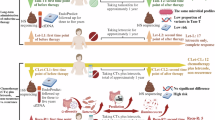
Abstract
Tamoxifen is essential in treating estrogen receptor-positive (ER+) breast cancer, primarily through its active metabolite, endoxifen. Emerging research suggests potential interactions between tamoxifen and gut microbiota. This study investigates the effects of tamoxifen on gut microbiota composition in postmenopausal ER+ and human epidermal growth factor receptor 2 negative (HER2−) breast cancer patients and explores correlations between gut microbiota and endoxifen plasma levels. This prospective observational study included postmenopausal ER+/HER2− breast cancer patients. Fecal and blood samples were collected before and during 6–12 weeks of tamoxifen therapy. Gut microbiota composition was analyzed using 16S rRNA amplicon sequencing of the hypervariable V4 gene region, and plasma endoxifen levels were measured using liquid chromatography-mass spectrometry. Changes in microbial diversity and composition were assessed, with correlations to endoxifen levels. A total of 62 patients were included. Tamoxifen significantly increased microbial richness (p = 0.019), although overall community structure remained consistent between pre- and during-treatment samples. Notable changes were observed in specific microbial taxa, with significant increases in genera such as Blautia (padjusted = 0.003) and Streptococcus (padjusted = 0.010), and decreases in Prevotella_9 (padjusted = 0.006). No significant correlations between gut microbiota and endoxifen levels were identified after multiple comparisons. Tamoxifen therapy increases gut microbial diversity in postmenopausal ER+/HER2− breast cancer patients, though overall microbial community structure remains stable. The absence of significant correlations with endoxifen levels suggests that while tamoxifen affects the gut microbiota, its role in endoxifen metabolism requires further study. More comprehensive research is needed to understand the relationship between tamoxifen, gut microbiota, and therapeutic outcomes.
Similar content being viewed by others
Introduction
Anti-hormonal therapy with tamoxifen is a cornerstone in the treatment of estrogen receptor-positive breast cancer. Tamoxifen and its active metabolites compete with estrogen for binding sites in tissues that express estrogen receptors (ER), such as the mammary epithelium, thereby selectively modulating receptor binding of estrogen1,2. Tamoxifen has a complex metabolism, but generally, it is broken down by different enzymes in the liver into two main parts: N-desmethyl-tamoxifen (NDM-tamoxifen) and 4-hydroxy-tamoxifen. These metabolites are then further converted into the primary and active metabolite endoxifen3. In the adjuvant setting, tamoxifen is given continuously in premenopausal patients or for 2–3 years before switching to aromatase inhibitors in postmenopausal patients3. Although tamoxifen significantly improves survival rates for patients with ER-positive breast cancer, responses to tamoxifen can vary4. The variability in patient responses to tamoxifen has been partially attributed to inter-individual differences in levels of endoxifen as endoxifen has a much greater affinity for ER compared to tamoxifen3. Previous research indicates that approximately 11–24% of patients do not achieve the proposed target endoxifen plasma concentration required for optimal efficacy5,6. Besides treatment response, variability in endoxifen levels is also associated with side effects, as indicated by a study by Lorizio et al.7. The most common side effects of tamoxifen include hot flashes (64%), vaginal dryness (35%), sleep problems (36%), weight gain (6%), and depression, irritability, or mood swings (6%)7. Inter-individual variability of endoxifen levels is influenced by individual differences in drug metabolism and its complex metabolic pathway6.
Recent research has suggested that tamoxifen may also have impact on the gut microbiota8,9. The gut microbiota plays a crucial role in human health, modulating the metabolism of xenobiotics and nutrients and interacting with the immune system10,11,12. Despite this vital role, our understanding of how xenobiotic compounds, such as therapeutic drugs, affect the composition and function of gut microbial communities is still limited. Even less is known about how gut microbiota influences the metabolism of specific drugs like tamoxifen, an area of research known as pharmacomicrobiomics that is still in its early stages13. A study using Caenorhabditis elegans models has shown that gut bacteria can modulate the body’s response to tamoxifen by altering fatty acid metabolism, thereby affecting the drug’s toxicity and efficacy14. By demonstrating that different bacterial species can lead to varying levels of tamoxifen toxicity through distinct metabolic pathways, this research underscores the critical role of the gut microbiota in influencing drug action and could be important for understanding individual responses to medications in humans. Given the current understanding, it is plausible that the diversity and activity of the gut microbiota could influence tamoxifen metabolism, potentially impacting both its therapeutic efficacy and the manifestation of side effects. However, this hypothesis remains preliminary, necessitating further research to explore this potential relationship.
To the best of our knowledge, studies investigating the relationship between tamoxifen and the gut microbiota are scarce, with observational human studies particularly lacking15. Nevertheless in vitro and in vivo studies highlight the impact of tamoxifen on the gut microbiota. Maier et al.9 demonstrated that tamoxifen administration significantly inhibited the growth of specific bacterial strains, including (non-toxigenic) Bacteroides fragilis, Clostridium saccharolyticum, Streptococcus salivarius, and Eubacterium eligens, when tested in vitro. A study by Li et al.8 found that in a breast cancer xenograft mouse model, tamoxifen therapy significantly reduced the levels of the bacterial genera Lachnospiraceae_UCG-006, Anaerotruncus, Alistipes, and Eubacterium compared to those in a control group. Additionally, in the same study tamoxifen therapy was associated with an increase in cytokines related to inflammation.
Furthermore, Alam et al.16 demonstrated that gut microbiota play a crucial role in tamoxifen pharmacokinetics. Their study found that β-glucuronidase (GUS) enzymes produced by gut bacteria hydrolyze glucuronidated tamoxifen metabolites, facilitating their reabsorption into systemic circulation. They also observed inter-individual variability in gut microbiota composition and enzymatic activity, which may contribute to differences in tamoxifen metabolism and drug response. Notably, their findings suggest that the gut microbiome may influence the enterohepatic recirculation of tamoxifen metabolites, underscoring the need to consider microbiome composition as a potential factor in tamoxifen therapy.
Research into the influence of tamoxifen, and consequently endoxifen, on the gut microbiota is still in its early stages. Therefore, it would be worthwhile to explore the association between the gut microbiota and endoxifen levels, as well as the bi-directional relationship between tamoxifen and gut microbiota.
More knowledge concerning the interactions between tamoxifen and the gut microbiota is required. This explorative observational study aims to investigate if tamoxifen impacts the gut microbiota in postmenopausal ER-positive (ER+) and human epidermal growth factor receptor 2 negative (HER2−) breast cancer patients, and if gut microbiota composition is correlated with endoxifen plasma levels. We hypothesized that tamoxifen therapy in postmenopausal ER+/HER2− breast cancer patients would alter gut microbiota diversity and composition.
Materials and methods
Inclusion of patients
Between November 2017 and November 2022, breast cancer patients were prospectively enrolled in three Dutch hospitals. Eligible patients were postmenopausal women with histologically proven ER+ (expression level of 10% or higher) and HER2− breast cancer scheduled for adjuvant tamoxifen therapy17. Exclusion criteria included distant metastasis, previous therapeutic antibiotics use within three months before fecal sampling, and previous chemotherapy within one month before fecal sampling.
The study was registered in the Overview of Medical Research in the Netherlands (OMON) under NL6141 and at ToetsingOnline under NL61646.068.17. The study was approved by the Medical Ethics Committee of azM/UM and conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines. All patients provided written informed consent.
Tamoxifen therapy
Patients were administered 20 mg of tamoxifen orally daily. Patients received the adjuvant endocrine therapy as part of standard care according to the Dutch guidelines for 2–3 years, followed by aromatase inhibitor therapy18. These guidelines are in line with the ESMO and ASCO guidelines19,20. The treatment of postmenopausal ER + /HER2− breast cancer depends on the clinical and pathological tumor stage but primarily consists of initial surgery, with or without chemotherapy administered before or after surgery, followed by anti-hormonal therapy. Radiotherapy was permitted for all patients according to guidelines though not all required it.
Sample collection and preprocessing
Patients collected fecal samples and completed questionnaires at two time points: before starting tamoxifen therapy (T0) and after 6–12 weeks of tamoxifen therapy. Additionally, blood samples were drawn during the second time point to measure endoxifen concentrations. This timeframe allowed sufficient time for tamoxifen and its active metabolites, such as endoxifen, to reach steady-state blood levels, ensuring that measurements accurately reflect treatment exposure. After fecal sample collection, samples were immediately stored in a freezer and then transported to the hospital in a cooled transport container (Sarstedt) to preserve the cold chain. Upon arrival at the hospital, the samples were initially stored at − 20 °C and later transferred to − 80 °C for long-term storage. Blood samples with a clot activator underwent centrifugation. The resulting serum was then carefully transferred into aliquots, which were stored at − 80 °C prior to the measurement of endoxifen levels. The questionnaires collected data on general medical characteristics, including weight, height, history of abdominal surgery, smoking habits, alcohol usage, diabetes, medication usage, and the use of pro- and prebiotics. They also covered reproductive history, nutritional status, and general performance and well-being. Baseline characteristics recorded included the Karnofsky Performance Score (KPS), nutritional status assessed with the Malnutrition Universal Screening Tool (MUST), and details on previous systemic therapy. Information on prophylactic or therapeutic antibiotic administration, prebiotic/probiotic use, exogenous estrogen use, and the use of nutritional supportive drinks was also registered.
Fecal microbiota data generation
Metagenomic DNA was isolated using the Ambion MagMax™ Total Nucleic Acid Isolation Kit (Thermo Fisher Scientific). This process involved a manual pre-processing step followed by automated purification of nucleic acids using the KingFisher FLEX system (Thermo Fisher Scientific), as described earlier21. To monitor potential contamination, DNA extraction blanks were included and processed alongside the fecal samples.
In more detail, to extract metagenomic DNA, 250 mg of the frozen fecal samples were homogenized in phosphate-buffered saline (PBS) and centrifuged for 1 min at 900 rpm. Cell lysis was achieved through a combination of chemical, mechanical, and thermal disruption methods. Zirconia beads were utilized for mechanical lysis. A lysis buffer containing 1 M Tris–HCl, 0.5 M EDTA, 5 M sterile NaCl, and SDS (final concentration 4%) was added to bead tubes from the Ambion MagMaxTM Total Nucleic Acid Isolation Kit (Thermo Fisher Scientific) and mixed with 175 µl of fecal supernatant in PBS. Mechanical disruption was carried out using a bead-beating procedure with the Fastprep™ Homogenizer (5.5 ms for 3 × 1 min, with 1-min rests in between, MP Biomedicals). The samples were then incubated for 15 min at 95 °C with gentle shaking. After centrifugation for 5 min at 11,000 rpm, the supernatant was transferred to an Eppendorf tube. A second round of bead beating, and incubation followed, after which the supernatants were pooled and stored at − 20 °C until further analysis. 200 µl of the supernatants were placed into a KingFisher 96-well deep well plate (Thermo Fisher Scientific), along with the bead mix from the Ambion MagMaxTM Total Nucleic Acid Isolation Kit (Thermo Fisher Scientific), isopropanol, and lysis buffer. Other plates were filled with wash buffers, elution buffer (with RNAse), and 96-tips for DW magnets (Thermo Fisher Scientific). The prepared plates were then processed in the KingFisher system according to the manufacturer’s standard protocol (Thermo Fisher Scientific). Following the removal of the plates from the system, the plate containing purified nucleic acids was incubated for 15 min at 37 °C to degrade RNA.
The V4 hypervariable region of the 16S rRNA gene was then amplified in triplicate using the 515F/806R barcoded primer pair, as previously described22. Pooled amplicons from the triplicate reactions were purified using the AMPure XP purification kit (Agencourt) according to the manufacturer’s instructions and eluted in 25 μl of 1 × low TE (10 mM Tris–HCl, 0.1 mM EDTA, pH 8.0). The amplicons were quantified using the Quant-iT PicoGreen dsDNA reagent kit (Invitrogen) and measured with a Victor3 Multilabel Counter (Perkin Elmer, Waltham, USA). The amplicons were then mixed in equimolar concentrations to ensure equal representation of each sample and sequenced on an Illumina MiSeq instrument (MiSeq Reagent Kit v3, 2 × 300 cycles, 10% PhiX), producing paired-end reads of 250 bases (~ 25,000 reads/sample)23.
Microbiota sequencing data processing
Forward and reverse primers were removed using Cutadapt v4.724. Subsequently, paired-end sequences were processed with DADA2 (v1.28.0), where they were filtered and trimmed (maxEE = 2, truncLen = 240/210 bp), denoised, merged (minOverlap = 10, maxMismatch = 0), and used to construct a sequence table25. Chimeras were identified and removed using the ‘consensus’ method. After denoising and merging, Amplicon Sequence Variants (ASV) shorter than 350 bases or longer than 500 bases were discarded. ASVs were then annotated to the genus level using the DADA2 implementation of the naïve Bayesian classifier, using the SILVA v138.1 reference database26. Species-level annotations were added with the DADA2 addSpecies() function. If an ASV could not be uniquely classified at a particular rank, they were aggregated together for the taxonomically aggregated statistical analyses under the name of the lowest classified rank, e.g. “Enterobacteriaceae family”.
Analysis of z-endoxifen
A liquid chromatography-mass spectrometry assay was used for the quantification of z-endoxifen as previously described27. In short, plasma samples were prepared with protein precipitation. The analyses utilized a triple quadrupole mass spectrometer, which operated in both positive and negative ion modes. The assay was validated for z-endoxifen concentrations ranging from 1 to 20 ng/mL.
Statistical analyses
Statistical analyses were performed using R Studio (R version 4.3.3)28. For the baseline characteristics, the mean, standard deviation, and range were calculated for each numeric variable.
Rarefied data were used for alpha-diversity analysis, while all other microbiota analyses utilized non-rarefied data. Alpha-diversity (observed richness and Shannon diversity) measurements were calculated at the ASV level using the phyloseq package29. The change in ASV richness (Δ ASV richness) was calculated by subtracting the richness at T0 from the richness at T1. The same was done for the change in Shannon index (Δ Shannon index). Assumptions of normality were tested using the Shapiro–Wilk test. Paired t-tests or Wilcoxon signed rank tests were used for paired samples. Otherwise, independent sample t-tests or Mann–Whitney U tests were used. Multivariable linear regression analysis was conducted to examine the associations between specific patient characteristics (age, BMI, and pathologic tumor stage) and Δ ASV richness or Δ Shannon index. To examine the associations between Δ ASV richness, Δ Shannon index, and the clinical variables of interest, Spearman correlation was performed using the corr.test function from the psych package (version 2.4.3) on a data frame containing all relevant variables30. P-values were adjusted for multiple comparisons using the false discovery rate (FDR) method, following the Benjamini–Hochberg procedure31. The correlations were visualized using the corrplot package (version 0.92), based on the correlation matrix generated by corr.test32.
The R packages phyloseq29, vegan33, microbiome34, dplyr35, ggplot236, and microViz37 were used for ordination and visualization of taxonomic composition. ASVs present in fewer than five samples were filtered out before these analyses. Unconstrained ordination was performed using Principal Component Analysis (PCA) based on centered-log-ratio transformed abundances at both the ASV and genus levels. Permutational multivariate analysis of variance (PERMANOVA) was used to analyze changes in overall microbiota composition, based on Aitchison distances. For the differential abundance analysis, only genera with a prevalence of at least 10% and a total abundance of at least 10,000 reads across all samples were included. Bacterial relative abundances were log10 transformed using the microbiome package, with zero counts replaced by half of the minimum non-zero value in the dataset as a pseudocount34. A correlation heatmap was generated to assess Spearman correlations between plasma endoxifen concentrations and various clinical variables, including antibiotic use between T0 and T1, BMI, and the relative abundances of bacterial taxa at the genus level. These analyses were performed using the microViz package (version 0.12.1)37. P-values were adjusted for multiple comparisons using the FDR method as mentioned above. An alpha of 0.05 was used as a threshold for statistical significance.
Results
A total of 62 ER+/HER2− postmenopausal breast cancer patients were included. The mean age was 66 years, and the mean BMI was 26 kg/m2 (Table 1). None of the patients used prebiotics in the last year. Four patients (7%) used probiotics in the last year, but not within the three months prior to study inclusion. In the last year preceding study inclusion, 63% of the patients used prophylactic or therapeutic antibiotics, but no therapeutic antibiotics were used in the three months prior to study inclusion.
Early-stage (stage I) breast cancer was found in 34 patients (55%). Most tumors were of the ductal type (68%), followed by lobular (26%) and mucinous (7%) types. All tumors were ER+/HER2−, according to the inclusion criteria. All patients had undergone breast surgery previously, with 48% of them receiving prophylactic antibiotics during surgery. (Neo)adjuvant chemotherapy was given in 14 patients (23%) and 73% of the patients had received radiotherapy. Of the patients receiving radiotherapy, tamoxifen therapy was started before (26.6%), during (66.7%), or after the radiotherapy (6.7%).
Possible factors influencing baseline microbiota richness, diversity, and composition
Different tests were performed to determine if there were already differences in microbiota richness, diversity, and composition at baseline (T0) based on factors such as previous chemotherapy, antibiotics during surgery, and surgery type. There were no significant differences in ASV richness and Shannon index between patients based on previous chemotherapy (richness: p = 0.299, Shannon: p = 0.656), antibiotics during surgery (richness: p = 0.168, Shannon: p = 0.365), or type of surgery (mastectomy vs. breast-conserving surgery, richness: p = 0.6801, Shannon: p = 0.8318) (See Supplementary Figs. 1 and 2). Unconstrained ordination using PCA showed no clustering of baseline samples based on previous chemotherapy, antibiotic use during surgery, or type of surgery (mastectomy vs. breast-conserving surgery). PERMANOVA supported these findings by showing no significant differences in microbial composition at either the genus or ASV levels. Specifically, no significant differences were observed based on previous chemotherapy (genus: p = 0.266; ASV: p = 0.151), antibiotics during surgery (genus: p = 0.297; ASV: p = 0.450), or type of surgery (genus: p = 0.636; ASV: p = 0.579). These results are visualized in Supplementary Figs. 3 and 4.
Microbiota richness, diversity, and composition during tamoxifen therapy: Significant differences in ASV richness between T0 and T1
In total, 122 fecal samples were collected as two patients provided samples at only one timepoint, resulting in 60 paired samples. ASV richness was significantly different between T0 and T1 (p = 0.0188), whereas the Shannon index did not differ (p = 0.166) (Fig. 1A and B) . The differences in ASV richness (Δ ASV-richness) between the two time points for paired samples showed an increase in ASV richness during tamoxifen therapy (Fig. 1B and D), whereas the Shannon index remained unchanged (Fig. 1A and C). Multivariable linear regression analysis revealed that none of the specific patient characteristics (age, BMI, and pathologic tumor stage) were significantly associated with Δ ASV richness (p > 0.05 for all variables) (Supplementary Tables 1 and 2).
(A, B) Violin plots illustrating the distribution of alpha-diversity measurements at two different time points (T0 and T1), highlighting whether alpha-diversity increases (green line) or decreases (purple line) between paired samples. Each plot shows the density of values with individual data points overlaid as black dots. (A) Shannon index. (B) ASV richness. (C, D) Violin plots illustrating the distribution of the shifts in alpha-diversity measurements (C Δ Shannon; D Δ ASV-richness) between T0 and T1 for paired samples. The dashed line in the plot indicates zero (no change), serving as a reference point to easily identify increases or decreases in alpha-diversity.
Additional analyses were performed to assess the potential effects of other factors that could explain the increase in ASV richness. Neither Δ ASV richness nor Δ Shannon index was associated with previous antibiotic exposure during surgery, and neither was associated with surgery type. Δ Shannon index was significantly higher in the group of patients who had undergone chemotherapy for breast cancer (p = 0.018). However, this was not the case for Δ ASV richness (Supplementary Figs. 5 and 6).
Correlation analyses revealed that antibiotic use between T0 and T1 was negatively correlated with the change in gut microbiota diversity (Δ Shannon index: rho = − 0.483, padjusted = 0.001, and Δ ASV richness: rho = − 0.369, padjusted = 0.019), suggesting that antibiotic use negatively affects alpha diversity (Supplementary Fig. 7). Additionally, there was no correlation between the number of days from the last surgery, chemotherapy, or antibiotic use to baseline (T0, start of tamoxifen therapy) and the change in gut microbiota diversity (Δ Shannon index or Δ ASV richness) between T0 and T1.
Principal component analysis (PCA) indicated no clustering of samples collected at T0 or T1. Similarly, PERMANOVA analysis showed no statistically significant differences in overall microbial community structure at both the genus level (p = 0.1656) and the ASV level (p = 0.1656) between T0 and T1 (Fig. 2A and B). The within-subject dissimilarity was calculated to evaluate the shifts in microbial community structure within individuals over time. The median dissimilarity between T0 and T1 was 20.6, with an interquartile range (IQR) of 7.6, indicating moderate variability in dissimilarity scores. When within-subject dissimilarity was compared to between-subject dissimilarity, the within-subject dissimilarity was significantly lower (Supplementary Fig. 8). This suggests that while there was some variability, the microbial communities within subjects remained relatively stable over time. Additionally, many of the patients with higher within-subject dissimilarity (i.e., least stable microbial communities) appeared to have had antibiotics administered between T0 and T1 (Fig. 2C).
Ordination plots derived from unconstrained principal components analysis (PCA) based on centered-log-ratio transformed abundances, showing the composition of the microbial community at ASV (A) and genus (B) levels for T0 and T1. P-values from the PERMANOVA analysis are indicated on the plots. (C) Violin plot illustrating the distribution of within-subject dissimilarity between T0 and T1 for paired samples (Aitchison distances calculated at ASV-level). The width of the plot at different levels indicates the frequency of observations. The dashed line represents the median within-subject dissimilarity.
Differential abundance analyses revealed notable changes in the abundance of specific taxa between T0 and T1 (Fig. 3). Specifically, ten genera showed a significantly increased abundance, including Blautia (padjusted = 0.003), Sellimonas (padjusted = 0.024), [Eubacterium] halli group (padjusted = 0.028), Oscillospiraceae Family (padjusted = 0.013), Ruminococcaceae Incertae Sedis (padjusted = 0.029), Ruminococcaceae UBA1819 (padjusted = 0.036), Christensenellaceae R-7 group (padjusted = 0.036), Eggerthellaceae Family (padjusted = 0.047), Family XIII AD3011 group (padjusted = 0.010), and Streptococcus (padjusted = 0.010). Two genera exhibited a significant decrease, including Prevotella_9 (padjusted = 0.006), and Subdoligranulum (padjusted = 0.036).
Differential abundant taxa between T0 and T1. Log10-transformed relative abundances of the 24 taxa that were statistically different in abundance between the timepoints before (T0) and during tamoxifen therapy (T1). The thick line represents the mean value, whereas thin lines represent shifts in abundance over time in individual patients.
Correlations between gut microbiota and plasma endoxifen levels
To investigate possible associations between plasma endoxifen levels (at T1) and gut microbiota composition or diversity (at T0 or T1), Spearman correlation analyses were performed. BMI at T0 and antibiotic use between T0 and T1 were also included in these analyses to provide a basis for comparison with the endoxifen results (Supplementary Table 3). At T1 (Fig. 4), two genera were associated with antibiotic use after FDR correction, Erysipelotrichaceae UCG-003 (rho = − 0.483, padjusted = 0.02) and Christensenellaceae R-7 group (rho = − 0.506, padjusted = 0.02). Antibiotic use was also associated with ASV richness (rho = -0.418, padjusted < 0.01) and Shannon diversity (rho = − 0.443, padjusted < 0.01) at T1. Several further correlations were observed between antibiotics and genus abundances at T1, and between baseline BMI and Bifidobacterium, but these were not significant after FDR correction for multiple testing. There were several positive correlations between gut genera and plasma endoxifen levels at T1, and two negative correlations with p-values < 0.05, but no correlations remained significant after FDR correction.
Heatmap of Spearman correlation coefficients for the correlations between the relative abundance of bacterial taxa on genus level and alpha-diversity measurements at T1 and plasma levels of endoxifen at T1, BMI, and antibiotic use between T0 and T1 (AB_T0_T1). An asterisk indicated p < 0.05 and a circle indicates FDR-corrected p < 0.05.
The genera Coprococcus (rho = 0.270, padjusted = 0.271), Marvinbryantia (rho = 0.295, padjusted = 0.244), Lachnospiraceae NK4A136 group (rho = 0.301, padjusted = 0.244), CAG-56, (rho = 0.267, padjusted = 0.271) and Intestinibacter (rho = 0.271, padjusted = 0.271) showed positive correlations with endoxifen concentrations. Conversely, Streptococcus (rho = − 0.275, padjusted = 0.271) and Eggerthella (rho = − 0.292, padjusted = 0.244) showed negative correlations with endoxifen concentrations. There were no correlations found between alpha-diversity variables and endoxifen concentrations.
Endoxifen levels at T1 were not correlated with microbiota composition or diversity at T0, after FDR correction (Supplementary Fig. 9). Nor was Antibiotic use between T0 and T1, and nor was baseline BMI.
Discussion
Our study demonstrated that alpha-diversity, in terms of ASV richness, significantly increased in postmenopausal ER+/HER2− breast cancer patients and the abundance of specific microbial taxa significantly changed during tamoxifen therapy. No significant differences were found in overall microbial community structure before and during tamoxifen therapy.
The increase in ASV richness during tamoxifen therapy suggests that tamoxifen may promote a more diverse gut microbiota. Although the Shannon index did not show differences between the two timepoints, indicating that the overall diversity—considering both richness and evenness—remained stable, the number of unique microbial taxa increased significantly. The change in ASV richness during treatment was not significantly associated with patient characteristics such as age, BMI, or pathological tumor stage. Additionally, there was no correlation between the number of days from the last surgery, chemotherapy, or antibiotic use to baseline (start of tamoxifen therapy) and the change in gut microbiota diversity between T0 and T1. This indicates that the effect of tamoxifen on alpha-diversity might be primarily due to the therapy itself. Given our findings of an increase in ASV richness during tamoxifen therapy compared to before therapy, two hypotheses were formulated. Firstly, the observed increase in diversity might potentially be caused by the therapy’s direct impact on inhibiting the growth of specific bacteria, thereby allowing other (normally underrepresented or absent) bacteria to take over the niches and expand. Similar observations were made in the study of Wu et al.38 that showed that alpha-diversity increased during neoadjuvant chemotherapy and minimally increased during adjuvant chemotherapy in breast cancer patients. Alternatively, it prompts consideration of whether an already inherently low alpha-diversity in breast cancer patients could be improved by the therapeutic anti-cancer effects of tamoxifen and/or the removal of the tumor. This is in line with most previous studies that have investigated the gut microbiota in breast cancer patients39,40,41,42,43,44,45. Although the results on alpha-diversity and used indices vary, most studies found a decrease in alpha-diversity in breast cancer patients compared to healthy controls40,43,45. If breast cancer treatment-naïve patients already exhibit lower alpha-diversity, it is possible that, depending on the type of treatment administered, their microbial diversity may recover or return to normal levels during the course of treatment. This potential for recovery aligns with the idea that gut microbiota can either revert to their original community structure or form a new balance following drug exposure46. The capacity for recovery or the formation of a new community structure depends on the resilience of the microbiota and the specific nature of the treatment, such as the administration of tamoxifen in this case.
Among the studies investigating the gut microbiota and breast cancer, only three studies also examined postmenopausal breast cancer patients39,40,41. Postmenopausal status can be an important factor in investigating the gut microbiota because hormonal shifts during menopause may alter the gut microbiota, although results on differences in alpha-diversity between post and premenopausal women are inconsistent47. Following up on this, another study showed that the alpha-diversity of HER2+ breast cancer patients was lower than that of HER2− breast cancer patients, suggesting that molecular subtype could be another possible influencing factor42. Given the variations in alpha-diversity among breast cancer patients and the potential influence of molecular subtype and menopausal status, there is a compelling need for more controlled studies with homogeneous breast cancer patient groups. To the best of our knowledge, there are currently no other observational cohort studies in humans that have investigated the effect of tamoxifen on the gut microbiota, and studies investigating this in mice are scarce. A study by Li and Gao et al. examined tamoxifen-induced alterations in the gut microbiota and inflammation using a breast cancer xenograft mouse model8. They found no significant differences in alpha-diversity but observed a trend towards lower Chao1, Shannon, and Simpson indices in the group of mice with breast cancer receiving a placebo compared to the group of mice with breast cancer treated with tamoxifen. This trend towards an increase in alpha-diversity during tamoxifen is in line with our results and suggests that tamoxifen may have a potential role in maintaining or enhancing microbial diversity in the gut, although further studies are needed to confirm these findings and understand the underlying mechanisms.
Besides the increase of alpha-diversity, the abundance of specific microbial taxa changed during tamoxifen therapy in our cohort. In total ten genera were found to increase during tamoxifen therapy, amongst others Blautia, [Eubacterium] halli group, Ruminococcaceae UBA1819, Eggerthellaceae Family, and Streptococcus. Two genera exhibited a significant decrease, including Prevotella_9, and Subdoligranulum. Comparing our findings with those of Li and Gao et al., both indicate significant microbial shifts due to tamoxifen therapy. Li et al. reported that in breast cancer mice treated with tamoxifen, the genera Bacteroides, Clostridium, Escherichia-Shigella, Ruminococcus, Prevotellaceae_UCG-001, and Akkermansia were significantly increased, whereas Lachnospiraceae_UCG-006, Anaerotruncus, Alistipes, and Eubacterium were significantly decreased8. Notably, our study aligns with Li et al.'s findings in the context of the increase of Ruminococcaceae and the decrease of Eubacterium, though the specific species differ. Furthermore, the study by Maier et al. provided insights into the in-vitro growth inhibition effects of tamoxifen on specific bacterial strains9. They observed growth inhibition amongst others in Eubacterium eligens, Eubacterium rectale, Prevotella copri, and Streptococcus salivarius. However, no inhibition was noted for Eggerthella lenta and Streptococcus parasanguinis. These findings resonate with our observation of decreased Prevotella_9, suggesting a potential inhibitory effect of tamoxifen on certain Prevotella species. Interestingly, while Maier et al. noted inhibition of some Streptococcus strains, our data indicated an overall increase in Streptococcus abundance, possibly highlighting strain-specific responses to tamoxifen within this genus.
Our exploratory analysis of correlations between gut microbiota and plasma endoxifen levels did not yield significant results after FDR correction. However, observed trends suggest potential interactions that warrant further investigation. The positive correlations between endoxifen concentrations and genera such as Coprococcus and Lachnospiraceae NK4A136 group, as well as the negative correlations with Streptococcus and Eggerthella, may indicate that there is an interaction between tamoxifen and the gut microbiota, where tamoxifen may influence the microbiota, and vice versa.
Notably, the negative correlation with Streptococcus is particularly interesting, as this genus was increased during tamoxifen treatment. This suggests that Streptococcus, of which some species might be able to bloom under tamoxifen therapy, may influence tamoxifen metabolism, resulting in lower concentrations of endoxifen. These are preliminary observations, and further research is necessary to explore these potential interactions. The lack of robust correlations highlights the complexity of these interactions and the need for larger, more detailed studies to elucidate these relationships.
In light of these findings, it is worth noting a recent study by Wasiak et al., that demonstrated that postbiotics derived from Lactiplantibacillus plantarum and Lactobacillus rhamnosus can enhance tamoxifen’s anticancer effects by promoting apoptosis and inhibiting the proliferation of breast cancer cells48. This suggests that the metabolites of microbial taxa could interact with tamoxifen, potentially influencing tamoxifen metabolism and treatment outcomes. Therefore, future research should not only focus on the direct effects of tamoxifen on gut microbiota but also explore how bacterial metabolites might be involved in tamoxifen metabolism, potentially offering new avenues for enhancing breast cancer treatment.
This study has several strengths and limitations. One strength is that it is the first to investigate the impact of tamoxifen on gut microbiota and its correlation with endoxifen levels in postmenopausal ER+/HER2− breast cancer patients. The relatively large, homogeneous cohort of 62 patients enhances the reliability of the findings. Using 16S rRNA gene sequencing provided detailed microbiota analysis before and during tamoxifen treatment, and correlating gut microbiota with endoxifen levels added valuable pharmacokinetic insights. However, the observational design limits causal conclusions, and whilst 16S rRNA gene amplicon sequencing gives reliable estimates of genus level taxonomic abundances, shotgun metagenomic sequencing would provide a higher taxonomic resolution and information on the functional capacity of the gut microbiota, which may be important for understanding the interaction of tamoxifen with the microbiota.
A key limitation of our study is the absence of CYP2D6 genotyping data, which plays a crucial role in tamoxifen metabolism. As a result, we cannot rule out the possibility that inter-individual variability in endoxifen levels was influenced by CYP2D6 metabolizer status and co-medication, which may have confounded our correlation analyses. Incorporating this genetic factor in future research could provide a more comprehensive understanding of the interplay between CYP2D6, gut microbiota, and tamoxifen efficacy49.
Additionally, our findings may not be generalizable to other populations, such as premenopausal women or different cancer subtypes. Future research with larger, diverse cohorts, longitudinal designs, and advanced sequencing techniques is needed to validate these preliminary findings and explore their broader applicability.
In conclusion, this explorative observational study suggests that tamoxifen therapy significantly increases ASV richness and alters specific microbial taxa in the gut microbiota of postmenopausal ER+/HER2− breast cancer patients. Despite the increase in ASV richness, no significant changes were found in overall microbial community structure and composition. Our results suggest that tamoxifen may foster a more diverse gut microbial environment, independent of patient-specific factors such as age, BMI, or treatment history with chemotherapy, surgery or antibiotics. Additionally, while correlations between gut microbiota and endoxifen plasma levels were not significant, observed trends indicate potential microbial interactions with tamoxifen metabolism that warrant further investigation. Future research with larger cohorts, incorporating CYP2D6 genotype, other influencing factors such as co-medication, and advanced sequencing, is needed to confirm these findings and uncover underlying mechanisms.
Data availability
Sequencing data is available from the European Nucleotide Archive (ENA), under study accession number PRJEB80483. The Additional data used and/or analyzed are available from the corresponding author on reasonable request.
References
Clusan, L., Ferrière, F., Flouriot, G. & Pakdel, F. A basic review on estrogen receptor signaling pathways in breast cancer. Int. J. Mol. Sci. 24(7), 6834 (2023).
Yang, G., Nowsheen, S., Aziz, K. & Georgakilas, A. G. Toxicity and adverse effects of Tamoxifen and other anti-estrogen drugs. Pharmacol. Ther. 139(3), 392–404 (2013).
Sanchez-Spitman, A. B. et al. Clinical pharmacokinetics and pharmacogenetics of tamoxifen and endoxifen. Expert. Rev. Clin. Pharmacol. 12(6), 523–536 (2019).
Davies, C. et al. Relevance of breast cancer hormone receptors and other factors to the efficacy of adjuvant tamoxifen: Patient-level meta-analysis of randomised trials. Lancet 378(9793), 771–784 (2011).
Fox, P. et al. Dose Escalation of tamoxifen in patients with low endoxifen level: Evidence for therapeutic drug monitoring-the TADE study. Clin. Cancer Res. 22(13), 3164–3171 (2016).
Braal, C. L. et al. Factors affecting inter-individual variability in endoxifen concentrations in patients with breast cancer: Results from the prospective TOTAM trial. Breast Cancer Res. Treat. 195(1), 65–74 (2022).
Lorizio, W. et al. Clinical and biomarker predictors of side effects from tamoxifen. Breast Cancer Res. Treat. 132(3), 1107–1118 (2012).
Li, H. et al. Potential risk of tamoxifen: Gut microbiota and inflammation in mice with breast cancer. Front. Oncol. 13, 1121471 (2023).
Maier, L. et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature 555(7698), 623–628 (2018).
Pope, J. L., Tomkovich, S., Yang, Y. & Jobin, C. Microbiota as a mediator of cancer progression and therapy. Trans. Res. J. Lab. Clin. Med. 179, 139–154 (2017).
Quigley, E. M. Gut bacteria in health and disease. Gastroenterol. Hepatol. 9(9), 560–569 (2013).
Jandhyala, S. M. et al. Role of the normal gut microbiota. World J. Gastroenterol. 21(29), 8787–8803 (2015).
Haiser, H. J. & Turnbaugh, P. J. Is it time for a metagenomic basis of therapeutics?. Science 336(6086), 1253–1255 (2012).
Diot, C. et al. Bacterial diet modulates tamoxifen-induced death via host fatty acid metabolism. Nat. Commun. 13(1), 5595 (2022).
Aarnoutse, R. et al. The clinical link between human intestinal microbiota and systemic cancer therapy. Int. J. Mol. Sci. 20(17), 4145 (2019).
Alam, Y. et al. Variation in human gut microbiota impacts tamoxifen pharmacokinetics. mBio 16(1), e0167924 (2025).
Wolff, A. C. et al. Human epidermal growth factor receptor 2 testing in breast cancer: American society of clinical oncology/college of american pathologists clinical practice guideline focused update. J. Clin. Oncol. 36(20), 2105–2122 (2018).
Richtlijnendatabase Borstkanker. In.: Federatie Medisch Specialisten (2024).
Cardoso, F. et al. Early breast cancer: ESMO clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 30(8), 1194–1220 (2019).
Burstein, H. J., Lacchetti, C. & Griggs, J. J. Adjuvant endocrine therapy for women with hormone receptor-positive breast cancer: ASCO clinical practice guideline focused update. J. Oncol. Pract. 15(2), 106–107 (2019).
Aarnoutse, R. et al. Changes in intestinal microbiota in postmenopausal oestrogen receptor-positive breast cancer patients treated with (neo)adjuvant chemotherapy. NPJ Breast Cancer 8(1), 89 (2022).
Caporaso, J. G. et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme J. 6(8), 1621–1624 (2012).
Galazzo, G. et al. Faecal microbiota dynamics and their relation to disease course in crohn’s disease. J. Crohns. Colitis 13(10), 1273–1282 (2019).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1), 10–12 (2011).
Callahan, B. J. et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13(7), 581–583 (2016).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41(Data base issue), 590–596 (2013).
van Nuland, M. et al. Development and validation of an UPLC-MS/MS method for the therapeutic drug monitoring of oral anti-hormonal drugs in oncology. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1106–1107, 26–34 (2019).
R: A language and environment for statistical computing. (R Foundation for Statistical Computing, Vienna). https://www.R-project.org/
McMurdie, P. J. & Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One 8(4), e61217 (2013).
psych: Procedures for Personality and Psychological Research, Northwestern University, Evanston, Illinois, USA. https://CRAN.R-project.org/package=psychVersion=2.2.5.
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B (Methodological) 57(1), 289–300 (1995).
R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84) [https://github.com/taiyun/corrplot]
vegan: Community Ecology Package. R package version 2.5-6. [https://CRAN.R-project.org/package=vegan]
microbiome R package [http://microbiome.github.io]
Wickham, H., François, R., Henry, L. & Müller, K. dplyr: A Grammar of Data Manipulation (R package version 1.0.9. In, 2022).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer, 2016).
Barnett, D., Arts, I. & Penders, J. microViz: an R package for microbiome data visualization and statistics. J. Open Sour. Softw. 6(63), 3201 (2021).
Wu, A. H., Vigen, C., Tseng, C., Garcia, A. A. & Spicer, D. Effect of chemotherapy on the gut microbiome of breast cancer patients during the first year of treatment. Breast Cancer (Dove Med Press) 14, 433–451 (2022).
Aarnoutse, R. et al. Intestinal microbiota in postmenopausal breast cancer patients and controls. Cancers (Basel) 13(24), 6200 (2021).
Goedert, J. J. et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J. Natl. Cancer Inst. https://doi.org/10.1093/jnci/djv147 (2015).
Zhu, J. et al. Breast cancer in postmenopausal women is associated with an altered gut metagenome. Microbiome 6(1), 136 (2018).
Wu, A. H. et al. Gut microbiome associations with breast cancer risk factors and tumor characteristics: A pilot study. Breast Cancer Res. Treat. 182(2), 451–463 (2020).
Byrd, D. A. et al. Associations of fecal microbial profiles with breast cancer and nonmalignant breast disease in the Ghana Breast Health Study. Int. J. Cancer 148(11), 2712–2723 (2021).
Yang, P., Wang, Z., Peng, Q., Lian, W. & Chen, D. Comparison of the gut microbiota in patients with benign and malignant breast tumors: A pilot study. Evol. Bioinform. Online 17, 11769343211057572 (2021).
Ma, Z., Qu, M. & Wang, X. Analysis of gut microbiota in patients with breast cancer and benign breast lesions. Pol. J. Microbiol. 71(2), 217–226 (2022).
de la Cuesta-Zuluaga, J., Boldt, L. & Maier, L. Response, resistance, and recovery of gut bacteria to human-targeted drug exposure. Cell Host. Microbe 32(6), 786–793 (2024).
Schreurs, M. P. H., De Vos van Steenwijk, P. J., Romano, A., Dieleman, S. & Werner, H. M. J. How the gut microbiome links to menopause and obesity, with possible implications for endometrial cancer development. J. Clin. Med. 10(13), 2916 (2021).
Wasiak, J. et al. Lactic acid bacteria-derived postbiotics as adjunctive agents in breast cancer treatment to boost the antineoplastic effect of a conventional therapeutic comprising tamoxifen and a new drug candidate: An aziridine-hydrazide hydrazone derivative. Molecules 29(10), 2292 (2024).
Hoskins, J. M., Carey, L. A. & McLeod, H. L. CYP2D6 and tamoxifen: DNA matters in breast cancer. Nat. Rev. Cancer 9(8), 576–586 (2009).
Acknowledgements
First, we would like to thank all the patients who participated in our study and donated fecal samples. We’re also grateful to the medical oncologists and research nurses from Catharina Hospital, VieCuri Medical Centre, Elkerliek Hospital, and Maastricht University Medical Center+ for their help with patient inclusion and sample collection. A special thanks to Marjan Laven, Maaike van Dam, Ramon Bax, Eva de Jong, Ilona van Rooij-Tieleman, Wendy Heuts, Monique Vercoulen, and Sandra Silvis for their dedicated support. Furthermore, we are grateful to Christel Driessen and Erik Beuken for their support in the lab work and for conducting 16S rRNA amplicon sequencing. Additionally, we wish to acknowledge Jessica Herpertz, Lisa Coolen, Janneke Waelen, Bob Bindels, Milou Stevens, and Emma Russ for their assistance with patient inclusion, sample collection, and sample processing. Lastly, we would like to thank the Bioanalytical Laboratory of the Department of Pharmacy & Pharmacology, the Netherlands Cancer Institute, Amsterdam, The Netherlands, for performing the endoxifen analyses.
Author information
Authors and Affiliations
Contributions
LEH: Conceptualization, Methodology, Validation, Formal Analysis, Investigation, Data curation, Writing—Original draft, Writing—Review and Editing, Visualization. DJMB: Conceptualization, Methodology, Validation, Formal Analysis, Writing—Review and Editing, Visualization. JZ: Conceptualization, Methodology, Validation, Investigation, Data curation, Writing—Review and Editing. RA: Conceptualization, Methodology, Validation, Investigation, Data curation, Writing—Review and Editing. JVG: Conceptualization, Methodology, Resources, Data curation, Writing—Review and Editing, Supervision. RG: Conceptualization, Methodology, Writing—Review and Editing. MB: Resources, Data curation, Writing—Review and Editing. YEAR: Resources, Data curation, Writing—Review and Editing. JV: Resources, Data curation, Writing—Review and Editing. JP: Conceptualization, Methodology, Validation, Recourses, Writing—Review and Editing, Supervision. MLS: Conceptualization, Methodology, Validation, Writing—Review and Editing, Supervision, Funding acquisition.
Corresponding author
Ethics declarations
Competing interests
LEH, JZ, and MLS have received research funding from Danone Global Research & Innovation Center, outside the submitted work. Additionally, MLS, RA, and JVG have received funding from Servier, and MLS from Illumina, all outside the submitted work. JVG has served as a consultant for Amgen, AstraZeneca, MSD, Pierre Fabre, and Servier, all outside the submitted work. JP has received research funding from Friesland Campina outside the submitted work. All other authors have no competing interests. All other authors have no competing interests.
Ethics approval and consent to participate
The study was approved by the Medical Ethics Committee of azM/UM and conducted in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines. All patients provided written informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hillege, L.E., Barnett, D.J.M., Ziemons, J. et al. The gut microbiota during tamoxifen therapy in patients with breast cancer. Sci Rep 15, 7874 (2025). https://doi.org/10.1038/s41598-025-91734-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91734-1
Keywords
This article is cited by
-
Impact of Gut Microbiota on Drug Metabolism and Absorption
Current Pharmacology Reports (2025)