into two clusters using the “ConsensusClusterPlus” algorithm31, based on the expression patterns of hub genes. We observed that expression levels of HSPB3, SLC16A1, SRI, PLEKHG6, and PADI2 were downregulated in cluster B, whereas those of GBP1 and TRIM22 were upregulated (Fig. 6D). Subsequent ssGSEA showed that the immune infiltration level of cluster A was comprehensively higher than that of cluster B, especially in MDSCs, neutrophils, activated B cells, Tregs, and Th2 cells (Fig. 7D). Further, more immune-related functions and pathways were enriched in cluster (A) These results suggest that different immunoregulation patterns exist in different clusters. A previous study demonstrated that the Th2 response contributes to sustaining homeostasis of the intestinal mucosa and also participates in pro-inflammatory pathways43. Moreover, in UC tissues, Th2 cells were aberrantly elevated, along with multiple cytokines produced, including IL-23 and TNF-α, which was found to be crucial for the development of UC. Patients might thus benefit from the anti-cytokine (IL-23) therapy, such as ustekinumab, which targets IL23p40 and shows an anti-inflammatory effect in patients with IBD6. Therefore, owing to the significant differences in immune cell infiltration and immune-related pathways between the two clusters, we further investigated the candidate drugs for each cluster using cMAP analysis based on DEGs between cluster A and cluster (B) Among those targeted biological pathways, cluster A and cluster B were distinct, indicating that hub LLPS-RGs could be used to guide the stratification of patients with UC and provide potential small-molecule drugs to achieve personalized treatment.
Conclusion
Bioinformatics analysis revealed the relationship between immune response disorder and LLPS-RGs in UC patients. Seven signatures were identified by machine learning algorithms for non-invasive diagnosis of UC patients, and demonstrated to be strongly related to immune infiltration and cytokines. And hub LLPS-RGs could guide the classification of UC patients and offer target small molecular drugs. Our study firstly offered a novel insight into the pathogenesis of UC that LLPS could regulate the immune response in UC, which may contribute to precise treatment of UC patients.
Data availability
Data supporting the present study are available upon request by contact with the corresponding author.
Abbreviations
- LLPS:
-
liquid–liquid phase separation
- UC:
-
Ulcerative colitis
- GSEA:
-
Gene set enrichment analysis
- ssGSEA:
-
Single sample GSEA
- GO:
-
Ontology enrichment
- KEEG:
-
Kyoto encyclopedia of genes and genomes pathway analysis
- LASSO:
-
Least absolute shrinkage and selection operator
- SVM:
-
Support vector machine
- RF:
-
Random forest
- DEGs:
-
Differential expression genes
- DE-LLPS-RGs:
-
Differential expression LLPS-related genes
- NETs:
-
Neutrophil extranuclear traps
- RRA:
-
Robust rank aggregation
- IBD:
-
Inflammatory bowel diseases
- ScRNA-seq:
-
Single cell RNA sequence
- t-SNE:
-
t-distributed stochastic neighbor embedding method (t-SNE)
- BCR:
-
B cell receptor
- DC:
-
Dendritic cell
- Tregs:
-
Regulatory T cells
- cMAP:
-
Connectivity map
References
Zhang, Y. Z. & Li, Y. Y. Inflammatory bowel disease: Pathogenesis. World J. Gastroenterol. 20 (1), 91–99 (2014).
Popov, J., Caputi, V., Nandeesha, N., Rodriguez, D. A. & Pai, N. Microbiota-Immune interactions in ulcerative colitis and colitis associated cancer and emerging microbiota-based therapies. Int. J. Mol. Sci. 22(21), 11365 (2021).
Kobayashi, T. et al. Ulcerative colitis. Nat. Rev. Dis. Prim. 6(1), 74 (2020).
Leppkes, M. & Neurath, M. F. Cytokines in inflammatory bowel diseases: Update 2020. Pharmacol. Res. 158, 104835 (2020).
Nakase, H., Sato, N., Mizuno, N. & Ikawa, Y. The influence of cytokines on the complex pathology of ulcerative colitis. Autoimmun. Rev. 21(3), 103017 (2022).
Parigi, T. L., Iacucci, M. & Ghosh, S. Blockade of IL-23: What is in the Pipeline? J. Crohn’s Colitis 16 (Supplement_2), ii64–ii72 (2022).
Mutua, V. & Gershwin, L. J. A review of neutrophil extracellular traps (NETs) in disease: Potential Anti-NETs therapeutics. Clin. Rev. Allergy Immunol. 61 (2), 194–211 (2021).
Mao, Y. S., Zhang, B. & Spector, D. L. Biogenesis and function of nuclear bodies. Trends Genet. 27 (8), 295–306 (2011).
Ow, Y. P., Green, D. R., Hao, Z. & Mak, T. W. Cytochrome c: Functions beyond respiration. Nat. Rev. Mol. Cell Biol. 9 (7), 532–542 (2008).
West, A. P. & Shadel, G. S. Mitochondrial DNA in innate immune responses and inflammatory pathology. Nat. Rev. Immunol. 17 (6), 363–375 (2017).
Wilson, E. B. & THE STRUCTURE OF PROTOPLASM. Sci. (New York NY) ;10(237):33–45. (1899).
Wang, B. et al. Liquid–liquid phase separation in human health and diseases. Signal. Transduct. Target. Therapy 6(1), 290 (2021).
Noda, N. N., Wang, Z. & Zhang, H. Liquid–liquid phase separation in autophagy. J. Cell Biol. 219(8), e202004062 (2020).
Mo, Y. et al. Liquid–liquid phase separation in cardiovascular diseases. Cells 11(19), 3040 (2022).
Sun, L. et al. Identification of molecular subtypes based on liquid–liquid phase separation and cross-talk with immunological phenotype in bladder cancer. Front. Immunol. 13, 1059568 (2022).
Xiao, Q., McAtee, C. K. & Su, X. Phase separation in immune signalling. Nat. Rev. Immunol. 22 (3), 188–199 (2021).
Bunnell, S. C. et al. T cell receptor ligation induces the formation of dynamically regulated signaling assemblies. J. Cell Biol. 158 (7), 1263–1275 (2002).
Oellerich, T. et al. The B-cell antigen receptor signals through a preformed transducer module of SLP65 and CIN85. EMBO J. 30 (17), 3620–3634 (2011).
Shelby, S. A., Holowka, D., Baird, B. & Veatch, S. L. Distinct stages of stimulated FcεRI receptor clustering and immobilization are identified through superresolution imaging. Biophys. J. 105 (10), 2343–2354 (2013).
Leek, J. T. Svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 42 (21), e161 (2014).
Ning, W. et al. DrLLPS: a data resource of liquid–liquid phase separation in eukaryotes. Nucleic Acids Res. 48 (D1), D288–d95 (2020).
Ritchie, M. E. et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43 (7), e47 (2015).
Kanehisa, M. & Goto, S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28 (1), 27–30 (2000).
Yu, G., Wang, L. G., Han, Y. & He, Q. Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics: J. Integr. Biol. 16 (5), 284–287 (2012).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 33 (1), 1–22 (2010).
Sanz, H., Valim, C., Vegas, E., Oller, J. M. & Reverter, F. SVM-RFE: Selection and visualization of the most relevant features through non-linear kernels. BMC Bioinform. 19 (1), 432 (2018).
Gu, Z., Gu, L., Eils, R., Schlesner, M. & Brors, B. Circlize implements and enhances circular visualization. Bioinf. (Oxford England) 30 (19), 2811–2812 (2014).
Robin, X. et al. pROC: An open-source package for R and S + to analyze and compare ROC curves. BMC Bioinform. 12, 77 (2011).
Newman, A. M. et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 12 (5), 453–457 (2015).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184 (13), 3573–87e29 (2021).
Wilkerson, M. D. & Hayes, D. N. ConsensusClusterPlus: A class discovery tool with confidence assessments and item tracking. Bioinf. (Oxford England). 26 (12), 1572–1573 (2010).
Lamb, J. et al. The connectivity map: Using gene-expression signatures to connect small molecules, genes, and disease. Science (New York, NY) 313(5795), 1929–1935 (2006).
Subramanian, A. et al. A Next Generation Connectivity Map: L1000 platform and the first 1,000,000 profiles. Cell 171 (6), 1437–52e17 (2017).
Hu, Y., Jiang, Q., Zhai, X., Liu, L. & Hong, Y. Screening and validation of the optimal panel of reference genes in colonic epithelium and relative cancer cell lines. Sci. Rep. 13 (1), 17777 (2023).
Hong, Y. et al. circPPP2R4 promotes colorectal cancer progression and reduces ROS production through the miR-646/FOXK1 axis. Mol. Carcinog. (2023).
Ye, B. & Lu, Z. Role of TRIM22 in ulcerative colitis and its underlying mechanisms. Mol. Med. Rep. 26(2), 249 (2022).
Ordás, I., Eckmann, L., Talamini, M., Baumgart, D. C. & Sandborn, W. J. Ulcerative colitis. Lancet 380 (9853), 1606–1619 (2012).
Gaffen, S. L., Jain, R., Garg, A. V. & Cua, D. J. The IL-23-IL-17 immune axis: From mechanisms to therapeutic testing. Nat. Rev. Immunol. 14 (9), 585–600 (2014).
Noviello, D. et al. The IL23-IL17 Immune axis in the treatment of ulcerative colitis: Successes, defeats, and ongoing challenges. Front. Immunol. 12, 611256 (2021).
Bunte, K. & Beikler, T. Th17 cells and the IL-23/IL-17 Axis in the pathogenesis of periodontitis and immune-mediated inflammatory diseases. Int. J. Mol. Sci. 20(14), 3394 (2019).
Ito, R. et al. Involvement of IL-17A in the pathogenesis of DSS-induced colitis in mice. Biochem. Biophys. Res. Commun. 377 (1), 12–16 (2008).
Sugihara, T. et al. The increased mucosal mRNA expressions of complement C3 and interleukin-17 in inflammatory bowel disease. Clin. Exp. Immunol. 160 (3), 386–393 (2010).
Kałużna, A., Olczyk, P. & Komosińska-Vassev, K. The role of innate and adaptive immune cells in the pathogenesis and development of the inflammatory response in ulcerative colitis. J. Clin. Med. 11(2), 400 (2022).
Kamdar, K., Nguyen, V. & DePaolo, R. W. Toll-like receptor signaling and regulation of intestinal immunity. Virulence 4 (3), 207–212 (2013).
DePaolo, R. W. et al. A specific role for TLR1 in protective T(H)17 immunity during mucosal infection. J. Exp. Med. 209 (8), 1437–1444 (2012).
Anitha, M., Vijay-Kumar, M., Sitaraman, S. V., Gewirtz, A. T. & Srinivasan, S. Gut microbial products regulate murine gastrointestinal motility via toll-like receptor 4 signaling. Gastroenterology 143 (4), 1006–16e4 (2012).
Wang, X. et al. Ulcerative colitis is characterized by a decrease in regulatory B cells. J. Crohn’s Colitis 10 (10), 1212–1223 (2016).
Ferrer-Picón, E. et al. Intestinal inflammation modulates the epithelial response to butyrate in patients with inflammatory bowel disease. Inflamm. Bowel Dis. 26 (1), 43–55 (2020).
Ning, Y. et al. Pyroptosis-related signature predicts the progression of Ulcerative Colitis and Colitis-Associated Colorectal Cancer as well as the Anti-TNF therapeutic response. J. Immunol. Res. 2023, 7040113 (2023).
Salvador-Martín, S. et al. Whole transcription profile of responders to Anti-TNF drugs in pediatric inflammatory bowel disease. Pharmaceutics 13(1), 77 (2021).
Lissner, D. et al. Monocyte and M1 macrophage-induced barrier defect contributes to chronic intestinal inflammation in IBD. Inflamm. Bowel Dis. 21 (6), 1297–1305 (2015).
Van De Walle, J., Hendrickx, A., Romier, B., Larondelle, Y. & Schneider, Y. J. Inflammatory parameters in Caco-2 cells: Effect of stimuli nature, concentration, combination and cell differentiation. Toxicol. Vitro: Int. J. Publish. Assoc. BIBRA 24 (5), 1441–1449 (2010).
Kappeler, A. & Mueller, C. The role of activated cytotoxic T cells in inflammatory bowel disease. Histol. Histopathol. 15 (1), 167–172 (2000).
Xue, G. et al. Aberrant alteration of follicular T helper cells in ulcerative colitis patients and its correlations with interleukin-21 and B cell subsets. Medicine 98 (10), e14757 (2019).
Yao, D., Dong, M., Dai, C. & Wu, S. Inflammation and inflammatory cytokine contribute to the initiation and development of ulcerative colitis and its associated cancer. Inflamm. Bowel Dis. 25 (10), 1595–1602 (2019).
Nakase, H., Sato, N., Mizuno, N. & Ikawa, Y. The influence of cytokines on the complex pathology of ulcerative colitis. Autoimmun. Rev. 21 (3), 103017 (2022).
Leppkes, M. & Neurath, M. F. Cytokines in inflammatory bowel diseases: Update 2020. Pharmacol. Res. 158, 104835 (2020).
Acknowledgements
We acknowledge the public database GEO and GSEA. And we appreciate the Editage (https://www.editage.cn/) for language service.
Funding
No funding.
Author information
Authors and Affiliations
Contributions
Y.H., L.L., and Q.Z. prepared the idea. Z.H. and Y.H. performed the experiments and manuscript. S.F., H.N., and J.Z. performed data analysis. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
All human biopsies were collected with informed permission from all subjects, and the Ethics Committee at Zhongnan Hospital of Wuhan University (No. 2024062 K).
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hong, Z., Fang, S., Nie, H. et al. Identification of the immune infiltration and biomarkers in ulcerative colitis based on liquid–liquid phase separation-related genes. Sci Rep 15, 4484 (2025). https://doi.org/10.1038/s41598-025-89252-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89252-1
Keywords
This article is cited by
-
Aberrant Enhancer Regulation, Phase Separation, and Autoimmune Diseases
Clinical Reviews in Allergy & Immunology (2025)