Abstract
Inbreeding can have negative effects, such as increasing the expression of deleterious alleles or reducing fitness. A method based on Hidden Markov Models (HMM) was developed to determine the probability of an individual genome in a homozygous-by-descent state (HBD). As a result of an experiment of divergent selection for birth weight environmental variability two lines were created: high variability line (H-Line) and low variability line (L-Line). The L-Line demonstrated a better performance in traits related with robustness than the H-Line. From a selection period of 20 generations, a total of 655 individuals from the H-Line and 675 individuals from the L-Line were genotyped with a high-density SNP array. We used a predefined multiclass HMM with a total of 9 age related HBD classes and 1 non HBD class. The sum of the probabilities of each HBD class was defined as the total HBD inbreeding (FHBD). In addition, FHBD was divided into age related groups as recent and ancient. Moreover, recent pedigree inbreeding (FPEDR) was defined using different generation thresholds (4 to 14). The evolution of FHBD across generations was similar in both selected lines. However, the distribution in each age-related class was different between lines in more recent generations. The H-Line presented twice as much FHBD by ancestors from 8 generations ago than the L-Line. Moreover, the correlations between recent FHBD and FPEDR obtained with different generation thresholds were greater in the H-Line when very recent FHBD was calculated from classes related with ancestors from 1 to 8 generations ago. However, in the L-Line, considering more than 4 generations ago to define very recent inbreeding did not affect the correlations with FPEDR. The HBD was the first methodology that could detect differences in the inbreeding pattern between the selected lines that could be related with the divergent selection, despite being under the identical mating policy and similar intensity of selection.
Similar content being viewed by others
Introduction
In livestock species, most breeds are selected and in a quantitative breeding scheme, reproducers that are selected are likely to be related. Inbreeding, resulting from the mating of related individuals, is associated with deleterious effects in fitness traits. This is known as inbreeding depression1,2. Therefore, the selection process leads to an inevitable increase in inbreeding over the generations and breeders have to deal with inbreeding depression on traits of interest3. It has been hypothesized that a purging process naturally occurs with the elimination of deleterious alleles throughout the generations and deleterious alleles are mostly young and maintained at low frequency4. Thus, the presence of homozygosity caused by ancient ancestors could probably be possible if alleles undergo a purging process and, therefore, ancient identity by descent is more likely to be beneficial.
Numerous genomic estimators of realized inbreeding have been proposed. Of these, runs of homozygosity (ROH) were shown to be useful estimators of realized inbreeding4. Moreover, the length of ROH was inversely related to the number of generations that have elapsed from the common ancestor and, therefore, ROH detection can distinguish between recent and ancient inbreeding4,5,6. However, ROH were highly dependent on the parameters setting to be detected4,7,8. Other authors proposed other likelihood-based approaches that relied on Hidden Markov Models (HMM) to identify homozygous by descent segments (HBD). This HMM were modelled as a function of marker allele frequencies, the genotyping error rates and intermarker genetic distances9. The HMM was initially proposed with some constrains: all the autozygosity appeared from a single event; all the HBD segments originated in the same generation; and all these segments had an identical expected length10. However, in natural populations the historical demographic events were complex. Therefore, new HMM methods have been developed with multiple age-based classes9. In these models, the length of the HBD segments have distinct expected distributions related with the number of generations that passed from the common ancestor2,9,11.
Current animal breeding programmes are implementing new selection objectives related with uniformity of some productive selection traits as significant variation around the optimal value of a trait can have a negative impact on production performance12. Moreover, selection for uniformity would result in animals that are more robust and better prepared to face environmental challenges. This uniformity is directly related to higher profitability and improvement in animal welfare13,14. A divergent selection experiment for birth weight variability in mice carried out over 33 generations demonstrated that it was possible to modify the genetic control of the birth weight environmental variability15. Following this experiment, two divergent lines were created: high variability line (H-Line) and low variability line (L-Line). The L-Line presented advantages in production, animal welfare, heritability, and traits related with robustness such as feed efficiency or longevity5,6,7,8. Furthermore, the L-Line showed higher response to selection than the H-Line20.
It is hypothesized that the residual variance decreases when the number of homozygous loci increases21,22,23. In previous studies performed in the mice lines divergently selected for birth weight variability, it was shown that there were no differences in the global level of homozygosity between the lines that could explain the differences in performance24,25. This same level of homozygosity could be expected as both lines were subjected to the same population size under the same selection intensity and under the same optimized mating policy, also avoiding sharing grandparents between mated animals17. However, differences were detected in the distribution of inbreeding across chromosomes between lines24.
The objective of this analysis was to examine the HBD patterns in both lines of mice divergently selected for birth weight environmental variability across generations in terms of ancient and recent inbreeding in each line. HBD ability to discriminate patterns of homozygosity increase in this population was evaluated.
Methods
Data
The pedigree and the genotype data were originated from an experiment of divergent selection for birth weight environmental variability in mice17. The housing and management conditions of the animals were in accordance with the Spanish legislation RD 53/2013 on the basic rules for the protection of animals used in experiments and other scientific purposes (Boletín Oficial del Estado 2013) and approved by the Animal Experimentation Committee (PROEX 224/18). Pedigree data contained 5,054 individuals, including the 1,340 individuals used for the study and their ancestors up to five generations of pedigree of the founder population of the experiment. Optimized matings during the selection experiment were restricted to animals not sharing grandparents with a selection intensity equivalent to around 30%. In each selection generation 43 females were mated with 43 males having a maximum of two parturitions. More details could be found in Formoso-Rafferty et al.17. Animals belonging to the five generations carried out before selection were randomly mated.
A total of 1,340 individuals from 20 generations of selection from generation 6 to generation 26 of the selection experiment (655 of H-Line and 675 of L-Line) were genotyped using the high density Affymetrix Mouse Genotyping Array. All the individuals had a call rate higher than 97%. single nucleotide polymorphisms (SNPs) with more than 3% missing data and SNPs mapped to sex chromosomes were removed. After that, 545,656 SNPs were kept. A minor allele frequency filter was not applied as pruning for low MAF can ignore large homozygous regions in the genome26. This subset of SNPs was used to detect ROH segments and estimate HBD probabilities. We used genotypes of individuals from generation 6 and not from the start of the experiment to ensure that individuals had a pedigree deep enough to compute recent pedigree inbreeding.
Methods to compute HBD probabilities
Under HMM the individual genome is described as a mosaic of multiple HBD and non HBD states. In the multiple age-based models, these HBD states are expected to arise from different generation of ancestors that are classified by the method in different age-related classes9. Each age-related class was defined by its rate Rk, defined by the authors that developed this algorithm as the length of the inbreeding loop or approximately half the age of the underlying ancestor9.The mean expected length of the HBD segments in this class was 1/Rk Morgans. Therefore, the number of generations from the common ancestors in which HBD segments appeared was approximately 0.5*Rk.
We calculated the HBD probabilities using two alternatives of HMM with multiple age-based models.
-
1.
Model with predefined Rk or mixKR. We used a total of 10 predefined classes. The Rk of HBD classes was set in a 2-exponential distribution: 21, 22, 23, 24, 25, 26, 27, 28, 29 and of one non HBD class was set to 29. In this analysis, only one non HBD state was used as recommended2,9,11. These values of Rk were defined to have a constant and limited degree of overlap between the exponential distribution9,11. The age related classes were defined based on previous works9,27. Thus, for example, the probability that a marker belongs to an HBD class 23 (8) is approximately equal to the probability that this marker is identity by descent from 4 generations ago.
-
2.
Models without predefined Rk or KR. With this model, the Rk was calculated in each individual and the number of classes could be selected according to the best (lowest) BIC in the population. The Rk of each class could be obtained by the median value of the Rk of all the individuals9. We fitted six models with a different number of classes: 2 (KR2), 3 (KR3), 4 (KR4), 5 (KR5), 6 (KR6) and 8 (KR8).
These two types of models were chosen because the first allows a better calculation of individual autozygosity and identification of HBD segments that can be captured by marker density9,11. The second model, was chosen because inbreeding coefficients were estimated with respect to the same reference population and offer more resolution on the contribution of the ancestral generation to total autozygosity9,11.
Estimation of genomic and pedigree inbreeding
The HMM with multiple age-based provided the calculation of the contribution of each class to the overall level of individual inbreeding by averaging the probabilities of each SNP being in each class over the whole genome. Global HBD genomic inbreeding (FHBD) was obtained by summing up the contribution of HBD in all HBD classes. In the KR models, only the global FHBD was calculated for each model namely FHBD_KR. In addition, from the mixKR model, global autozygosity was computed namely FHBD as the summed of autozygosity contribution of each HBD K(FHBD_2, FHBD_4, FHBD_8, FHBD_16, FHBD_32, FHBD_64, FHBD_128, FHBD_256 and FHBD_512)9. The proportion of FHBD in each class was calculated in the intermediate generations (6, 7, and 9) and the most recent generations of the experiment of selection (23, 24, 25 and 26) in both lines to compare the evolution of the age on ancestors contributing to total FHBD.
Other groups of HBD classes were formed from the mixRK model regarding the number of generations that have elapsed from the common ancestor based on the results obtained from the distribution in the classes of FHBD within each selected line: very recent HBD genomic inbreeding, recent and ancient.
The ROH genomic inbreeding was calculated as \({F}_{ROH}=\frac{\sum LROH}{LAUTO}\)28 where LROH and LAUTO are respectively the total length of all ROH segments and the autosomal genome (2,456,563 kb). ROH were detected using the sliding windows (SW) algorithm29. The parameters set for SW were 50 SNPs per window; 1 heterozygote allowed in a window; 5 missing SNPs allowed in a window; the minimum length of a ROH was 1000 kb; the minimum number of homozygous SNPs in a ROH was set to 100; the minimum density was set at 1 SNPs/50 Kb; window threshold was set to 0.5 and the minimum distance between two SNPs was 1000 kb. More details can be found in Ojeda-Marín et al.24.
Third, genealogical inbreeding (FPED) defined as the probability that two alleles from an individual were identical by descent, was computed following Meuwissen and Luo30. Furthermore, FPED was decomposed in recent and ancient inbreeding as shown by Wright31. Recent pedigree inbreeding coefficients (FPEDR) were calculated using the classical method32. Thresholds of the generations to consider FPEDR were essayed considering different thresholds checking from 4 to 14 generations back.
Pearson correlation coefficient was calculated between different genomic and pedigree inbreeding estimators.
The R package RZooRoH was used to analyse HBD segments9,27. PLINK v1.9 was used to detect ROH29, ENDOG v4.9 was used to compute FPED33 and our own FORTRAN code was developed to determine FPEDR. Correlations were calculated with the R function “cor”.
Results
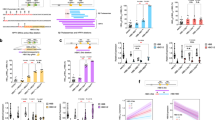
The evolution of FHBD, FROH and FPED is shown in Fig. 1. The trend was positive for the three inbreeding coefficients in both selected lines. The trend of FHBD and FROH was nearly parallel during the analysed twenty generations. However, FROH was greater than FHBD: in the 26th generation when the FROH was 0.68 in the H-Line and 0.67 in the L-Line and FHBD was 0.58 in the H-Line and 0.57 in the L-Line. The lowest values were observed for FPED, being 0.35 in both selected lines.
Evolution across selection generations of genomic inbreeding calculated from homozygosity by descent probabilities obtained from a multi-class Hidden Markov Model (FHBD), runs of homozygosity (FROH) and pedigree inbreeding (FPED) in high variability line (H-Line) and in low variability line (L-Line). FHBD and FROH are represented on the left axis and FPED is represented on the right axis.
Figure 2 shows the proportion of genomic inbreeding captured by the mixKR model in each predefined class in both selected lines in intermediate and most recent generations of the selection experiment. The difference in the HBD pattern between lines was different in intermediate generations (6, 7, 8 and 9) from recent generations (23, 24, 25 and 26). In the intermediate generations, the highest contribution was from FHBD_32 in both selected lines, followed by FHBD_16 and FHBD_64. However, in the most recent generations the distribution of FHBD shifted to younger HBD classes in both lines. The contribution of FHBD_8 was similar in both selected lines in most recent generations (0.17). However, the contribution of FHBD_16 was almost twice as much in the H-Line (0.17) than in the L-Line (0.10).
Proportion of genomic inbreeding captured by homozygosity by descent segments computed with a predefined multi-class Hidden Markov Model (FHBD) in each class. The proportion of FHBD in each HBD class was computed in individuals from the high variability line (H-Line) and the low variability line (L-Line). The proportion of FHBD is represented in intermediate generations (6, 7, 8 and 9) and in most recent generations of the selection experiment (23, 24, 25 and 26). FHBD_2: genomic inbreeding coefficient obtained from homozygosity by descent probabilities (FHBD) of age class 2. FHBD_4: FHBD of age class 4. FHBD_8: FHBD of age class 8. FHBD_16: FHBD of age class 16. FHBD_32: FHBD of age class 32. FHBD_64: FHBD of age class 64. FHBD_64: FHBD of age class 64. FHBD_128: FHBD of age class 128. FHBD_256: FHBD of age class 256. FHBD_512: FHBD of age class 512.
Therefore, given the distribution of FHBD in the different age-related classes, we defined the genomic inbreeding regarding the number of generations elapsed from the common ancestor in each line in two ways to check the impact of considering, more or less, generations back. In the alternative 1 recent FHBD was defined between Rk 2 to Rk 16 (FHBD_R1), and the ancient FHBD was defined between Rk 32 to Rk 512 (FHBD_A1) regarding the distribution of HBD in the H-Line. In the alternative 2 very recent FHBD was defined between Rk 2 to Rk 8 (FHBD_R2) and ancient FHBD was defined between Rk 16 to Rk 512 (FHBD_R2) regarding the distribution of HBD in the L-Line. Figure 3 represents the evolution of recent and ancient HBD inbreeding in both selected lines across generations. In the case of very recent and recent inbreeding, was calculated from the distribution of HBD in the H-line (Fig. 3a) and in the L-Line (Fig. 3b). Both FHBD_R1 and FHBD_R2 showed a positive evolution in both selected lines. The FHBD_R1 was 0.19 in both selected lines in generation 6 and 0.34 in both selected lines in generation 26. The FHBD_R2 was 0.08 at generation 6 in both selected lines and 0.22 in the H-Line and 0.24 in the L-Line at generation 26. The evolution of FHBD_A1 was positive in the L-Line and negative in the H-Line. The evolution of FHBD_A2 was negative in both selected lines.
Evolution across the selection generations of genomic inbreeding calculated from homozygosity by descent probabilities divided into recent inbreeding, and ancient inbreeding in the high variability line (H-Line) and in the low variability line (L-Line). (a) Recent inbreeding (FHBD_R1) and ancient inbreeding (FHBD_A1) calculated depending on the distribution of HBD in the H-Line. (b) Recent inbreeding (FHBD_R2) and ancient inbreeding (FHBD_A2) calculated depending on the distribution of HBD in the L-Line.
The correlations between different inbreeding coefficients are shown in Table 1. In both selected lines, the total genomic inbreeding obtained from total HBD probabilities presented higher correlations with the other inbreeding coefficients than FHBD calculated from different HBD age classes. In the H-Line the correlation between FPED and FHBD was 0.78, while the correlation between FPED and FROH was 0.82. Nevertheless, in the L-Line both the correlations FPED-FHBD and FPED-FROH were 0.83. The correlations between FHBD and FROH were 0.97 in the H-Line and 0.98 in the L-Line. When FHBD were divided into age related subtypes, FHBD_R1 and FHBD_R2 showed the greatest correlation with FHBD, FPED and FROH , while most of the correlations with FHBD_A1 and FHBD_A2 were negative or close to zero. Moreover, in the H-Line the correlations between FHBD_R1 and FHBD (0.71), FPED (0.62), and FROH (0.68) were greater than the correlations between FHBD_R2 and FHBD (0.58), and FROH (0.52). However, in the L-Line the correlations between FHBD_R1 and the rest of the inbreeding coefficients were only slightly lower than for FHBD_R2, except for FPED, where the correlation was slightly higher for FHBD_R1 (0.55) than for FHBD_R2 (0.52) but also quite similar.
Figure 4 shows the correlations between FHBD_R1 and FHBD_R2 with FPEDR calculated with different generation thresholds (4 to 16) in both selected lines. In this figure, the correlations between FPEDR and FHBD calculated from classes between 2 to 32 and between total FHBD are shown to test whether the correlations change with different age thresholds in the calculation of FHBD. The correlations with FHBD_R1 were in general higher than with FHBD_R2 in both selected lines. The greatest correlation was detected earlier with FHBD_R2 (when a threshold of 11 generations was used to calculate FPEDR) than with FHBD_R1 (when a threshold of 13 generations was used to calculate FPEDR) in the L-Line. In the H-Line the greatest correlation was observed at the same generation threshold (9 generations) in both alternatives. Correlations between FHBD≤32 and FPEDR were not better than with FHBD_R1 and FHBD_R2 in either line. The correlations between FHBD and FPEDR in the H-Line increased up to generation 9 and then appeared to be stable. In the L-Line, the correlations between FHBD and FPEDR tend to increase when older generations were used as a threshold for calculating FPEDR.
Correlation between recent pedigree inbreeding (FPEDR) computed with different generation thresholds (4 to 16 generations) and different inbreeding coefficients computed from homozygosity by descent probabilities (HBD) calculated from a multi-class Hidden Markov Model. (a) Correlations between different FPEDR and different genomic inbreeding coefficients computed from HBD in high variability line. (b) Correlations between different FPEDR and different genomic inbreeding coefficients computed from HBD in low variability line. FHBD_R1: genomic inbreeding coefficient obtained from homozygosity by descent probabilities (HBD) by summing class 2 to class 16. FHBD_R2: genomic inbreeding coefficient obtained from HBD by summing class 2 to class 8. FHBD≤32: genomic inbreeding coefficient obtained from HBD by summing class 2 to class 32. FHBD: genomic inbreeding coefficient obtained from HBD by summing all classes.
Table 2 shows the comparison between the fitted KR models with different number of classes in both selected lines. The model with the best BIC for most of the individuals was the model with two HBD classes for both lines: 624 individuals in the H-Line and 623 individuals in the L-Line. The median of the Rk for the best KR model was similar between lines for both HBD classes, around 15 for the first HBD class and 160 for the second HBD class. The mean FHBD_KR in the H-Line was between 0.52 and 0.53 for all the models in the H-Line and 0.53 in the L-Line. The correlation between FHBD_KR and FHBD was 1.00 for all the models and in both selected lines. When the number of classes increased, the last class, that corresponded to the oldest common ancestors, was older in the L-Line than in the H-Line.
Discussion
Inbreeding is linked to an increase of expression of deleterious alleles. A purging process produces deleterious alleles to be mostly inherited due to recent inbreeding and thus maintained at a low frequency in populations15. Moreover, it has been described that the homozygous tracts of the genome are non-randomly distributed across the genome and could appear due to selection34. The lines of mice used in this analysis was selected for the same trait, birth weight environmental variability, but in opposite directions. In both lines, animals to be mated were not allowed to share grandparents. This prevented the possibility of very recent inbreeding. Moreover, it has been hypothesised that selection for homogeneity could result in more homozygous individuals21,23. Previous studies performed in this population showed that there were no differences in global inbreeding between lines but there were differences in the distribution of homozygosis across the genome24,25. Therefore, this population of mice divergently selected for environmental birth weight provided an opportunity to study the power of genomic inbreeding assessed using HBD probabilities, which can determine the number of generations that elapsed from the common ancestor to produce homozygous segments of a specific length9,35.
The authors who developed the multi class HBD model recommended using this kind of model because it can determine which generations of ancestors contribute to the present autozygosity. Thus making it possible to compare an individual’s feasibility because the same number of age classes is used in all of them9. Moreover, the results of the present analysis showed a correlation of 1.00 between each fitted genomic inbreeding obtained from the KR model (FHBD_KR) and with the mixKR model (FHBD). The total levels of FHBD in both lines were high. Another study performed in the European Bison also detected high levels of FHBD: mean 0.31 in one of the analysed populations and 0.40 in the other population2. Moreover, Solé et al.25 detected a mean of genomic inbreeding coefficient higher than 0.30 in Belgian Blue Beef Cattle. Other authors reported high levels of homozygosity in sheep and dog populations9. These studies reported different likely origins of the high FHBD detection as intense selection11 or severe bottlenecks2,9. In this mice population the high levels of genomic inbreeding could be explained by the relatively low effective population size (around 32) and the high intensity of selection17,25.
The evolution of total pedigree and genomic inbreeding was highly similar across the selection generations. This excluded the possibility that the difference in performance between lines was produced because of a higher level of total homozygosity in one of the lines, as what was described in the previous studies performed in this population using other approaches to detect genomic inbreeding24,25. The trend of FROH and FHBD was almost parallel across generations, FROH was greater than FHBD in all the generations. One explanation of this observation could be that FROH and FHBD are not being measured with the same unit, FROH is defined as the proportion of genome covered by ROH28 and FHBD is the sum of the probability across the genome of be in HBD state9. Moreover, it has been described that the detection of ROH and HBD are dependent on the user defined parameters9,11. In the present study only one set of parameters was used and these could be affecting the scale of both FHBD and FROH. Another possible explanation could be the reference generation set to consider FHBD (≤ Rk 512). In another study, the total value of FHBD was greater than FROH but Rk ≤ 8192 was considered to calculate FHBD11. However, in the present study the minimum length applied to consider a ROH was 1 Mb which is almost equivalent to consider homozygous segments that appeared approximately 50 generations ago36. Moreover, Alemu et al.10 detected that the mean value of FROH was always under the different values obtained with different generation thresholds of FHBD. These results might suggest that in the divergently selected lines the smallest segments could not be detected with the HMM or are masked by more recent segments11. Furthermore, when the FHBD was subdivided in coefficients related with the distance of the common ancestor, FHBD_R1 and FHBD_R2 trend was positive across the selection generations. This could be explained by the change in the mating policy since the start of the experiment that results in lower effective population size as shown in other studies25. This might explain the higher correlations with the other inbreeding coefficients compared to FHBD_R1 and FHBD_R2 which showed a more stable evolution. Solé et al.25 observed that the effect of using or not ancient segments to calculate the correlations between total FHBD and accumulative FHBD was almost non-existent. This implied that the recent FHBD classes were probably more relevant to manage because it explained most of the variation in the individual genomic inbreeding.
In general, the correlations detected between FHBD and the other inbreeding coefficients were high in both selected lines. Other authors, detected high correlations between FHBD and other genomic inbreeding coefficients but lower correlations between FHBD and FPED (0.46)11. Nevertheless, their study was not done in a divergent selection experiment in mice that could result in faster and more efficient than in cattle, which is usually not that intense nor divergent. However, Alemu et al.1 detected high correlations between FHBD and FPED that were greater when only most recent HBD classes were used to calculate FHBD. Moreover, correlations with FPED were also high with FROH. These three coefficients matched with the definition of IBD and FROH and FHBD had been demonstrated at capturing IBD more efficiently than other genomic inbreeding measures1. Furthermore, the comparison of molecular inbreeding measures with FPED is not straightforward because FPED cannot necessarily be considered as the golden standard1. Nevertheless, the correlations between FHBD and the other inbreeding coefficients were lower compared to what was observed in other studies in this population24,25. In these studies, the high correlations were attributed to the high number of animals belonging to different generations and to a good quality of the pedigree. In the present study, a lower number of generations were used to ensure that all the individuals had enough pedigree depth to calculate the FPEDR.
This study was designed to check whether the selection process had differently affected the lines, but using a methodology that allowed to distinguish the number of generations back where homozygosis appeared24,25. We showed that the distribution of FHBD in age-related classes was different between lines and changed across generations. In both selected lines, most of the increase was associated with FHBD_8, which represented HBD probabilities from 4 generations ago. This might be related to the design of the experiment, where the mated females and males did not share grandparents17. However, FHBD_16 in the H-Line only made a greater or equal contribution than FHBD_8. Moreover, when this distribution was shown to determine the boundaries to calculate recent and ancient HBD inbreeding in the selected lines, the correlations in the H-Line improved for very recent inbreeding with the rest of inbreeding coefficients and with FPEDR calculated with different generation thresholds. In addition, the correlations between total FHBD and the different recent FHBD suggest that the realized autozygosity was represented by older pedigree inbreeding in the L-Line than in the H-Line. This different distribution between lines could be due to the opposite direction of the selection. The selection for homogeneity has been hypothesized to increase the homozygosity. However, Holland et al.37 reported a decrease or maintenance of the genetic variance in that case. Moreover, none of the previous analyses done in the mice experiment found any differences between lines in terms of inbreeding or homozygosity as expected according to the strictly additive genetic model used for selection24,25. However, these results show that the main origin of inbreeding in the L-line was explained by ancestors 4 generations back, whilst in the H-line it seems to have come from ancestors 8 generations back, thus showing a partial effect of selecting to modify the variability. In the Blue Beef Cattle most of the FHBD was explained by HBD segments produced less than 16 generations ago11. Furthermore, in the European Bison differences in the distribution of HBD were detected in each analysed line: the Lowland line presented a higher proportion of the genome in more younger classes than the Lowland-Caucasian line that was consistent with the higher genetic drift sustained by the Lowland line2. Nevertheless, in other studies in sheep or humans, the youngest HBD classes represented the smaller percentage of total genome-wide autozygosity9,38.
These differences between lines in the distribution of the proportion of the genome in HBD classes were reflected in the correlations between recent FHBD and FPEDR calculated with different generation thresholds. The H-Line presented the maximum of correlation earlier than the L-Line. A study performed in the Pura Raza Española horses39 showed that FHBD calculated with the Viterbi algorithm9 divided by generations presented the highest correlations with recent pedigree inbreeding from a similar number of generations. Therefore, the results obtained in this study suggested that, contrary to expectations, the H-Line had higher percentage of recent genomic autozygosity than the L-Line, despite being under the same mating policy and similar intensity of selection.
In the present study, the KR model that better fit in both selected lines was the 2 HBD classes and 1 non-HBD class. Other authors11 detected that for a high density SNP array the model that better fitted was a KR model with 3 HBD classes and 1 non HBD class while the 2 HBD classes and 1 non-HBD classes was better for a medium density SNP array. A possible explanation is that the best model in both lines could be a reflection of the particular structure of this population created through selection and with the particular constraints in the mating policy17. However, the medians obtained with the 2KR model of the first and the second class in Sole et al.20 (15 and 198, respectively) were similar to the medians detected for both lines in the present study. Moreover, other authors that performed HBD analysis in whole genome sequence data suggested that the HBD algorithm could better detect shorter segments than the ruled based methods i.e. the sliding windows algorithm to detect ROH. The mentioned authors also emphasised the need for a good quality control of the SNPs to obtain the best results in this type of analysis40. Lavanchy et al.41 determined that with a marker density between 22 SNPs/Mb and 2 SNPs/Mb was enough to estimate FHBD and FROH correctly. In the selected mouse lines, the marker density was approximately 9 SNPs/Mb when only the polymorphic markers were considered.
The availability of genotypes from different generations enabled the study of FHBD evolution across a wide number of generations. However, in other livestock populations, it is common for the generation interval to be longer, and only a few generations could be genotyped. This could complicate the comparison of FHBD across different generations.
Conclusions
Differences in robustness between lines would not be attributed to global pedigree and genomic inbreeding. However, the distribution of FHBD in different HBD classes was different between lines. The H-Line presented a higher contribution of ancestors from 4 to 8 generations ago than the L-Line, in which most of the recent inbreeding were produced 4 generations ago, despite being under the same mating policy and similar intensity of selection.
Availability of data and materials
The data that support the findings of this study are not openly available and are available from the corresponding author upon reasonable request.
References
Alemu, S. W. et al. An evaluation of inbreeding measures using a whole-genome sequenced cattle pedigree. Heredity 126, 410–423 (2021).
Druet, T. et al. Genomic footprints of recovery in the European Bison. Shaapiro B, editor. J. Hered. 2020 ;esaa002.
Leroy, G. Inbreeding depression in livestock species: review and meta-analysis. Anim. Genet. 45, 618–628 (2014).
Caballero, A., Fernández, A., Villanueva, B. & Toro, M. A. A comparison of marker-based estimators of inbreeding and inbreeding depression. Genet. Sel. Evol. 27, 54–82 (2022).
Pemberton, T. J. et al. Genomic patterns of homozygosity in worldwide human populations. Am. J. Hum. Genet. 91, 275–292 (2012).
Kirin, M. et al. Genomic runs of homozygosity record population history and consanguinity. PLoS One. 5, e13996 (2010).
Villanueva, B. et al. The value of genomic relationship matrices to estimate levels of inbreeding. Genet. Sel. Evol. 53, 42 (2021).
Peripolli, E. et al. Runs of homozygosity: Current knowledge and applications in livestock. Anim. Genet. 48, 255–271 (2017).
Druet, T. & Gautier, M. A model-based approach to characterize individual inbreeding at both global and local genomic scales. Mol. Ecol. 26, 5820–5841 (2017).
Leutenegger, A. L. et al. Estimation of the inbreeding coefficient through use of genomic data. Am. J. Hum. Genet. 73, 516–523 (2003).
Solé, M. et al. Age-based partitioning of individual genomic inbreeding levels in Belgian Blue cattle. Genet. Sel. Evol. 49, 92 (2017).
Marjanovic, J., Mulder, H. A., Khaw, H. L. & Bijma, P. Genetic parameters for uniformity of harvest weight and body size traits in the GIFT strain of Nile tilapia. Genet. Sel. Evol. 48, 41 (2016).
Bolet, G. et al. Genetic homogenisation of birth weight in rabbits: Indirect selection response for uterine horn characteristics. Livest. Sci. 111, 28–32 (2007).
Poigner, J., Szendrö, Z., Lévai, A., Radnai, I. & Biró-Németh, E. Effect of birth weight and litter size on growth and mortality in rabbits. World. Rabbit. Sci. 8, 17–22 (2000).
Formoso-Rafferty, N., Cervantes, I., Ibáñez-Escriche, N. & Gutiérrez, J. P. Modulating birth weight heritability in mice. J. Anim. Sci. 95, 531 (2017).
Formoso-Rafferty, N., Cervantes, I., Sánchez, J. P., Gutiérrez, J. P. & Bodin, L. Effect of feed restriction on the environmental variability of birth weight in divergently selected lines of mice. Genet. Sel. Evol. 51, 27 (2019).
Formoso-Rafferty, N., Cervantes, I., Ibáñez-Escriche, N. & Gutiérrez, J. P. Genetic control of the environmental variance for birth weight in seven generations of a divergent selection experiment in mice. J. Anim. Breed. Genet. 133, 227–237 (2016).
Formoso-Rafferty, N., Cervantes, I., Ibáñez-Escriche, N. & Gutiérrez, J. P. Correlated genetic trends for production and welfare traits in a mouse population divergently selected for birth weight environmental variability. Animal. 10, 1770–1777 (2016).
Formoso-Rafferty, N., Gutiérrez, J. P., García-Álvarez, A., Pérez, T. & Cervantes, I. Impact of selection for birth weight variability on reproductive longevity: A mice model. J. Anim. Breed. Genet. 139, 370–379 (2022).
Formoso-Rafferty, N., Chavez, K. N., Ojeda, C., Cervantes, I. & Gutiérrez, J. P. Selection response in a divergent selection experiment for birth weight variability in mice compared with a control line. Animals (Basel). 26, 10–920 (2020).
Lerner, I. M. Genetic Homeostasis (Oliver and Boyd, 1954).
Lewontin, R. C. Selection in and of populations. In Ideas in Modern Biology (ed. Moore, J. A.) 299–311 (Natural History Press, 1964)
Zhivotovsky, L. A. On the difference between mean and optimum of quantitative characters under selection. Evol. 46, 1574–1578 (1992).
Ojeda-Marín, C., Gutiérrez, J. P., Formoso-Rafferty, N., Goyache, F. & Cervantes, I. Differential patterns in runs of homozygosity in two mice lines under divergent selection for environmental variability for birth weight. J. Anim. Breed. Genet. 141, 193–206 (2024).
Ojeda-Marín, C., Cervantes, I., Formoso-Rafferty, N. & Gutiérrez, J.P. Genomic inbreeding measures applied to a population of mice divergently selected for birth weight environmental variance. Front. Genet.14 (2023).
Meyermans, R., Gorssen, W., Buys, N. & Janssens, S. How to study runs of homozygosity using PLINK? A guide for analyzing medium density SNP data in livestock and pet species. BMC Genomics. 21, 94 (2020).
Bertrand, A. R., Kadri, N. K., Flori, L., Gautier, M. & Druet, T. RZooRoH: An R package to characterize individual genomic autozygosity and identify homozygous-by-descent segments. Methods Ecol. Evol. 10, 860–866 (2019).
McQuillan, R. et al. Runs of homozygosity in European populations. Am. J. Hum. Genet. 83, 359–372 (2008).
Chang, C. C. et al. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience. 4, 7 (2015).
Meuwissen, T. & Luo, Z. Computing inbreeding coefficients in large populations. Genet. Sel. Evol. 24, 305–313 (1992).
Wright, S. Systems of mating. I. The biometric relations between parent and offspring. Genetics. 6, 111–23 (1921).
Falconer, D. S. & Mackay, T. F. C. Introduction to Quantitative Genetics. 4th edn (Pearson Longman, 1996).
Gutierrez, J. P. & Goyache, F. A note on ENDOG: a computer program for analysing pedigree information. J. Anim. Breed. Genet. 122, 172–6 (2005).
Bosse, M. et al. Regions of homozygosity in the porcine genome: consequence of demography and the recombination landscape. PLoS. Genet. 8, e1003100 (2012).
Ceballos, F. C., Joshi, P. K., Clark, D. W., Ramsay, M. & Wilson, J. F. Runs of homozygosity: Windows into population history and trait architecture. Nat. Rev. Genet. 19, 220–234 (2018).
Curik, I., Ferenčaković, M. & Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 166, 26–34 (2014).
Holland, J. B. et al. Recurrent selection in oat for adaptation to diverse environments. Euphytica. 113, 195–205 (2000).
Álvarez, I. et al. Ancient homozygosity segments in West African Djallonké sheep inform on the genomic impact of livestock adaptation to the environment. Animals (Basel). 10, 1178 (2020).
Perdomo-González, D.I et al. Fine-tuning genomic and pedigree inbreeding rates in equine population with a deep and reliable stud book: the case of the Pura Raza Española horse. J Anim Sci Biotechnol. 13, 127 (2022).
Berghöfer, J., Khaveh, N., Mundlos, S. & Metzger, J. Simultaneous testing of rule- and model-based approaches for runs of homozygosity detection opens up a window into genomic footprints of selection in pigs. BMC Genomics. 23, 564 (2022).
Lavanchy, E. & Goudet, J. Effect of reduced genomic representation on using runs of homozygosity for inbreeding characterization. Mol. Ecol. Resour. 23, 787–802 (2023).
Acknowledgements
The genotyping service was carried out at CEGEN-PRB3-ISCIII; which is supported by grant PT17/0019, of the PE I+D+i 2013-2016, and funded by ISCII and ERDF.
Funding
This study was funded by a grant from the Ministry of Science, Innovation, and Universities; Grant Number: PGC2018-096198-A-I00.
Author information
Authors and Affiliations
Contributions
C.O-M. and I.C. and J.P.G. conceptualized the analysis. C.O-M. developed the formal analysis. C.O-M.; C.O-M., J.P.G., and I.C. investigated; N.F., J.P.G. and I.C. provided the resources. C.O-M. curated the data; C.O-M. wrote the original draft. C.O-M., N.F., J.P.G. and I.C. wrote, revised and edited the manuscript. J.P.G. and I.C. supervised all the development of the research. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ojeda-Marín, C., Gutiérrez, J.P., Formoso-Rafferty, N. et al. Performance of homozygosity by descent in two mice lines divergently selected for birth weight environmental variability. Sci Rep 15, 5511 (2025). https://doi.org/10.1038/s41598-025-89254-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89254-z