Abstract
Head and neck squamous cell carcinoma (HNSC) is a common malignant tumor with high incidence and mortality rates. ETS variant transcription factor 4 (ETV4), an important transcription factor, plays a key role in various cancers. However, the role of ETV4 in HNSC remains unclear. This study aimed to explore the potential prognostic value and oncogenic effects of ETV4 in HNSC. We analyzed ETV4 expression in HNSC patients’ data from the TCGA database, alongside clinical pathological characteristics. The STRING and GEPIA databases were utilized to explore ETV4’s interaction proteins and expression related genes. Gene Set Enrichment Analysis (GSEA) was performed on the stratified TCGA-HNSC cohort based on ETV4 expression levels. The correlation between ETV4 expression and immune cells, immune checkpoints, immune regulatory genes was further analyzed using R packages and TISIDB database. Finally, knockdown ETV4 in nasopharyngeal carcinoma cells (NPCs) using siRNA and evaluate cell proliferation, migration, and invasion using CCK-8, wound healing, clone formation, and Transwell assays. ETV4 was significantly overexpressed in HNSC and closely related with clinical pathological characteristics and prognosis. GSEA enrichment analysis showed significant enrichment of ETV4 in multiple immune suppression pathways. Further immune-related analysis indicated that ETV4 negatively correlated with most immune cells, immune checkpoints, tumor-infiltrating lymphocyte type characteristic molecules, immunoinhibitors, immune activators and MHC molecules. Knocking down ETV4 significantly inhibited the proliferation, migration and invasion of NPCs. ETV4 may serve as a prognostic biomarker and immunotherapy target in HNSC. High expression of ETV4 may have a negative regulatory effect on the immune level, matrix components and immune regulatory molecules.
Similar content being viewed by others
Introduction
Head and neck squamous cell carcinoma (HNSC), a malignant tumor originating from the oropharynx, nasopharynx, larynx, and oral cavity, is currently the sixth most common cancer worldwide1. Despite some advancements in the diagnosis and treatment of HNSC in recent years, the incidence and mortality rates remain high2. Therefore, identifying early and effective prognostic biomarkers, and exploring and discovering new therapeutic targets, are of significant importance for the diagnosis and prognosis of HNSC patients. In this context, transcription factors have emerged as important regulators of cancer progression and potential therapeutic targets. Among the various transcription factors, ETS variant transcription factor 4 (ETV4), a member of the ETS family of proteins, has garnered attention for its role in multiple cancer types. ETV4 regulates the transcription of multiple genes, impacting cell proliferation, apoptosis, differentiation, and tumorigenesis3. Studies have reported that ETV4 can promote the malignant progression of colon adenocarcinoma by activating the transcription of the HES1 gene and the phosphorylation of the STAT3 gene4; it can promote the proliferation, migration, and invasion of hepatocellular carcinoma through the activation of the B3GNT3-mediated TGF-β signaling pathway5; and it can also promote the epithelial-mesenchymal transition process of non-small cell lung cancer by activating MMP13, thereby promoting the invasion and metastasis of lung cancer cells6. Additionally, ETV4 can activate cell cycle regulatory genes such as cyclin D1, promoting the rapid transition of pancreatic cancer cells from the G1 to S phase of the cell cycle7. In pancreatic ductal adenocarcinoma, overexpression of ETV4 has also been reported to be associated with an increased risk of distant metastasis of the tumor8. ETV4 also plays a crucial role in angiogenesis in hepatocellular carcinoma by upregulating MMP14 expression, and knocking down ETV4 can significantly reduce vascular formation in hepatocellular carcinoma9. Moreover, it has been found that the expression of ETV4 in various tumors is significantly correlated with immune cell infiltration, tumor heterogeneity, mismatch repair gene expression, DNA methylation, and tumor stemness10. Knocking out ETV4 can also promote autophagy-dependent apoptosis in glioblastoma cells by reducing the transcriptional activation of EMP1, and this effect is achieved by inhibiting the activity of the PI3K/AKT/mTOR signaling pathway11. It is evident that ETV4 plays an important role in various types of tumors.
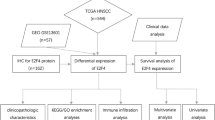
However, the prognostic value, functional role, and potential mechanisms of ETV4 in HNSC are not yet clear. Accordingly, we conducted bioinformatics analysis and experimental validation to examine its expression and prognostic value, co-expressed genes, and functions in HNSC, and explored its correlation with immune infiltration. Furthermore, to validate its functional role, we selected a common subtype of HNSC, nasopharyngeal carcinoma (NPC) cells, for functional verification. NPC, with its distinct biological behaviors and therapeutic challenges, serves as a valuable model for studying cancer aggressiveness and potential therapeutic targets. By focusing on NPC cells, we aim to provide informative insights for the treatment of more aggressive HNSC. In addition, we knocked down the expression of ETV4 in NPC cell lines using siRNA interference technology and analyzed its effects on the growth, invasion, and migration of nasopharyngeal cancer cells through a series of experiments, including CCK-8, Transwell migration and invasion, clone formation, and cell scratch assays. The design and workflow of this study, which outlines the steps from data collection to experimental validation, are detailed in Fig. 1. The results of this study will help to further understand the role of ETV4 in HNSC and provide new clues and targets for the diagnosis and treatment of HNSC.
Materials and methods
Data collection
Expression profile and associated clinical information from the TCGA-HNSC cohort were downloaded and organized from The Cancer Genome Atlas Program database (TCGA) (https://cancergenome.nih.gov/). After standardization using the log2 (TPM + 1) method, the data were used for subsequent analysis. This dataset includes 546 HNSC patients who were divided into high and low expression groups based on the median expression value of ETV4. Various analysis results were visualized using R packages for data visualization.
Reagents and materials
Human immortalized nasopharyngeal epithelial cell line NP69, nasopharyngeal carcinoma cell lines CNE1 and CNE2 (Pricella, China), total RNA extraction kits (Solarbio, China), BeyoRT cDNA first-strand synthesis kit (Beyotime, China), Hiff™ qPCR SYBR® Green Master Mix (Yeasen, China), DMEM/F-12 medium (Gibco, USA), 1640 medium (Pricella, China), fetal bovine serum (Thermo Fisher, China), Anti-ETV4 antibody (1:1000, 10684-1-AP, Proteintech, Wuhan, China), Anti-GAPDH antibody (1:10000, 10494-1-AP, Proteintech, Wuhan, China), Goat Anti-Rabbit Secondary Antibody (H + L) (1:8000, RGAR001, Proteintech, Wuhan, China), CCK-8 cell proliferation assay kit (UElandy, China), BD Matrigel™ Basement Membrane Matrix (matrix gel) (Corning, USA), 4% tissue cell fixative (Solarbio, China), transwell chambers and 0.1% crystal violet staining solution (Jet Bio-Filtration, China) were the main materials of this study. siRNA, RNA transfection reagent, and forward and reverse primers for ETV4 mRNA and GAPDH mRNA were all purchased from Shenggong Biotechnology (Shanghai) Co., Ltd., China.
Method
Expression analysis in pan-cancer
The expression analysis of ETV4 across various cancer types was conducted using R Studio (version 4.2.1). The “ggplot2” (version 3.3.6) package was utilized for generating visualizations, while statistical comparisons were performed using the “stats” (version 4.2.1) and “car” (version 3.1-0) packages. The Wilcoxon rank sum test was selected as the statistical method to evaluate the differential expression of ETV4. The analysis involved comparing ETV4 expression differences across various cancer tissues and corresponding normal tissues, cancer tissues and adjacent tissues, as well as cancer tissues and paired normal tissues.
Expression in HNSC and its relationship with clinical pathological characteristics
The expression levels of ETV4 mRNA in HNSC were analyzed in comparison with normal tissues and paired samples. The “ggplot2” package was utilized to generate box plots visualizing ETV4 expression across various clinical pathological characteristics, including age, sex, T stage, M stage, N stage, and pathological grade. To assess the statistical significance of the differences observed between groups defined by these clinical variables, a one-way ANOVA followed by Tukey HSD post-hoc test was conducted using the “stats” package. Additionally, the diagnostic accuracy of ETV4 was assessed via the construction of receiver operating characteristic (ROC) curves using the “pROC” (version 1.18.0) package, providing a comprehensive evaluation of ETV4 expression in relation to clinical and pathological characteristics in HNSC.
Prognostic value analysis
To evaluate the prognostic significance of ETV4 in HNSC, the “survival” (version 3.3.1) and “rms” (version 6.3-0) packages were employed for univariate and multivariate Cox regression analyses, assessing the influence of various patient characteristics such as gender, age, T/N/M staging, pathological grading, radiotherapy, and ETV4 mRNA expression levels on overall survival (OS). For a visual representation of the Cox regression outcomes, forest plots were generated using the “ggplot2” package. A prognostic nomogram was developed with the “rms” package to predict 1-, 3-, and 5-year survival probabilities for HNSC patients, incorporating risk factor maps to quantitatively assess the impact of each factor. Calibration curves were constructed using the “rms” package to evaluate the model accuracy by comparing predicted and actual survival rates across different time points. Kaplan-Meier survival analysis was performed using the “survminer” (version 0.4.9) package, and the log-rank test was applied to statistically assess differences in OS, disease-specific survival (DSS), and progression-free interval (PFI) between patient groups stratified by the median expression level of ETV4.
PPI and function enrichment analysis
The STRING database (https://www.string-db.org/) pinpointed ETV4-interacting proteins with a confidence level of 0.15 and a limit of 50 interactions, prioritizing “Textmining” and “Experiments”. From the GEPIA database (http://gepia.cancer-pku.cn/index.html), the top 100 genes correlated with ETV4 expression in HNSC were selected and their relationships were graphically represented using scatter plots. The combined data of proteins and genes underwent GO and KEGG12,13,14 enrichment analysis, with the top 10 results presented in bubble plots. The TCGA-HNSC samples were subjected to Gene Set Enrichment Analysis (GSEA) software (version 4.1.0) based on the median expression of ETV4, and the top 5 enrichment items with a normalized enrichment score (NES) rank less than 0 and an adjusted p-value (p.adjust) less than 0.05 were identified, indicating significant biological processes and pathways associated with ETV4 expression levels in HNSC.
Hub gene and activity pathway analysis
An interactive network of ETV4-related genes (a total of 150 genes) was constructed with the STRING database and visualized using Cytoscape software. The MCODE plugin was employed to identify core network modules, while the CytoHubba plugin pinpointed key genes, with the top 10 nodes ranked by MCC designated as hub genes. The expression correlations of these hub genes in HNSC were analyzed, and the findings were visualized as a heatmap using the “ggplot2” package. The active and inhibited pathways involving the hub genes were examined with the GSCALite database (http://bioinfo.life.hust.edu.cn/web/GSCALite/). The 10 hub genes were input into GSCALite, specifying the TCGA-HNSC cancer type and “Pathway Activity” to generate a pie chart of their associated pathways.
Immune infiltration analysis
To evaluate the relationship between ETV4 expression and immune cell infiltration in HNSC, the ssGSEA algorithm provided in the R-package GSVA (version 1.46.0) was employed to estimate the activity of 24 distinct immune cell types. The Spearman correlation coefficient was calculated to quantify the strength of associations, with the results being visualized through lollipop and scatter plots crafted using the “ggplot2 package”. For statistical comparison of immune cell enrichment between groups with high and low ETV4 expression, the Mann-Whitney U test was applied as facilitated by R’s stats package. Furthermore, the estimate algorithm, operational through the estimate package in R, was used to correlate ETV4 expression levels with ImmuneScore, StromalScore, and ESTIMATEScore, providing insights into the potential for non-tumor cell infiltration based on the expression profiles of immune and stromal cell markers.
Immune checkpoint and immune regulatory related genes analysis
The expression profile data from the TCGA-HNSC cohort was used to analyze the correlation between ETV4 expression levels and 16 common immune checkpoint-related genes, including TIGIT, PDCD1, TNFRSF4, LAG3, and CTLA4. The “ggplot2” package was utilized to generate a correlation heatmap, visually representing the relationships. The Spearman correlation method was employed to quantify the association strengths, with the results effectively displayed through the heatmap. Further insights were gained by querying the TISIDB database (http://cis.hku.hk/TISIDB) to analyze the correlation of ETV4 with immune-related characteristic molecules of 28 types of tumor-infiltrating lymphocytes (TILs), immunemodulation factors (including immunoinhibitors, immunostimulators, and MHC class-relatedimmunoinhibitors, immune activators, and MHC molecules), chemokines, and chemokines receptor-related molecules. This comprehensive analysis was performed using the “Lymphocyte”, “Immunomodulator”, and “Chemokine” modules available in the database. The scatter plots of the two cells/molecules with the highest correlation with ETV4 expression in HNSC are shown in the figure.
Cell culture and siRNA interference
CNE1, CNE2 and NP69 cells were cultured in vitro using DMEM/F-12 and 1640 medium with 10% serum, respectively. Total RNA was extracted from these cell lines, and first-strand cDNA was synthesized for subsequent qRT-PCR detection. Amplification was performed on the Roche LightCycler96 PCR instrumen, with pre-denaturation at 95 °C for 300 s; denaturation at 95 °C for 5 s; annealing/extension at 60 °C for 30 s; for a total of 40 cycles. The 2−ΔΔCt method was applied to calculate relative quantification values. Based on the ETV4 mRNA sequence, three siRNAs were designed and selected for transfection into NPCs. After transfection, the knockdown efficiency was detected by qRT-PCR. The primer sequences are shown in Table 1.
Western blotting
Following the qRT-PCR experiments, Western blotting analysis was also conducted on CNE1, CNE2, and NP69 cells, as well as on NPC cells that had been transfected with three types of siRNA. The process involved collecting the cells and lysing them with lysis buffer to extract total proteins. The protein samples were mixed with loading buffer, boiled to denature, and then separated by SDS-PAGE electrophoresis. The proteins were transferred onto a 0.45 μm PVDF membrane, which was subsequently blocked with a solution of 5% non-fat dry milk in TBST to reduce non-specific binding. The membrane was incubated with specific primary antibodies, washed, and then incubated with HRP-conjugated secondary antibodies. Finally, the proteins were detected by treating the membrane with a luminol-based reagent and exposing it to film, which allowed for the assessment of the expression changes of ETV4 protein in both tumor and normal cells, as well as the effects of siRNA transfection.
Cell viability assay (CCK8)
Transfected NPCs were seeded in 96-well plate at a density of 2000 cells per well. Each group had six replicates. At 0, 24, 48, and 72 h, 10 µL of CCK-8 reagent was added to each well, followed by a 2-hour incubation. The optical density (OD) at a wavelength of 450 nm was measured using a spectrophotometer. Growth curves were generated, and survival rates were calculated for each group.
Clone formation assay
Transfected NPCs were inoculated into six-well plates at 1000 cells per well and cultured in the incubator for 14 days. The culture medium was refreshed every 2–3 days, monitoring for clone formation. Once formed, cells were fixed with 4% tissue cell fixative for 30 min, stained with 0.1% crystal violet for 20 min, air-dried, photographed, and the images documented.
Wound healing assay
Transfected NPCs were seeded in six-well plates and cultured to reach approximately 90% confluence. Wounds were introduced by making 3–4 horizontal lines across the well using a 10 µL pipette tip. After removing detached cells with sterile PBS, fresh medium was added. The wounds were photographed under an inverted microscope at 0 and 24 h post-wounding.
Transwell migration and invasion assay
Transfected NPCs, after a 12-hour starvation period, were trypsinized and resuspended to a density of 4 × 104 cells/mL in serum-free medium. In the migration assay, 300 µL of the cell suspension was added to the upper chamber, and 600 µL of medium with 20% FBS was filled in the lower chamber. The invasion assay included the preliminary step of coating the upper chamber with 100 µL diluted Matrigel, allowed to solidify for 1–2 h at 37 °C. The cell suspension was then added, and both assays were conducted over 24 to 48 h. After the incubation, chambers underwent PBS washing, followed by cell fixation with 4% fixative for 30 min, and staining with 0.1% crystal violet for 20 min. After drying, the chambers were photographed under an inverted microscope and the pictures were saved.
Statistical analysis
The Wilcoxon test was employed to assess the significance of ETV4 expression between tumor and adjacent tissues. The Logrank test determined the significance of survival rate differences between groups with high and low ETV4 expression levels. The correlation of ETV4 with its co-expressed genes, as well as immune cell infiltration, was evaluated using Spearman correlation analysis. This analysis examined the correlation of ETV4 with tumor-infiltrating lymphocytes (TILs), immune regulatory factors, and inflammatory chemokines. Experimental data, including qRT-PCR and Western blotting results, were presented as mean ± standard deviation (Mean ± SD). The independent-sample t-test was utilized to compare the means of two normally distributed samples with homogeneous variances. For multiple comparisons, one-way analysis of variance (ANOVA) was used. Statistical significance was set at P < 0.05.
Results
Expression of ETV4 in pan-cancer
The expression analysis across pan-cancer showed that ETV4 is highly expressed in 24 cancer types compared to normal tissues. These include bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), cervical squamous cell carcinoma and endocervical adenocarcinoma (CESC), cholangiocarcinoma (CHOL), colon adenocarcinoma (COAD), diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), HNSC, kidney chromophobe (KICH), brain lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian serous cystadenocarcinoma (OV), pancreatic adenocarcinoma (PAAD), rectum adenocarcinoma (READ), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), testicular germ cell tumors (TGCT), thyroid carcinoma (THCA), thymoma (THYM), uterine corpus endometrial carcinoma (UCEC), and uterine carcinosarcoma (UCS) (Fig. 2A). Compared to adjacent tissues, ETV4 was upregulated in 15 cancer types, notably in HNSC, BLCA, CHOL, COAD, ESCA, GBM, and KICH (Fig. 2B). In paired comparisons of tumor and normal tissues, ETV4 was generally upregulated in cancers such as BLCA, BRCA, CHOL, COAD, ESCA, HNSC, LIHC, LUAD, LUSC, READ, STAD, THCA, and UCEC (Fig. 2C). The findings indicate a broad upregulation of ETV4 across various tumor tissues, including HNSC.
Expression of ETV4 in pan-cancer. Note: (A) Box plot illustrating the differential expression of ETV4 in 33 tumor tissues compared to normal tissues; (B) Box plot showing ETV4 expression differences between 33 tumor tissues and their corresponding adjacent tissues; (C) Comparative analysis of ETV4 expression in 18 tumor tissues and normal paired normal tissues.
ETV4 expression in HNSC and correlation with clinical pathological characteristics
Analysis of the TCGA-HNSC cohort data (Table 2) revealed a significant upregulation of ETV4 in HNSC tissues compared to normal or paired tissues (P < 0.001) (Fig. 3A, B). The diagnostic ROC curve demonstrated high diagnostic value for HNSC (AUC = 0.758, 95% CI: 0.698–0.817) (Fig. 3C). ETV4 expression was notably correlated with age, sex, pathological T stage, M stage, N stage, and clinical grade of HNSC patients, with higher levels observed in those aged 60 years or older, male, and patients with pathological stage T1 or higher, distant or lymph node metastasis, and pathological grade III or higher (Fig. 3D-I) .
Expression of ETV4 in HNSC and correlation with clinical variables. Note: (A) Box plot showing the expression difference of ETV4 between HNSC tumor tissues and normal tissues; (B) Paired sample comparison of ETV4 expression in HNSC tumor tissues and normal tissues; (C) ROC curve representing the diagnostic potential of ETV4 in HNSC; (D)-(I) Graphical representation of the association between ETV4 mRNA expression and various clinical variables in HNSC patients, including age (D), gender (E), T stages (F), M stages (G), N stages (H), and pathological grades (I).
Prognostic value of ETV4 in HNSC
Univariate and multivariate Cox regression analyses identified T3 & T4 stage tumors (P < 0.001, HR: 1.934 [1.413–2.649]), N2 & N3 stage (P < 0.001, HR: 2.296 [1.686–3.127]), M1 stage (P = 0.003, HR: 22.631 [2.830–180.948]), pathological stages III + IV ((P = 0.003, HR: 1.839 [1.236–2.737]), radiation therapy (P = 0.002, HR: 0.617 [0.455–0.837]), and high ETV4 expression (P = 0.002, HR: 1.539 [1.172–2.020]) as prognostic risk factors for HNSC (Fig. 4A). Independent risk factors included N2&N3 stage (P < 0.001, HR: 4.769 [2.489–9.107]), pathological stages III + IV (P = 0.034, HR: 4.759 [1.124–20.146]), and high ETV4 expression (P = 0.03, HR: 1.874 [1.061–3.307]), with radiation therapy (P < 0.001, HR: 0.121 [0.061–3.307]) as a protective factor (Fig. 4B). Constructed nomograms based on clinical features and ETV4 expression levels predicted the 1-, 3-, and 5-year survival of HNSC patients with good accuracy (Fig. 4C). The calibration curves also showed good agreement between the predicted and actual survival rates for HNSC patients at 1-year, 3-year, and 5-year survival (Fig. 4D-F). The risk factor plots depicted the correlation between ETV4 expression and survival outcomes, showing increased disease progression and metastatic risk with higher ETV4 expression (Fig. 4G). Patients with elevated ETV4 expression had significantly shorter OS, DSS, and PFI (P < 0.05), indicating ETV4 as a poor prognostic indicator for HNSC (Fig. 4H-J).
Prognostic value of ETV4 expression in HNSC. Note: (A) Univariate Cox regression analysis of clinical pathological characteristics in HNSC patients; (B) Multivariate Cox regression analysis of clinical pathological characteristics in HNSC patients; (C) Prognostic nomogram based on clinical variables and ETV4 expression to predict overall survival; (D)-(F) Calibration curves validating the nomogram’s predictive accuracy for HNSC patients survival of 1-, 3-, and 5-year; (G) Risk factor plot displaying the prognostic model’s risk score and stratification; (H)-(J) Kaplan-Meier survival curves comparing the overall survival (OS), disease-specific survival (DSS), and progression-free interval (PFI) for patients with different levels of ETV4 expression.
Functional enrichment of ETV4 interaction and expression related genes
The STRING database analysis identified 50 proteins, including COP1, EWSR1, TMPRSS2, SLC45A3, CD99, GDNF, Eppin, LPP, SCGB2A2, HIF1A, NCOA3, and others, interacting closely with ETV4 (Fig. 5A). The GEPIA database analysis of the top 100 genes associated with ETV4 expression in HNSC revealed significant positive correlations with ETV5, SLCO3A1, and SPAST (Fig. 5B-D). Intersection analysis uncovered SPRY2 as a common gene(Fig. 5E). Functional enrichment of these genes indicated involvement in processes like branching morphogenesis of an epithelial tube, intracellular steroid hormone receptor signaling pathway, and regulation of animal organ morphogenesis, with molecular functions including DNA-binding transcription factor binding, DNA-binding transcription activator activity, RNA polymerase ll-specific, and nuclear receptor binding (Fig. 5F). GSEA analysis of the TCGA-HNSC cohort, stratified by ETV4 expression, showed that high ETV4 expression samples were significantly enriched for GO terms related to lymphocyte mediated immunity, B cell receptor signaling pathway, antigen binding, immunoglobulin complex, and T cell receptor complex, while immune pathways such as antigen processing and presentation, natural killer cell mediated cytotoxicity, cytokine cytokine receptor interactions, and allograft rejection were significantly suppressed (Fig. 5G). This suggests a potential role for ETV4 in the tumor immune process, with a possible negative regulation of tumor immunity.
Functional enrichment analysis of ETV4 interaction and expression related genes. Note: (A) Protein-protein interaction (PPI) network of ETV4, where the lines connecting the proteins are color-coded to represent different types of evidence supporting these interactions: red for experimentally verified, blue for database-confirmed, green for co-expression based, purple for neighborhood based, yellow for gene fusion based, and black for co-occurrence based; (B)-(D) Expression correlation plots for the top 3 genes most strongly correlated with ETV4; (E) Venn diagram representing the intersection of ETV4 interaction and expression-related genes; (F) GO and KEGG enrichment analysis of ETV4 interaction and expression-related genes; (G) Gene Set Enrichment Analysis (GSEA) for HNSC patient cohorts stratified by ETV4 expression levels.
Hub genes in the ETV4 co-expression network and associated pathways
The ETV4 co-expression gene network was reconstructed using the STRING database and Cytoscape software, identifying 2 core sub-networks with the MCODE plugin (Fig. 6A, B). The CytoHubba plugin pinpointed 10 key genes (Hub genes) in the network, including EP300, ERBB2, ESR1, ETV4, HIF1A, JUN, MYCN, NFKB1, SOX2, and STAT1 (Fig. 6C). ETV4 was among these hub genes, which showed strong positive correlations in HNSC (Fig. 6D). Functional enrichment analysi showed their involvement in processes such as epithelial cell proliferation, positive regulation of transcription from RNA polymerase ll promoter, and cellular response to chemical stimulus, within cellular components like the transcription initiation complex, RNA polymerase ll transcription regulator complex, and euchromatin, with molecular functions including DNA-binding transcription activator activity, RNA polymerase ll-specific DNA-binding transcription activator activity, transcriptional coactivator binding, and transcription coregulator binding (Fig. 6E). Pathway enrichment highlighted the HIF-1 signaling pathway, Th17 cell differentiation, PD-L1 expression and PD-1 checkpoint pathway in cancer and thyroid hormone signaling pathway (Fig. 6E). Further analysis using the GSCALite database showed these hub genes were mainly involved in the regulation of cell apoptosis, cell cycle, DNA damage response, epithelial mesenchymal transition (EMT), androgen receptor (AR), estrogen receptor (ER) and other pathways. ETV4 was particularly implicated in the activation of cell cycle regulation, EMT induction, and the suppression of AR, ER, PI3K/AKT, and PAS/MAPK signaling pathways (Fig. 6F). This suggests a complex role for ETV4 and associated hub genes in the regulation of cancer-related pathways.
Hub gene analysis in the ETV4 co-expression network. Note: (A) ETV4 co-expression network constructed using the STRING database and Cytoscape software; (B) Core sub-networks identified using the MCODE plugin; (C) Top 10 hub genes based on the MCC ranking by the CytoHubba plugin; (D) Heatmap of the expression correlation of the 10 hub genes in HNSC; (E) GO and KEGG enrichment analysis of the 10 hub genes; (F) Active and inhibitory pathways involving the 10 hub genes.
Correlation of ETV4 expression with immune cell infiltration and checkpoint
The lollipop plot depicted a predominantly negative correlation between ETV4 expression and 24 immune cell types, with only a few showing positive correlation (Fig. 7A). The scatter plot detailed negative associations with immune cells such as CD56dim NK cells (R = -0.374, P < 0.001), cytotoxic cells (R = -0.283, P < 0.001), T cells (R = -0.294, P < 0.001), Th17 cells (R = -0.278, P < 0.001), CD8 T cells (R = -0.260, P < 0.001), antigen-presenting dendritic cells (aDC) (R = -0.269, P < 0.001), regulatory T cells (TReg) (R = -0.245, P < 0.001), immature dendritic cells (iDC) (R = -0.233, P < 0.001), and B cells (R = -0.204, P < 0.001), and a positive correlation with CD56bright NK cells (R = 0.216, P < 0.001) (Fig. 7B). Further analysis showed that ETV4 expression influenced the infiltration of 16 immune cell types, such as aDC, B cells, CD8 T cells, cytotoxic cells, eosinophils, CD56bright NK cells, iDC, neutrophils, with significantly lower proportions in the high ETV4 expression group (P < 0.01) (Fig. 7C). The Estimate algorithm revealed a negative correlation between ETV4 expression and scores like ImmuneScore, StromalScore, and ESTIMATEScore, indicating a potential suppressive effect of high ETV4 expression on the immune landscape and tumor microenvironment in HNSC (Fig. 7D). Additionally, ETV4 showed negative correlations with numerous immune checkpoint genes, including TIGIT, TNFRSF4, LAG3, CTLA4, TNFSF4, ICOS, LAIR1, and BTLA (P < 0.05) (Fig. 7E).
Correlation of ETV4 expression with immune cell infiltration and immune checkpoint in HNSC. Note: (A) Lollipop plot of the correlation between ETV4 expression and 24 immune cell types infiltration; (B) Scatter plots showing the correlation between ETV4 expression and various immune cells; (C) Dot plots of immune infiltration scores for high and low expression of ETV4; (D) Scatter plots of the correlation between the ETV4 and ImmuneScore, StromalScore, and ESTIMATEScore; (E) Heatmap of the correlation between ETV4 and 16 common immune checkpoint-related genes.
Correlation of ETV4 with immunoregulatory genes such as immunoinhibitors, immunostimulators and MHC class-related molecules
Analysis using the TISIDB database across 33 tumor types revealed that in HNSC, ETV4 expression was negatively correlated with immune-related characteristic molecules of TILs, including Act CD8, Tem CD8, Tfh, Th1, Imm B, NK, MDSC, NKT, iDC, with Imm B and Tem CD8 showing the highest correlation (Fig. 8A). ETV4 expression was also negatively correlated with immunoinhibitor-related genes like BTLA, CD244, CD96, CSF1R, TIGIT, LAG3, and IL10, with CD96 and CSF1R being the top 2 correlated genes (Fig. 8B). Similarly, a negative correlation was observed with immunostimulator-related genes, including CD27, CD28, CD40LG, CD48, CD80, ICOS, KLRC1, TMEM173, and TNFRSF9, with KLRC1 and TNFRSF9 showing the highest correlation (Fig. 8C). MHC class-related molecules such as B2M, HLA-B, HLA-C, HLA-DMA, HLA-DMB, HLA-DOA, HLA-DPA1, HLA-DRA also showed a negative correlation with ETV4 expression, with HLA-DPA1 and HLA-DRA being the most correlated (Fig. 8D). Chemokine analysis indicated a negative correlation with ETV4 expression for CCL2, CCL4, CCL5, CCL8, CCL14, CCL19, CCL28, and CXCL9, and a positive correlation for CCL26, CXCL5, CXCL8, with CXCL9 and CCL26 being the most strongly correlated, respectively (Fig. 8E). Additionally, ETV4 expression also negatively correlated with chemokine receptors CCR1, CCR2, CCR4, CCR5, CCR6, CCR8, CXCR3, CXCL6, and CX3CR1, with CCR6 and CX3CR1 showing the highest correlation (Fig. 8F). These correlations suggest that ETV4 may serve as a potential target in cancer immunotherapy due to its general negative association with immune regulatory genes.
Correlation of ETV4 expression with immunoregulatory genes.Note: (A) Correlation of ETV4 expression with tumor-infiltrating lymphocytes (TILs); (B)-(D) Correlation of ETV4 expression with three types of immune modulators (including immunoinhibitors immunostimulators, and MHC class-related molecules); (E)-(F) Correlation of ETV4 expression with chemokines and their receptors. Also on the right side of each figure, the two cells/molecules with the highest degree of correlation (positive or negative) with ETV4 expression are displayed.
Verification of ETV4 expression and its impact on biological behavior of NPC
Compared with NP69, both qRT-PCR and Western blotting experiments revealed significantly increased levels of ETV4 mRNA and protein in CNE1 and CNE2 (P < 0.01) (Fig. 9A). Among them, CNE1 cells were chosen for knockdown experiments. All three siRNAs effectively reduced ETV4 expression, as demonstrated by qRT-PCR results, with si-ETV4-1 and si-ETV4-2 showing the greatest efficacy (Fig. 9B). CCK8 assays indicated a significant decrease in the proliferative capacity of NPCs following ETV4 knockdown (P < 0.01) (Fig. 9C). Similarly, colony formation assays revealed a marked reduction in NPC proliferation/cloning ability (P < 0.001) (Fig. 9D). Wound healing assays showed a significant decrease in the migration capacity of NPCs (P < 0.001) (Fig. 9E). Transwell migration and invasion assays further confirmed the role of ETV4 in cell migration and invasion, demonstrating a substantial decrease in the number of migrating and invading cells following ETV4 knockdown (P < 0.001) (Fig. 9F, G). These results underscore the role of ETV4 in the proliferation, migration and invasion of NPCs.
Validation of ETV4 expression and its impact on the proliferation, migration, and invasion of nasopharyngeal carcinoma cell. Note: (A) The expression levels of ETV4 mRNA and protein in nasopharyngeal carcinoma cell lines CNE1 and CNE2; (B) The knockdown efficiency of ETV4 by three siRNAs assessed using quantitative real-time PCR (qRT-PCR) and Western blotting; (C) CCK-8 assay was used to evaluate cell proliferation; (D) Colony formation assay was conducted to assess the proliferation/cloning ability; (E) Wound healing assay was performed to determine cell migration capability; (F)-(G) Transwell migration and invasion assay were used to evaluate the migratory and invasive capabilities of cell, respectively.
Discussion
Transcription factors play a crucial role in cellular differentiation, development, and stress responses, and their dysregulation or mutation may lead to altered cellular metabolic processes, immune responses, and even trigger oncogenic signaling cascades15. More and more studies have emphasized the central role of transcription factors in cancer biology and have identified transcription factors as one of the potential strategies for targeted therapy16. The ETS family of transcription factors, comprising multiple members, often leads to oncogenic changes when their activity is disrupted17. ETV4, as a member of the ETS transcription factor family, has been found to play an important role in various cancers, especially as a tumor initiator promoting tumor growth, invasion, and metastasis3. However, the role and underlying mechanisms of ETV4 in HNSC are not yet clear. Our study found that ETV4 is highly expressed in various tumors, including HNSC, and that high expression of ETV4 is closely associated with the clinical pathological characteristics and prognosis of HNSC patients. This suggests that ETV4 likely plays a key role in the development of HNSC. In-depth investigation of the function and regulatory mechanisms of ETV4 may provide new insights and approaches for the treatment and prevention of HNSC. We found that more than 50 proteins, such as COP1, EWSR1, TMPRSS2, SLC45A3, CD99, GDNF, EPPIN, LPP, SCGB2A2, HIF1A, and NCOA3, interact closely with ETV4. These proteins are known to be closely related to the occurrence and development of other tumors. For example, COP1 has been found to promote the proliferation, invasion, and migration of glioma cells by ubiquitinating other proteins18; overexpression of EWSR1 (Ewing sarcoma breakpoint region 1/EWS RNA binding protein 1) has been recognized as an oncogenic molecule capable of promoting the development of multiple myeloma19; CD99 has been reported as an overexpressed transmembrane protein in various malignancies and is also considered a potential target for immunotherapy20. Therefore, the interaction of ETV4 with these proteins may form a complex regulatory network affecting the occurrence and development of HNSC. When analyzing the genes associated with ETV4 expression in HNSC, we found that some genes, such as ETV5, SLCO3A1, and SPAST, were significantly and positively correlated with the expression of ETV4. ETV5 has been demonstrated to activate the transcription of Slug genes to promote the malignant progression of HNSC21; SLCO3A1 has also been reported as a prognostic model gene for HNSC, exerting a significant impact on the prognosis of HNSC22. It is evident that these interacting proteins and expression related genes may have a synergistic effect, jointly influencing the developmental process of HNSC.
Further, we conducted enrichment analyses on the functions and pathways they participate in, and found that they were primarily involved in the branching morphogenesis of an epithelial tube, intracellular steroid hormone receptor signaling pathway, and regulation of animal organ morphogenesis, etc. And these processes we know are important for tumor development. During the process of tumorigenesis, the normal structure and morphology of epithelial tissues are likely to be disrupted or altered, resulting in the appearance of abnormal branching morphology23. Such abnormal tissue development is likely to promote tumor growth and spread. Studies have also reported that the abnormal activation of steroid hormone receptor signaling pathways may lead to abnormal proliferation and invasion of tumor cells24. Therefore, the involvement of ETV4 in the regulation of these biological processes may also have a significant impact on the development of HNSC.
To gain a more comprehensive understanding of the functions and pathways involved in the ETV4 gene, we performed a single-gene enrichment analysis (GSEA) based on the stratified TCGA-HNSC cohort by ETV4 expression levels. The results revealed that in samples with high ETV4 expression, the top five significantly enriched GO terms were lymphocyte mediated immunity, B cell receptor signaling pathway, antigen binding, immunoglobulin complex, and T cell receptor complex. Immune pathways such as antigen processing and presentation, natural killer cell mediated cytotoxicity, cytokine cytokine receptor interactions, and allograft rejection were significantly suppressed. This suggests that ETV4 is likely involved in the immune processes of HNSC and may negatively regulate tumor immunity in HNSC. Previous studies have reported that ETV4 can mediate immune escape through programmed death ligand-1 (PD-L1) and programmed death ligand-2 (PD-L2), and the genetic knockout of ETV4 can significantly reduce the expression of PD-L1 and PD-L225. PD-L1 and PD-L2 constitute an important anti-tumor immune response26, and their overexpression in the tumor microenvironment are usually associated with immune escape and tumor immune tolerance. Therefore, by regulating the expression of PD-L1 and PD-L2, ETV4 may affect the interaction between HNSC tumor cells and immune cells, thereby modulating the intensity and effectiveness of the immune response. Thus, the role of ETV4 in HNSC may not only be limited to promoting tumor cell proliferation and invasion, but may also influence the immune response of HNSC tumors by regulating immune escape pathways.
To further understand the relationship between ETV4 and HNSC tumor immunity, we performed correlation analyses of ETV4 with 24 immune cell types, 16 common immune checkpoints, 28 types of tumor-infiltrating lymphocytes (TILs), and various immune regulatory genes. These results demonstrated an overall negative correlation between ETV4 expression levels and the presence and activity of these immune components, highlighting ETV4’s potential as a modulator of the immune landscape in HNSC. Especially regarding immune cells, we found a negative correlation between ETV4 and several key types of immune cells, including CD56dim NK cells, cytotoxic cells, T cells, Th17 cells, CD8 T cells, aDC, TReg, iDC, and B cells. These findings suggest that high expression of ETV4 may lead to impaired quantity and function of immune cells in HNSC patients, thereby weakening the immune surveillance and clearance capacity against tumors27. Some immune cells secrete various cytokines and chemokines, which can also promote tumor growth28. For instance, cytokines such as IL-6 and IL-10 have been reported to suppress tumor specific immune responses and alter the differentiation and function of immune/inflammatory cells through the NF-κB and STAT3 signaling pathways29. Therefore, ETV4 may indirectly facilitate the growth and development of HNSC by negatively regulating them.
Additionally, immune checkpoints play a critical role in maintaining immune balance by modulating the activation and function of immune cells. However, tumor cells can exploit immune checkpoint signaling pathways to evade immune surveillance and attack30. In our study, we observed a negative correlation between ETV4 and various immune checkpoint-related genes, such as TIGIT, TNFRSF4, LAG3, CTLA4, TNFSF4, ICOS, LAIR1, and BTLA. The combinations of TNFRSF4, CTLA4, ICOS, and BTLA with PD-L1 can predict overall survival in patients with oral squamous cell carcinoma31; whereas TIGIT is also considered to play a key role in immune suppression and is associated with a favorable prognosis in HNSC32. These findings suggest that the negative correlation of ETV4 with these immune checkpoints may further emphasize its potential role in tumor immune evasion. High expression of ETV4 may suppress the expression or function of these immune checkpoints, affecting T cell activation and effector functions, thereby disrupting the immune response. Furthermore, we also found that ETV4 was negatively correlated with various immune-related characteristic molecules and immune regulatory genes. In summary, these findings underscore the multifaceted influence of ETV4, potentially revealing it as a key node in the immunosuppressive mechanisms of HNSC. Understanding its interactions with them could be crucial for developing strategies to counteract tumor immune evasion and enhance immunotherapeutic responses.
To further validate the functional role of ETV4, we utilized nasopharyngeal carcinoma (NPC) cells for functional validation. NPC, with its distinct biological behaviors and therapeutic challenges, serves as a valuable model for studying aggressiveness and potential therapeutic targets in cancer, especially in the context of HNSC, as highlighted in recent reviews33. By focusing on NPC cells, which are among the most common subtypes of HNSC in certain populations and possess distinctive epidemiological and molecular characteristics, we aim to provide insights beneficial for treating more aggressive forms of HNSC. Our study ultimately found that knockdown of ETV4 led to a reduction in NPC cell proliferation, migration, and invasion, which aligns with the role of ETV4 in promoting other malignant tumors3. This provides a solid foundation for understanding the role of ETV4 in HNSC and offers crucial information for future research, particularly in highly invasive and difficult-to-treat cases of HNSC. Of course, our study also has certain limitations. Future research will expand to include other subtypes of HNSC, such as oral cavity and laryngeal cancers, to provide a more comprehensive understanding of ETV4’s role across the full spectrum of HNSC.
Conclusion
The study establishes ETV4 as a significant prognostic biomarker and potential immunotherapy target in HNSC. Its elevated expression correlates with key clinical features and is indicative of a poorer prognosis. The study suggests that high ETV4 levels may exert a negative regulatory effect on immune levels, matrix components, and the overall tumor microenvironment. These insights support the potential of ETV4 as a therapeutic target, offering new directions for HNSC treatment strategies aimed at modulating immune responses.
Data availability
All data generated or analysed during this study are included in this published article.
References
Peng, Q. et al. Clinical significance and integrative analysis of the cuproptosis-associated genes in head and neck squamous cell carcinoma. Aging (Albany NY). 15, 1964–1976 (2023).
Yu, W., He, X., Zhang, C. & Huangfu, H. Transcriptomics data mining to uncover signature genes in head and neck squamous cell carcinoma: a bioinformatics analysis and rna-sequencing based validation. Am. J. Cancer Res. 13, 5513–5530 (2023).
Jiang, W., Xu, Y., Chen, X., Pan, S. & Zhu, X. E26 transformation-specific variant 4 as a tumor promotor in human cancers through specific molecular mechanisms. Mol. Ther. Oncolytics. 22, 518–527 (2021).
Yao, D., Bao, Z., Qian, X., Yang, Y. & Mao, Z. Etv4 transcriptionally activates hes1 and promotes stat3 phosphorylation to promote malignant behaviors of colon adenocarcinoma. Cell. Biol. Int. 45, 2129–2139 (2021).
Zhou, Z. et al. Etv4 facilitates proliferation, migration, and invasion of liver cancer by mediating tgf-beta signal transduction through activation of b3gnt3. Genes Genomics. 45, 1433–1443 (2023).
Qian, X. J., Wang, J. W., Liu, J. B. & Yu, X. The mediating role of mir-451/etv4/mmp13 signaling axis on epithelialmesenchymal transition in promoting non-small cell lung cancer progression. Curr. Mol. Pharmacol. 17, e269821484 (2024).
Tyagi, N. et al. Etv4 facilitates cell-cycle progression in pancreatic cells through transcriptional regulation of cyclin d1. Mol. Cancer Res. 16, 187–196 (2018).
Gao, X. et al. Etv4 promotes pancreatic ductal adenocarcinoma metastasis through activation of the cxcl13/cxcr5 signaling axis. Cancer Lett. 524, 42–56 (2022).
Su, H. et al. Etv4 facilitates angiogenesis in hepatocellular carcinoma by upregulating mmp14 expression. Biochem. Biophys. Res. Commun. 684, 149137 (2023).
Zhang, R. et al. Oncogenic role and drug sensitivity of etv4 in human tumors: a pan-cancer analysis. Front. Oncol. 13, 1121258 (2023).
Wang, J. et al. Knockdown of etv4 promotes autophagy-dependent apoptosis in gbm cells by reducing the transcriptional activation of emp1. Oncol. Lett. 23, 41 (2022).
Ogata, H. et al. Kegg: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 27, 29–34 (1999).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. Kegg for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587–D592 (2023).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Hasan, A., Khan, N. A., Uddin, S., Khan, A. Q. & Steinhoff, M. Deregulated transcription factors in the emerging cancer hallmarks. Semin Cancer Biol. 98, 31–50 (2024).
Jeong, D. W., Lee, S. & Chun, Y. S. How cancer cells remodel lipid metabolism: strategies targeting transcription factors. Lipids Health Dis. 20, 163 (2021).
Wang, Y., Huang, Z., Sun, M., Huang, W. & Xia, L. Ets transcription factors: multifaceted players from cancer progression to tumor immunity. Biochim. Biophys. Acta Rev. Cancer. 1878, 188872 (2023).
Zhang, W. & Wu, Z. Cop1 facilitates the proliferation, invasion, and migration of glioma cells by ubiquitination of dlg3 protein. Neurol. Res. 45, 858–866 (2023).
Nishiyama, D. et al. Ewsr1 overexpression is a pro-oncogenic event in multiple myeloma. Int. J. Hematol. 113, 381–394 (2021).
Cardoso, L. C. et al. Cd99 expression in glioblastoma molecular subtypes and role in migration and invasion. Int. J. Mol. Sci. ; 20. (2019).
Yu, S. et al. Etv5 facilitates tumor progression in head-neck squamous cell carcinoma. Oral Dis. (2023).
Ge, Y., Xia, J., He, C. & Liu, S. Identification of circrna-lncrna-mirna-mrna competitive endogenous rna networks as prognostic bio-markers in head and neck squamous cell carcinoma. Technol. Health Care. 31, 2193–2212 (2023).
Kahata, K., Maturi, V. & Moustakas, A. Tgf-beta family signaling in ductal differentiation and branching morphogenesis. Cold Spring Harb Perspect. Biol. ; 10. (2018).
Groner, A. C. & Brown, M. Role of steroid receptor and coregulator mutations in hormone-dependent cancers. J. Clin. Invest. 127, 1126–1135 (2017).
Ma, P. et al. Super-enhancer receives signals from the extracellular matrix to induce pd-l1-mediated immune evasion via integrin/braf/tak1/erk/etv4 signaling. Cancer Biol. Med. 19, 669–684 (2021).
Zeynep, O. et al. Pd-l1 and pd-l2 expression in colorectal cancer. Indian J. Pathol. Microbiol. 66, 31–37 (2023).
Mukherjee, N. et al. Klrf1, a novel marker of cd56(bright) nk cells, predicts improved survival for patients with locally advanced bladder cancer. Cancer Med. 12, 8970–8980 (2023).
Lu, C., Liu, Y., Ali, N. M., Zhang, B. & Cui, X. The role of innate immune cells in the tumor microenvironment and research progress in anti-tumor therapy. Front. Immunol. 13, 1039260 (2022).
Li, L. et al. Effects of immune cells and cytokines on inflammation and immunosuppression in the tumor microenvironment. Int. Immunopharmacol. 88, 106939 (2020).
Dulal, D. et al. Tackling of immunorefractory tumors by targeting alternative immune checkpoints. Cancers (Basel) ; 15. (2023).
Xu, X. et al. Etv4 potentiates nuclear yap retention and activities to enhance the progression of hepatocellular carcinoma. Cancer Lett. 537, 215640 (2022).
Wen, J., Mao, X., Cheng, Q., Liu, Z. & Liu, F. A pan-cancer analysis revealing the role of tigit in tumor microenvironment. Sci. Rep. 11, 22502 (2021).
Lee, A. W., Ma, B. B., Ng, W. T. & Chan, A. T. Management of nasopharyngeal carcinoma: current practice and future perspective. J. Clin. Oncol. 33, 3356–3364 (2015).
Acknowledgements
We appreciate the databases of TCGA, STRING, GEPIA, TISIDB and GEO for the availability of the data.
Author information
Authors and Affiliations
Contributions
Y. T. has contributed to the experimental design, supervision and writing of the manuscrip; L. T. reviewed and edited the manuscript; W. X., T. H., C. H., W. Y., S. W. and F. L. participated in the experiment, data processing and visualization.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tang, Y., Xie, W., Hu, T. et al. Identification of ETV4 as a prognostic biomarker and correlates with immune cell infiltration in head and neck squamous cell carcinoma. Sci Rep 15, 7044 (2025). https://doi.org/10.1038/s41598-025-90731-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-90731-8
Keywords
This article is cited by
-
Plasma epigenomic profiling reveals treatment-emergent squamous transformation in prostate cancer
npj Precision Oncology (2025)