Abstract
The COVID-19 pandemic highlighted the need for improved epidemic spread forecasting, a critical precursor for developing optimal control measures for spread mitigation. Well-recognized shortcomings in computing basic and effective reproduction numbers (\(\mathscr {R}_0\), \(\mathscr {R}_e\))-fundamental metrics for forecasting-underscore the need for new methods for estimating them from available data. We present a novel computational framework for estimating reproduction numbers from empirical spread data. The framework is derived from a mechanistic, spatiotemporal, Partial Differential Equation (PDE) model of epidemic spread utilizing mathematical results from PDE epidemic models. Forecasts of spatiotemporal effective reproduction number \(\mathscr {R}_e\) using the framework are found to be in excellent agreement with COVID-19 spread trends for Hamilton County, Ohio, USA, for three distinct periods. Furthermore, the forecasts are shown to align with corresponding reproduction numbers computed independently using the Wallinga-Teunis and Cori retrospective methods used in epidemiology. In summary, the results establish the validity of the framework and indicate applicability to future epidemics-especially for regions such as counties and for timeframes extending in weeks-even during dynamic phases when obtainable real-time infection spread data will likely be sparse.
Similar content being viewed by others
Introduction
Accurately forecasting the spatiotemporal spread of an epidemic based on currently available, often sparse, data—whilst arduous even for smaller geographical regions and shorter periods—is essential for developing countermeasures for mitigation and containment of spread. The challenges in this endeavor were brought into sharp relief during the COVID-19 pandemic when inadequate predictability held catastrophic consequences across geographical scales, as public health systems across the world scrambled to implement interventional measures for quelling the spread. Therefore, advancing the science of forecasting spatiotemporal epidemic dynamics emerges as a critical aspect of preparedness against future epidemics, especially in the aftermath of our collective, global COVID-19 experience. This imperative fundamentally motivates this article, where we propose a novel approach to forecasting epidemic spread based on a computational framework developed from a compartmental PDE epidemic model. Moreover, the predictive efficacy of the proposed framework is established using a case study wherein empirical COVID-19 infection spread data from Hamilton County, Ohio, United States, is used to explicitly validate the numerical results obtained from the new approach. Adding yet another layer of validation, the spread forecasts obtained from the framework are shown to be well-aligned with corresponding results independently obtained by applying two distinct, widely used retrospective methods in epidemiology: the Wallinga–Teunis (W–T) method and the Cori method to the aforementioned data.
In our quest for improved forecasting, we focus on the fundamental metrics used to characterize the intensity of epidemic spread in theoretical epidemiology as well as in public health policy development—the basic reproduction number (\(\mathscr {R}_0\)) and the effective reproduction number (\(\mathscr {R}_e\))1,2,3. By definition, \(\mathscr {R}_0\) describes the average expected number of susceptible individuals that a person infected with a given pathogen, e.g. SARS-CoV-2, infects in a fully susceptible population, typically at the onset of an epidemic or pandemic. In contrast, \(\mathscr {R}_e\) represents an active epidemic’s potential for propagation across varying time-frames, accounting also for the effects of interventional control measures4,5. More specifically, \(\mathscr {R}_e\) is the number of people in a population who can be infected by an individual at any specific time after the onset of an epidemic. The \(\mathscr {R}_e\) value changes as the population becomes increasingly immunized, either by individual immunity acquired following infection or vaccinations and population change owing to individuals succumbing to the infection. Both \(\mathscr {R}_0\) and \(\mathscr {R}_e\) for an epidemic are typically expressed either as individual values or as residing within a specified low-to-high range. The interpretation is generally straightforward: an \(\mathscr {R}\) value > 1 implies that the disease is likely to propagate, with each infected individual on average infecting more than one other person, thus sustaining the outbreak. In contrast, an \(\mathscr {R}\) value < 1 indicates a probable decline in the outbreak, as the average number of new infections generated by each infected individual is less than one, leading to a gradual reduction in case numbers1,6. We note that during the height of the COVID-19 pandemic, daily reports of \(\mathscr {R}_0\) and \(\mathscr {R}_e\) were used to estimate the severity of the outbreak at different locations. These metrics were also used to determine the impositions of interventions as well as their relaxation. However, the salient point is that current methods of estimating \(\mathscr {R}_0\) and \(\mathscr {R}_e\) suffer from several significant limitations. For instance: (1) It is notoriously challenging to estimate them accurately and in real-time; (2) They are sensitive to the spatial and temporal oscillations in infection rates that profoundly influence the underlying epidemic dynamics7; (3) They depend not only on the biological characteristics of emerging pathogens (such as viruses) about which little is known at the start of a new outbreak (duration of infectivity after a person is infected, transmissibility, and so on) but also on understanding trends in—typically random—human behavior (e.g. contact rate). In particular, at the start of the COVID-19 pandemic, asymptomatic spread was not understood and this resulted in underestimated \(\mathscr {R}_0\) and \(\mathscr {R}_e\). In addition to the unknowns and tenuous assumptions involved in their computation, these metrics are usually estimated retrospectively from serial epidemiological data or alternately by using theoretical mathematical models. The most common method is to use cumulative incidence data (cumulative epidemic curve of cases)8. The implicit lag between the calculated \(\mathscr {R}_0\) and the time of onset of cases within a population leads to inaccuracies and discordant estimates of \(\mathscr {R}_0\). To summarize, the heavy reliance on \(\mathscr {R}_0\) and \(\mathscr {R}_e\) by public health agencies, by the mass media, and by the public at large often overlooks the scientifically well-recognized limitations associated with computations of these epidemiological parameters9. A promising pathway to improved forecasting of epidemic spread lies in developing a predictive methodology that can yield accurate a priori estimates of \(\mathscr {R}_0\) and \(\mathscr {R}_e\) readily computable from currently available data. We therefore now turn to the central question of developing such estimates, observing that the methodology is best anchored on robust, mechanistic, mathematical models of epidemic spread. Notably, prior research10 has mapped pathways for computing \(\mathscr {R}_0\) within ordinary differential equation (ODE)-based models. However, ODE-based models have significant limitations, as will be discussed in the next paragraph.
A fundamental class of mathematical epidemic models is the compartment model11,12 which has also been extensively invoked to analyze the COVID-19 pandemic (see, for instance,8). In the most basic formulation of compartmental models, a population is divided into compartments comprising Susceptible (density of population denoted by S), Infected (I), and Recovered (R) individuals. However, infections that trigger epidemics typically have a latency period (understood as the finite time elapsed before an infected individual becomes infectious) which may be taken into account by introducing a Latent (L) compartment into the model13. Thus, an epidemic progresses as susceptible individuals transition, first to the Latent, and then successively onto the Infected and Recovered compartments, all according to prescribed transition rules. The dynamics of the epidemic are represented in this framework by a system of coupled differential equations for the compartmental densities S, I, R. Whilst efforts focused on analyzing exclusively the temporal dynamics of epidemics continue to follow the historical precedent of treating these equations as ODEs with time as the only independent variable14, ODE models—by definition—are incapable of accounting for spatial dynamics. Therefore, consideration of spatiotemporal epidemic dynamics demands that the aforementioned population densities be treated as functions of both spatial and temporal variables (e.g. S(x, y, t) in two-dimensional Cartesian space, and time). It follows then that the coupled equations be formulated as PDEs9,15,16,17. In particular, parabolic PDEs of the reaction-diffusion type afford a natural setting to study epidemic dynamics18,19,20. Of note here is the role played by such reaction-diffusion PDEs in providing a probabilistic description of stochastic dynamic systems. While this role is well-recognized in statistical mechanics and its applications21,22, the key insight here is that epidemic models based on reaction–diffusion type PDEs can naturally characterize the dynamical relationships between randomness in human movement as well as pathogen transmissibility characteristics and stochastic epidemic spread. However, the complexity of epidemics as stochastic dynamical systems cannot be overemphasized, with multiple driving factors including geospatial dynamics, compliance with interventions (e.g. masking, vaccination), not to mention the appearance of new variants with different latency phases, transmissibility, and pathogenicity23.
Motivated by the above considerations, in recent work, we developed and validated the predictive efficacy of a reaction–diffusion type PDE epidemic model using COVID-19 spread data from both Hamilton County, Ohio. The methodology used was to first let the developed PDE model learn its defining parameters from the actual spread data spanning a period of 30 days, based on an optimal learning scheme. Prediction of spatiotemporal spread for the succeeding 15 days was then obtained by numerically solving the PDE model with the learned parameters. This prediction was validated against the actual spread data for those 15 days. Our objective here is to leverage these encouraging findings and develop a methodology to accurately estimate the reproduction numbers \(\mathscr {R}_0\) and \(\mathscr {R}_e\) into the future, from the PDE model trained with available data from the past. Here, we choose data from the geographical location of the county and the period of 15 days to validate the estimates of the reproduction numbers because our previous results indicate those choices to be adequately aligned with the modeling assumptions. Indeed, the accuracy of estimation will depend on these choices. For instance, assuming that the model parameters learned using data from the previous 30 days will be valid for an arbitrary period beyond a few weeks would likely lead to erroneous predictions, as would similar assumptions for larger geographical locations.
We now turn to the question of mathematically deriving reproduction numbers from epidemic models. The pioneering work in this context was reported for ODEs models where the population densities are considered exclusively to the functions of time. The key result is that \(\mathscr {R}_0\) is the principal eigenvalue (i.e. the spectral radius) of the Next Generation Matrix derived from the ODE framework. This idea first generalized to PDE frameworks in19,24, provides the analytical basis for the computations of reproduction numbers - a cornerstone of this article. The final aspect is to validate the results from the developed computational framework. This important step is accomplished by comparing the estimated \(\mathscr {R}_0\) and \(\mathscr {R}_e\) numbers with those obtained using the W-T and Cori methods; an established, yet retrospective method used in epidemiology. More specifically, the comparative methods facilitate the computation of reproduction numbers from existing spread data. As such, we compare the reproduction number estimates obtained from our PDE-based method applied to Hamilton County COVID-19 spread data with the corresponding estimates obtained by applying the W–T and Cori method to the same data. This exercise, which we repeat for data from multiple timeframes, allows us to validate our method.
To summarize the primary contributions of this article: (i) We propose an alternate method of computing \({\mathscr {R}_0}\) and \({\mathscr {R}_e}\) that leverages the results of a spatiotemporal PDE model developed and validated by the authors in a previous study of stochastic epidemic spread. In particular, the method advances similar methods proposed in the literature by introducing a specific, novel numerical framework for computing different model components. As a result, the proposed method provides a mechanism to predictively compute the \(\mathscr {R}_e\) values based on the PDE model that uses past data to predict the future evolution of an epidemic. The currently used approaches, such as the W–T method25 and the Cori method26, are retrospective and, as such, have not been used in a predictive manner to compute \(\mathscr {R}_e\) values. Furthermore, the proposed method, since it utilizes a spatiotemporal PDE model, also provides a natural mechanism to calculate the \(\mathscr {R}_e\) values at any desired level of spatial granularity. Lastly, while the W–T, Cori, and similar methods are entirely data-driven methods of computing \(\mathscr {R}_e\), the proposed method combines data with a mechanistic PDE model that captures the physics of the spread along with inherent uncertainties. As such, the proposed method is robust to uncertainties in data arising from the data-gathering process. (ii) The paper provides a numerical implementation and validation of the proposed approach using data obtained from Hamilton County, OH for three different time frames. A comparison with the W–T method25 and Cori method26 for each of the time frames demonstrates that the proposed method accurately captures the trends shown by the actual infection data even in certain cases where the comparative methods are found to be inadequate. To the best of our knowledge, this article presents the first computations of \(\mathscr {R}_e\) values on real-world epidemic data using a spatiotemporal PDE model.
The rest of the article is set as follows. We present the foundational PDE epidemic model in Section 2a, followed by a discussion of its discretized version essential for the numerical implementation, in Section 2b. The Next Generation Matrix (NGM) obtained from the PDE framework is discussed in Section 2c, followed by the mathematical derivation of the reproduction numbers as the spectral radius of the NGM. The corresponding derivation of the reproduction numbers in the discretized case is presented in Section . The essential details of the W–T and the Cori methods are provided in Section . Turning next to applying the developed framework to COVID-19 spread data from Hamilton County , the steps are detailed in “Case study”. Specifically, model parameter optimization is discussed in “Hamilton County, Ohio”, and the details of the model validation are presented in Section “Model validation”. We present and discuss the results of the validation in “Results”. The article concludes in in “Discussion and conclusions” with final remarks, including an outlook for further research.
Analytical framework
Partial differential equation (PDE) model
In this paper, we adopt a compartmental modeling approach that may be traced back to the seminal work of Kermack and Mckendrick27 in mathematical epidemiology. Essentially, this approach partitions a population subject to an epidemic outbreak into a distinct number of disjoint compartments; in the simplest models, these are the Susceptible (S), Infected (I), and Recovered (R) compartments. Infection spread is governed by a transition rule representing the mapping of individuals from the Susceptible to the Infected compartment (i.e. the process of susceptible individuals becoming infected). Recovery is likewise represented by a transition rule that maps individuals from the Infected to the Recovered compartment. Moreover, the overall dynamics of epidemic evolution are represented by a system of coupled differential equations for the population densities S, I, and R, with the coupling terms representing the corresponding compartmental transitions. As an epidemic evolves—typically through multiple outbreaks—I correspondingly records multiple local maxima to ultimately approach and stay bounded around a stable equilibrium value. Attainment of this endemic equilibrium marks the end of the epidemic outbreak. We note that the I need not necessarily vanish at this equilibrium but attain a value below the threshold required to trigger further outbreaks.
In the compartmental models discussed above, there exists a fundamental distinction between the two types of differential equations that characterize a model—ODE and PDE. ODE-based SIR models consider the compartmental density functions (S, I, R) exclusively as functions of time; consequently, such models only account for temporal epidemic dynamics. In other words, ODE models only track changes in the number of individuals in each compartment over time and, therefore, are unable to address the spatial dynamics of epidemic spread28. On the other hand, PDE models account for spatial variations that emerge from the interactions in a population and thereby capture the heterogeneity of disease spread across different locations. Specifically, the coupled PDEs in such a model describe how the distribution of individuals in each compartment changes not only over time but also across space29, thereby describing spatiotemporal disease transmission.
A typical characteristic of infection spread is the latency period when an individual is infected but not infectious yet30. Accounting for this feature motivates the introduction of a Latent (L) compartment into epidemic models. Yet another subtle but significant aspect related to latency and spatiotemporal spread is the spatial displacement of infectious individuals in whom the infection is latent. This aspect may be accounted for by the introduction of a convolution term in PDE models (see, for instance, Li et al.13).
Finally, it is remarkable that diffusive PDE models-viewed through the lens of statistical mechanics—can naturally provide a probabilistic interpretation of epidemic spread dynamics. The underpinnings of this interpretation lie in the fact that the probability density functions that describe diffusive stochastic processes satisfy PDEs similar to those that arise in PDE epidemic models. Given that uncertainties—due to random human behavior and random transmission characteristics of the pathogen that triggers an epidemic—play a critical role in dictating epidemic spread, the probabilistic interpretation further endows PDE models with immense power to analyze stochastic epidemic spread.
In light of the above discussion, we now consider the compartmental epidemic model described by the following coupled system of PDEs:
\(\lambda\) is the birth rate in the spatial domain. \(N_{x,y}\) is the total population in cell (x, y), remains constant throughout the specified time period of simulation. \(\eta _{S}\) is the diffusion coefficient representing the intensity of random motion of the Susceptible population. \(\eta _{L}\) is the diffusion coefficient representing the intensity of the random motion of the Latent population. \(\eta _{I}\) is the diffusion coefficient representing the intensity of random motion of the Infected population. \(\eta _{R}\) is the diffusion coefficient representing the intensity of random motion of the Recovered population. \(\nabla ^{a}\) is the Laplacian operator representing the Brownian motion of individuals in a system. \(\theta\) is the mortality rate due to natural deaths. \(\phi\) is the infection rate. \(\delta\) is the rate of individuals exiting the infected compartment due to deaths or recovery. \(\omega\) is the recovery rate of individuals. \(\epsilon\) is the fraction of the infected population that survives the latency period and enters the infected category. \(f_{a}(x,y)\) is the Gaussian kernel defining the extent of the mobility of the latent population. \(\tau\) is the latency period of the model. S, L, I & R is the population densities of Susceptible, Latent, Infected, and Recovered individuals at a specific location and time.
Focusing now on the details of the model equations—which are adapted from Li et al.13—firstly, we reiterate that the population is categorized into disjoint subsets representing the population densities of Susceptible (S), Latent (L), Infectious (I) and Recovered (R) individuals. Note that the population densities are simultaneously functions of the (Cartesian) spatial variables (x, y) as well as time t, with the spatiotemporal dynamics of an epidemic dictated by the system of PDEs as individuals transition through the Susceptible Latent, Infected, and Recovered compartments. Secondly, we note that the diffusion terms in the equations (the Laplacian operator \(\nabla ^a\)) represent the random spatial motion of individuals in the respective compartments. This feature is inherited from the fact that invoking the theory of Brownian motion, under appropriate normalization conditions, the population densities in the model may be interpreted as probability density functions describing diffusive stochastic processes. Thirdly, in our previous work, the efficacy of this PDE model to predict real-world epidemic spread was validated using empirical COVID-19 data from both a larger spatial domain (the State of Ohio, USA) as well as a smaller one (Hamilton County, Ohio, USA)31. Indeed, the present effort is motivated by and grounded upon the aforementioned validation analysis.
PDE model: numerical framework
Our objective of computing reproduction numbers from the PDE model involves numerically solving the PDEs. To this end, guided by our previous work31, we now discretize the system of PDEs using Euler’s forward algorithm as follows.
Here \(X^{T+1} _{x,y}\) denotes the value of variable X (\({X \in \{S,L,I,R\}}\)), on day \((T + 1)\) in the cell with discretized spatial coordinates (x, y) in the two-dimensional domain under consideration, as a function of the values of the corresponding variables on the previous day T. In the discretization, \(\mathscr {N}\) is the number of cells in each dimension of the domain, and \(\sigma ^{T}\) denotes the latent population in a cell on day (T). Two points regarding how latency is taken into account are in order. Firstly, \(\tau\) represents the latency period; therefore, \(\sigma ^{T}\) is computed for the day T based on \(I^{T-\tau }\) and \(S^{T-\tau }\)—the values of I and S from \((T-\tau )\) days ago. Secondly, we consider the realistic scenario of spatial migration of the infected population during the latency period contributing to the spread. This is represented by the convolution kernel \(f_a\) on the RHS of Eq. (9), the explicit form of which is given by the Gaussian distribution in Eq. (10). We note that the Gaussian distribution captures the diminishing probability of people moving to a farther location compared to a closer one13.
We consider no-flux boundary conditions in the numerical solutions of the PDEs. In other words, we assume no traffic of individuals in either direction at the boundary of the spatial domain. Moreover, this no-flux condition is enforced in the numerical solution by having“reflecting” boundaries for the grid, i.e. attempted transgression of the boundary will result in individuals bouncing back into the cells that define the boundary. This assumption allows us to maintain the law of conservation with respect to the population of the system.
Basic reproduction number: analytical formulation
The basic reproduction number \(\mathscr {R}_0\) is defined as the average expected number of secondary infections that occur in a fully susceptible population due to contact with an infectious individual. A disease spreads if the reproduction number is greater than 1; contrapositively, the spread ceases if the reproduction number is less than 1. The infection becomes an epidemic outbreak if \(\mathscr {R}_0\) stays persistently greater than 110. On the other hand, if \(\mathscr {R}_0\) stays consistently less than 1 for a considerable amount of time, it indicates a waning phase of an epidemic32.
A key idea in this work is to numerically compute the reproduction number \(\mathscr {R}_0\) and hence \(\mathscr {R}_e\) for a spatiotemporal domain from a discretized version of the PDE model. To establish the mathematical basis for this computation, we first note the fundamental result from ODE-based epidemic models that \(\mathscr {R}_0\) is the principal or largest eigenvalue (i.e., the spectral radius) of the next-generation matrix derived from an ODE model4,6,32,33. Whilst this result resists straightforward generalization to the PDE case, approaches such as employing a variational formula have been reported for basic Susceptible-Infected-Susceptible (S, I, S) models with diffusion (see, for instance, Allen et al.34). In particular, Wang et. al.19,35 established \(\mathscr {R}_0\) for reaction-diffusion PDE systems as the spectral radius of the next-infection operator, using the theory of principal eigenvalues. We find that this approach can be fruitfully adapted for our purposes and now provide an outline of the development, referring to the previous literature19,24,35 for details.
At the outset, we note that out of the four compartments in the model that we consider, the Latent and Infected compartments form a subsystem of infected states, whereas the Susceptible and Recovered compartments are the uninfected states. Also, note that the initial infection-free state corresponds to \(L = I = R = 0\).
Consider a compartmental model comprising n compartments, where n is the disjoint union of an arbitrary number of infected and susceptible compartments.
Denoting the population density in the \(i^{th}\) compartment by \(u_i(t,\textbf{x})\), let us collect the populations in all compartments in a column vector \(u(t,\textbf{x}) =(u_1(t,\textbf{x}), \dots , u_n(t,\textbf{x}))^T\), where each \(u_i\) is non-negative, \(\textbf{x}=[x, y]\) is the position vector in 2-dimensional space \(\textbf{R}^2\) represented using Cartesian coordinates, and T is the standard transpose operator. Next, the total number of compartments is categorized into two distinct (disjoint) groups: m infected compartments, denoted by \(i=1,\ldots ,m,\), and the remaining uninfected compartments, identified by \(i=m+1,\ldots ,n\). A generic reaction-diffusion system of PDEs (along with the necessary boundary conditions) representing the aforementioned compartmental model can now be written as:
In the above equation, \(u_i\) is the density of individuals in the \(i^{th}\) compartment, while \(d_i(\textbf{x})\) is the corresponding diffusion coefficient (representing the random movement of the population \(u_i\)). The terms \(\mathscr {F}_i\) and \(\mathscr {V}_i\) are the reaction terms in the \(i^{th}\) compartment (accounting for transitions between compartments induced by the disease spread). Specifically, \(\mathscr {F}_i(\textbf{x},u)\) characterizes the rate at which the newly infected population is introduced into a given compartment i. Also,
where \(\mathscr {V}^+\) denotes the transfer rate of individuals into this compartment through all means, and \(\mathscr {V}^-\) is the transfer rate of individuals out of the compartments. We note that the PDEs are defined in the spatial domain \(\Omega\) (a subset of \(\mathbb {R}^2\)), which is assumed to have a smooth (i.e. continuously differentiable) boundary \(\partial \Omega\). The symbol \(\nu\) represents the outward-facing unit normal vector along the boundary \(\partial \Omega\). The second of the equations 11 is the no-flux boundary condition and reflects the physical assumption that the boundary is impermeable to individual movement in both the inward and outward directions.
It is useful here to write the system of Eq. (11) in an equivalent form by expanding the first term of the RHS of the PDE using the product rule for differentiation as:
where,
Before proceeding to define the basic reproduction number \(\mathscr {R}_0\) corresponding to the PDE Eq. (12), we now briefly discuss the key ideas behind the Next Generation Operator defined in terms of the key operators \(\mathscr {F}_i\) and \(\mathscr {V}_i\). Firstly we note that these operators were first defined in the context of ODE models as matrices32. Accordingly, the Next Generation Matrix (NGM), is obtained in terms of the F and V matrices as \(FV^{-1}\). The F matrix is called the “new infection matrix” or the “transmission matrix”. It represents the rate at which new infections are generated by the currently infected individuals and is an \(m\times m\) matrix (considering only the infected group) with its \((p,q)^{th}\) entry given by:
Here \(u^0\) represents the disease-free equilibrium point. These matrices are evaluated at the Disease Free Equilibrium (DFE), where no infections are present, serving as a crucial starting point for determining \(\mathscr {R}_0\), which quantifies the potential for disease spread in a fully susceptible population. Moreover, each element of the matrix \(F_{pq}\) represents the expected number of new infections in the compartment p produced by an infected individual in compartment q over a single infectious period in a fully susceptible population. On the other hand, the V matrix, which is known as the “transition matrix” or “transfer matrix”, characterizes the movement of individuals between different compartments due to recovery, progression of the disease, or death. It is also an \(m\times m\) matrix (once again considering only the infected group) with its \((p,q)^{th}\) entry given by:
The basic reproduction number \(\mathscr {R}_0\) is then established as the spectral radius of the NGM. The underlying rationale is the fact that the largest absolute eigenvalue—which is the spectral radius—of the NGM matrix represents the peak average number of new infections produced by a single infected individual. This takes into account the rate of transfer among various stages of the disease within a single infection cycle. Essentially, this value reflects the most significant rate of disease transmission in a population, considering the detailed progression and the contributing interactions.
Refocusing our attention on the PDE, and referring to the discussion in19,24 for more details, consider the following vector-valued reaction–diffusion equation. We note that this is identical to the previous equations in 12, except that here only all the infected compartments are considered. Moreover, the corresponding population densities are collected in a column vector. The vector-valued PDE is:
Where \(u_\mathbb {I}\)= \((u_1,...,u_m)^T\) and \(d_\mathbb {I}(\textbf{x})= diag(d_1(\textbf{x}),\ldots , d_m(\textbf{x}))\) and \(c_\mathbb {I}(\textbf{x}) = diag(c_1(\textbf{x}),\ldots , c_m(\textbf{x}))\).
Now, let \(\mathscr {T}(t)\) be the operator that temporally propagates an initial spatial distribution that satisfies the PDE Eqn. 16. If we take the initial infection distribution (arrangement of infections at the beginning of the observation period) to be \(\zeta (\textbf{x})\)= \((u_1(t=0,\textbf{x}),\dots ,u_m(t=0,\textbf{x}))^T\), then, \(\mathscr {T}(t)(\zeta (\textbf{x}))\) represents the distribution of the infections for any \(t > 0\). Therefore, the spread of new infections at any time \(t > 0\) can be given as \(F\mathscr {T}(t)(\zeta (\textbf{x}))\). The distribution of total new infections can then be represented by the following equation:
Next, consider the NGM operator K that takes the starting distribution of infections and describes how it spreads to the total number of infected individuals over time, as explained in33. It can be defined as:
Following19,24,35, the spectral radius of K yields the basic reproduction number of the PDE model:
where \(\rho\) is the spectral radius of the matrix. Moreover, Theorem 3.1 of Wang et al.19, (which follows from Theorem 3.12 of Thieme et al.36), provides the following equation:
where \(\Gamma := \nabla . (d_\mathbb {I}\nabla V) - V\). The term \(\nabla . (d_\mathbb {I}\nabla V)\) represents the divergence of the gradient of V multiplied by a diffusion coefficient \(d_\mathbb {I}\).
Basic reproduction number: numerical formulation
We now present the numerical formulation—within a discretized setting—corresponding to the mathematical description of \(\mathscr {R}_0\) provided in “Basic reproduction number: analytical formulationR0spsanalytical”. Let \(\mu\) be an eigenvalue of K in Eq. (17) such that \(K(\zeta (\textbf{x}))= \mu \zeta (\textbf{x})\) with corresponding eigenvector \(\zeta (\textbf{x}) = (\zeta _1(\textbf{x}),..., \zeta _m(\textbf{x}))^T\). Then,
Suppose that \(V= diag(\nu _1,\ldots ,\nu _m)\), \(\psi (\textbf{x})\)= \(- \Gamma ^ {-1} (\zeta (\textbf{x}))\), where \(\psi (\textbf{x}) = (\psi _1(\textbf{x}),..., \psi _m(\textbf{x}))^T\), then \(-\Gamma (\psi (\textbf{x})) = \zeta (\textbf{x})\); i.e.,
For numerical computation, we consider a square grid of size \(\mathscr {N} \times \mathscr {N}\). We consider a sufficiently large integer \(\mathscr {N}>0\) that represents the grid size of the computational domain. In the formulation below, we compute the reproduction number for each row indexed by \(\texttt {y}\). For each row \(\texttt {y}\), we consider a one-dimensional discretized spatial domain (normalized) along the x-axis [0, 1]. Here, \(x_j = j/\mathscr {N}\), \(d_{ij} = d_i(x_j)\), \(c_{ij} = c_i(x_j)\), \(\psi _{ij} = \psi _i(x_j)\) and \(\zeta _{ij} = \zeta _i(x_j)\) for \(j= 1,\ldots ,\mathscr {N}\). By applying a central difference scheme, we rewrite Eq. (22) as:
for all \(1\le\)j\(\le \mathscr {N}\), with \(\psi _{i,1}= \psi _{i,2}, \psi _{i,\mathscr {N}}= \psi _{i ,\mathscr {N}-1}\) according to the no-flux, Neumann boundary conditions. By combining the \({\mathscr {N}}\) terms in a matrix, we obtain:
where \(A= diag(A_1, \ldots , A_m)\), and:
with \(b_{i,j}^+ = -\mathscr {N}(d_{i,j}\mathscr {N} + \frac{c_{i,j}}{2}), \quad b_{i,j}^- = -\mathscr {N}(d_{i,j}\mathscr {N} - \frac{c_{i,j}}{2}), \quad \text {and} \quad a_{i,j} = 2d_{i,j}\mathscr {N}^2 + \nu _i; for \quad all \quad 0\le j \le \mathscr {N}\).
From Theorems 3.1 and 3.2 in Wang et al.24, it follows that:
The above equation may also be written as:
where \(\mathscr {I}_{\mathscr {N}}\) is the identity matrix of dimensions \((\mathscr {N}) \times (\mathscr {N})\) and \(\bigotimes\) is the Kronecker product. The standard definition of the Kronecker product is as follows: Given an \(r \times s\) matrix M with elements \(m_{ij}\) and another matrix Q of size \(p \times q\),
Substituting \(\Psi \approx A^{-1}\Phi\) in the above equation yields:
Therefore, we can now state the following central result:
We note the introduction of row index \(\texttt {y}\) in the subscript above to emphasize that the above equation computes the reproduction number for row \(\texttt {y}\).
As discussed previously, at the onset of an epidemic, \(\mathscr {R}_0\) acts as a marker of the spread intensity, predicated on the assumption that the entire population is susceptible. However, as the epidemic progresses, this assumption becomes tenuous, owing to factors including the acquisition of immunity, behavioural modifications, and spatial heterogeneities. In other words, \(\mathscr {R}_0\) is an inadequate measure of the intensity of spread beyond the initial stages of an active epidemic37. This necessitates the introduction of \(\mathscr {R}_e\), which reflects the actual average number of secondary cases per infectious case at a given time and space for an evolving epidemic. Calculating \(\mathscr {R}_e\) is crucial for understanding the current state of epidemic spread at any point in time, as well as for assessing the impact of public health interventions and making informed decisions about future strategies38. It helps capture the dynamic nature of the epidemic by accounting for changes in susceptibility, contact patterns, and mobility, thus providing a more timely and context-specific metric. Referring to1,5,39, we note that:
where S = The total number of susceptible individuals at a point in time, and P = The total population at that time.
Reproduction number for the discretized SLIR model
In the light of the discussion in the previous section, we now focus on the reproduction number \(\mathscr {R}_e\) for the discretized version (Eqs. 5–8) of our underlying SLIR model. At the outset, we note that the two infected compartments in our model are the Latent and the Infected ones given in (6) and (7), respectively. We recall that when an individual gets infected, they first migrate into the Latent compartment (where they are considered to be infected but not infectious). Once they are fully infectious after a specific incubation period \((\tau )\), they transition into the Infected compartment.
Next, we recall that in the model, all new infections are accounted for in the F matrix. The F and V matrices can be derived by linearizing the discretized equations of the model; the linearization may be achieved by the standard analytical method of computing the Jacobian (first-order partial derivatives of a vector function) of each equation relative to the others at an infection-free equilibrium state denoted by \((S^0,L^0,I^0,R^0)= (S^0,0,0,0)\). The infection-free steady state, as discussed previously, represents the initial scenario when the entire population is susceptible (denoted by \(S^0\)), and, therefore, there are no individuals in the Latent, Infected or Recovered states. The linearization procedure facilitates the estimation of \(\mathscr {R}_0\) as described analytically in the previous sections. Next, we determine the F and V matrices for our underlying model, as outlined in the Eqs. (6) and (7). As the first step in this process, we compute the \(\mathscr {F}\) and \(\mathscr {V}\) matrices, defined in Eq. (12), as:
From Eqns. 14 and 15 , we obtain the matrices F and V as:
We note that the disease-free equilibrium, \(S^0\), represents a steady state where the number of individuals becoming susceptible (e.g., through birth, represented by \(\lambda )\) is balanced by the number of individuals leaving the susceptible state (e.g., through natural death, which is represented by \(\theta\)). The equilibrium value of \(S^0\) in Eq. (33) is therefore determined by the ratio of these rates10,24,40:
The numerical formulation presented in “Basic reproduction number: numerical formulation” is now used in a grid framework where x and y represent the row and column indices. \(L_{x,y}\) represents the Latent population for the grid element positioned at row x and column y. We now apply Eq. (23) to obtain the Latent population \(L_{x,y}\) and arrive at the following equation:
where \(V_L\), which represents the contributions to V (see Eqn: 34) from the Latent compartment, is given by:
We also note, in deriving 36, that we assume the diffusion coefficient for the Latent population to be spatially uniform and given by \(d_L\). From Eq. (23), we note that \(c_L\) vanishes under the above assumption. Therefore, the \(A_L\) matrix is obtained as:
where,
Similarly, we now determine the matrix \(A_I\). First, we compute the \(I_{x,y}\) using the Eq. (23):
where \(V_I\), which represents the contributions in V (see Eq. (34)) from Infected compartment, is given by:
In the derivation of Eq. (40), we note that the diffusion coefficient for the Infected compartment (\(d_I\)) is assumed to be spatially uniform. This assumption leads to the elimination of the term \(c_I\) in Eq. (23). Therefore, the \(A_I\) matrix is given by:
where,
Therefore, the matrix A is given by:
By substituting the obtained expressions for the matrix A (Eq. 44) and the matrix F (Eq. 33) into Eq. (28) and solving the eigenvalue problem described by Eq. (28), one obtains the principal eigenvalue \(\rho\) in the equation (Eq. 29). From there, we find the \(\mathscr {R}_{e(\texttt {y})}^{PDE}\) of the system by using Eq. (30) that provides the reproduction number of for row \(\texttt {y}\) at any specified time. The overall reproduction number of the entire region is then computed by averaging over all \(\mathscr {N}\) rows using:
where, ’\(\mathscr {N}\)’ is the grid size of the spatial domain, \(\mathscr {R}_{e(\texttt {y})}^{PDE}\) is the reproduction number for the row \((\texttt {y})\), and \(\mathscr {R}_e\) is the reproduction number for the entire domain.
The Wallinga–Teunis method
To carry out a comparative study with respect to established (albeit retrospective) methods for calculating the reproduction number, we first choose the Wallinga & Teunis (W*-T) method25. To determine the reproduction number for a primary case, the W–T method employs the probability distribution for the generation interval (also called the serial interval), which is the time from symptom onset in a primary case to symptom onset in a secondary case. By using this distribution, the method estimates the relative likelihood that a secondary case has been infected by a specific primary case, given the observed difference in their time of symptom onset. The effective reproduction number is then calculated by summing over all observed secondary cases, each weighted by its relative likelihood of being infected by the particular primary case. Our calculations applying the W–T method were carried out through the R package, \(EpiEstim v2.2-4\)26. This framework assumes that there is a complete recording of cases and that there are no asymptomatic cases. The W–T method is completely retrospective, meaning it contains a sliding window that takes in the data for the previous 30 days as input to provide the \(\mathscr {R}_e\) estimates for a given day.
We choose the Wallinga–Teunis (W–T) method as a validation tool for our study as it is a well-established method that has been widely used in the field of epidemiology and by public health agencies, including the Center for Disease Control (CDC), the World Health Organization (WHO), as well in academic research. In addition to being used in multiple studies during the COVID-19 pandemic (see, for instance,41,42), it was also used during the previous SARS epidemic and for other infectious respiratory disease epidemics to gain a comprehensive understanding of the transmission dynamics over time. For our purposes, one of the main strengths of the W–T method is that it does not assume a constant serial interval, allowing for dynamic changes over time resulting from interventions and changes in the pathogen (virus). It is also robust when the incidence is low, and it considers the entire epidemic curve, thus providing a good benchmark41,42. Moreover, it relies on retrospectively collected data. For those reasons, we employ the W–T method to validate the prediction performance of our framework.
Cori method
To further buttress the validation, we also employ the Cori method—yet another widely adopted, retrospective method in epidemiology for reproduction number computations26. The Cori method focuses on obtaining instantaneous values of\(\mathscr {R}_{e}\). This method assumes that the random spread of disease over time follows a Poisson process with mean \(R_t \sum _{s=1}^{t} I_{t-s} w_s\), where \(I_{t-s}\) is the incidence infected in time step \((t-s\)) and \(w_s\) is a probability distribution describing the average infectiousness profile after infection, approximated by the distribution of the serial interval.
The infectiousness profile of individuals-indicating how contagious they are after becoming infected-is modeled using a distribution of the serial interval. The model assumes that the transmissibility of the disease (i.e., the ease of spread) remains constant over a fixed period. To estimate the reproduction number (\(\mathscr {R}_{e}\)), this method follows a Bayesian framework. In this framework, prior information about transmissibility is represented by a Gamma prior distribution, which is based on previous knowledge or appropriate assumptions.
Post observing new data, the model updates its estimate of the reproduction number using this Bayesian estimation approach. The updated estimate (posterior distribution) for the reproduction number also follows a Gamma distribution but with new parameters that reflect the observed data.
Similar to the W–T method, the latest R package of \(EpiEstim v2.2-4\)26, developed by the Cori team at Imperial College, London, UK43 was employed. We ran the program on R4.2.2 (64 bit), using the function “estimate_R”for the Cori method. We calculated the \(\mathscr {R}_{e}\) values over sliding weekly windows in each timeframe.
Case study
Hamilton County, Ohio
We now apply the ideas and methodology presented in the previous sections to numerically compute reproduction numbers from COVID-19 data for Hamilton County, Ohio, USA, and also validate the results by comparing them to those obtained from the W-T and Cori methods. To this end, we first follow our previous work44 to predict the trajectory of the S, I, and R variables for Hamilton County for time frames of 15 days, based on actual data for the preceding 30 days. We refer to this actual dataset - provided by the Epidemiology Division of Hamilton County Public Health45 - as the“true dataset”. We note that this spatiotemporal dataset provides the daily count of infections and deaths in each zip code across the entire county, starting from April 20, 2020 (Day 1 in our study). We examined three distinct periods, each spanning 45 days, in our case study. The first timeframe, spanning from April 20, 2020, to June 3, 2020, represented the initial stage of the epidemic. The second timeframe analyzed data from October 15, 2020 to December 28, 2020, and lastly, the third timeframe focused on the time period February 15, 2021, to March 31, 2021. The considered timelines and the rationale supporting their selection are as follows:
1. Timeframe-1 (T1): April 20, 2020, to June 3, 2020. This period represents the initial phase of the pandemic for the County, coinciding with the commencement of lockdowns and other containment measures.
2. Timeframe-2 (T2): October 15, 2020 to December 28, 2020. This timeframe captures the onset of increase of infections trend and thereby offers a window into the rapid dynamics of the spread.
3. Timeframe-3 (T3): February 15, 2021, to March 31, 2021. This period corresponds to the decline of infections immediately following the peak, offering insights into the relaxation dynamics of the post-peak scenario.
We initiate the process by considering the entire population of the county as the initial value for S. When individuals in the S compartment become infected, they transition to the Latent (L) compartment, characterized as infected yet incapable of transmitting the disease. After this latency phase, guided by the convolution kernel’s dynamics, these individuals then progress to the infected compartment. For the duration of the latency period in this study, we reference the work of He et al.46. This period, indicated by the symbol \(\tau\), is assigned to be 5 days. For each subsequent day, we calculate the value of S by deducting the total number of cases, recoveries, and deaths from the previous day’s data. We emphasize that we have completely and comprehensively utilized all the available data as such for each specific time frame. Once we pre-process the dataset in this manner, we incorporate it into our discretized model.
For our spatial analysis in the study, the shapefile of Hamilton County was utilized, obtained specifically from the IPUMS National Historical Geographic Information System dataset47. The county’s geography was methodically divided into a structured cellular grid, comprising of 3600 cells in a \(60\times 60\) arrangement, cumulatively spanning roughly 410 square miles in area. The dimensions of this mesh grid were chosen by considering the following three pivotal factors:
-
1.
The precision necessary for the study’s accuracy,
-
2.
The computation cost, and
-
3.
The granularity of the available data, particularly the population statistics detailed at the zip code level.
A coarser mesh might simplify the computations but at the cost of missing details and increased discretization errors, while an excessively fine mesh could lead to computational loads. Our selected grid size thus represents an optimized compromise, aligning the model’s accuracy with feasible computational demands. Figure 1 provides a visual representation of the Hamilton County shapefile and the corresponding grid size distribution. Each cell within this grid is characterized by the following attributes:
-
(i)
A logical value indicating whether the cell belongs inside Hamilton County,
-
(ii)
a logical value indicating if the cell is part of the boundary,
-
(iii)
Initial values for Susceptible (S), Infected (I) and Recovered (R) population to each cell.
Figure 2 demonstrates the distribution of population in each cell, with respect to the population in the zip code to which the cell belongs. It’s important to note that our model operates in terms of population densities and the provided data from the shapefile of Hamilton county47 contains actual population numbers for each Zip-code in Hamilton county. To bridge this gap, we assume that the data is uniformly distributed within each zip code. By doing so, we calculate the actual values of S, I, and R for each cell in our grid. We achieve this by evenly distributing the true numbers of S, I, and R from the dataset across all cells representing that county. This results in an estimated population density for each category in each cell, which we then utilize in our model. It is to be noted that this model does not assume spatial uniformity over the entire county. For numerical computations, we divide the county (more generally, any spatial region) into a number of grid cells. We assume uniformity in population distribution in each of those cells. Moreover, we note that the finest granularity of publicly available population data for the county is at the level of zip-code. Hence, we are constrained to assume uniform population density for all cells that belong to a zip-code. The use of population demographic information (age, gender, ethnicity, and so on) may affect the model output if such factors are considered in the underlying PDE model equations. Such a study, though of interest, is beyond the scope of the present manuscript. We note that a case study on predicting spread dynamics across a larger spatial domain was reported in our previous article Majid et al.31,44, where we analyzed the PDE model predictions using COVID-19 data from entire state of Ohio, USA.
For our simulations, we consider a Neumann (no-flux) boundary condition, which treats the cells on the boundary as a solid wall, that the population can not cross. Any attempt to cross the boundary will result in the individual being reflected to their nearest neighboring cell, which is present within the boundary. This allows us to maintain population conservation in the domain (apart from the deaths).
To get the \(\mathscr {R}_e\) estimates for validation, we run the W-T and Cori methods on the Hamilton County COVID-19 dataset. We have used the parametric method to define the serial interval to have a Gamma prior distribution with a mean of 5 days and a standard deviation of 3 days46. The “wallinga_teunis” function in the R package“EpiEstim”was used to carry out the calculation with 100 simulations. The size of the window was set to 30 days to match the amount of training data used in our PDE model.
Model parameter optimization
Our objective is to obtain the model parameters that minimize the objective function representing the error between the ground truth and the model results. By comparing the model results with the ground truth data for each timeline, we aim to fine-tune the model parameters to better reflect the real-world infection dynamics during these periods. The objective function used in this study is given by:
\(L_m\) is the error between the model data and true data is calculated using the root-mean-square approach. \(N_z\) is the number of zip-codes \((N_z = 58)\). T is the total number of days in the training phase \((T=30)\). t is the current time index. z is the current zip-code index. X is the ground truth data of the \(z^{th}\) zip-code. \(\tilde{X}\) is the model output obtained by summing the values for all the cells belonging to the \(z^{th}\) Zip-code.
We initialize the values of the non-dimensional parameters under a reasonable range and start optimizing the parameters using the initial 30 days’true data. Adopting the methods described in these references:49,50,51, we employ a Genetic Algorithm (GA) to optimize the non-dimensional parameters in our model equations, ensuring a good fit to the actual data spanning 30 days (called training phase) from Hamilton County. Once we have obtained the parameter values that best match the true data, we then use them to predict the trajectory of infections for the next 15 consecutive days.
Employing GA, we heuristically optimize the 10 non-dimensional parameters in our discretized PDE compartmental model (Eqs. 5–8). The GA was chosen specifically for its ability to navigate complex, multi-dimensional parameter spaces effectively. The fitness function, derived from Eq. (46), quantitatively demonstrates how well the parameters fit the actual data. We initialized the GA with a random population of candidate solutions and utilized parallel processing to enhance computational efficiency. Furthermore, to preserve the biological relevance of these parameters, we enforce that all parameter values be positive-invariant, meaning they are constrained to remain strictly positive throughout the optimization process. This comprehensive approach ensures that the optimized parameters are robust and reliable for fitting the actual infection trajectories.
Model validation
We emphasize that during the numerical analysis, the model parameters were optimized using data from the initial 30 days of the true dataset, which served as the training phase. Throughout the analysis, we ensured that the parameter values remained within physically reasonable ranges.
Subsequently, we employed the optimized parameters obtained from the training phase to predict the values of S, I, and R for the following 15 days, which formed the testing phase.
The optimized parameters derived from the training data are presented in Table 1, and their consecutive training vs prediction graphs are presented in Figs. 3, 4 and 5. Here, Susceptible, Infected, and Recovered plots are obtained for each day by summing over the S, I, and R values for all the cells.
While the model prediction results demonstrate its ability to effectively capture the dynamic trends of the susceptible, infected, and recovered populations, it is essential to acknowledge potential sources of error. One limitation of the model is that it assumes constant parameter values throughout the simulation period and considers spatial uniformity throughout Hamilton County. However, in reality, parameter values may change over time due to different factors such as super-spreaders, the intensity of the epidemic, Non-Pharmaceutical Interventions, and they can also vary across different regions based on local government actions.
To address these limitations and improve the model’s efficacy, future work could involve using a longer training period, which may provide a more comprehensive understanding of the parameter variations over time. Alternatively, focusing on specific time periods when Non-Pharmaceutical Intervention (NPI) measures were consistently implemented as per public health policy can help enhance prediction accuracy.
Additionally, considering smaller spatial regions instead of assuming spatial uniformity could provide more localized insights and improve the precision of predictions. By incorporating these refinements, the model’s predictive capability can be enhanced, leading to more accurate representations of real-world epidemic dynamics.
Results
Hamilton County, Ohio
Next, we present the results, observing at the outset that they fall under two categories. Firstly, for each of the three timelines considered, we present the respective effective reproduction numbers (\(\mathscr {R}_e\)) and compare them against the corresponding reported case incidence data for Hamilton County. This comparison provides the basis to evaluate the efficacy of our method in accurately forecasting the trend of infection spread. Secondly, we independently compute the corresponding effective reproduction numbers (\(\mathscr {R}_e\)) using the W–T and Cori methods and directly compare the results obtained using both approaches. We reiterate and underscore here that the W–T method and Cori method are essentially retrospective methods and, as such, are incapable of forecasting \(\mathscr {R}_e\). However, as an established epidemiological framework, it validates the results obtained from our method. While we compare the results from our framework with those of the comparative methods, we need to consider the difference in how computations using the two methods are carried out. For calculating the \(\mathscr {R}_e\) value for any day, both the W–T method and Cori method use the previous 30 day data. However, the method based on our framework uses the first 30 day data for optimizing the model parameters and then uses the optimized parameters to calculate the \(\mathscr {R}_e\) values for the entire 45 day period in each of the three timeframes.
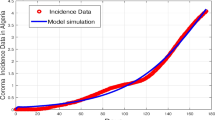
As the basis to understand and evaluate the results, in Fig. 6 we present the actual daily reported cases in Hamilton County for the entire period between 7 March 2020 to 15 September 2022. In addition, the three timelines T1, T2, and T3 chosen for the analysis are indicated in the figure, along with the points in time at which the Alpha, Delta, and Omicron variants of SARS-CoV-2 (the causative virus for COVID-19) are estimated to have emerged in circulation52. Next, the results corresponding to the first timeline T1 are presented in Fig. 7. The graph in the top part of this figure presents the effective reproduction numbers (\(\mathscr {R}_e\)) plotted against time. Specifically, here the ’Model training’ (Solid blue line) plot represents the learning period during which the model parameters are computed from actual data using the optimization scheme. The forecast reproduction numbers (i.e., the model predictions) are represented by the ‘Model testing’ (Dotted red line) plot. The corresponding reproduction numbers obtained from the W–T method are shown in the ‘\(\mathscr {R}_e\)-WT Method’ (Yellow line with circles at each time axis data point) plot. To facilitate easy comparison with these results, the graph in the bottom part of this figure presents the incidence rates during T1. Here, the ‘True Incident Cases’ plot (Magenta line with circles at each time axis data point) indicates the reported daily cases, whereas the ‘5-day Moving Average’ (Blue line) plot indicates the corresponding stated average.
The plot of the reproduction number calculated for T1 is demonstrated in Fig. 7. The numerical estimation of the reproduction number for the simulation in this timeframe initiates at approximately 0.95 and appears to decrease slightly as the simulation progresses, following a relatively stable trajectory with minor fluctuations. In contrast to our framework’s results, the results obtained from the W–T method and especially the Cori method appear to display a more pronounced change, starting from approximately 1.2 and rapidly moving down to match our results. This is common for both methods at the beginning of the pandemic. While the comparative methods demonstrate a higher reproduction number at the beginning of the simulation, it undergoes a linear decrease over time, ultimately converging with the output of our framework towards the end of the simulation.
The graph in Fig. 8 illuminates the computed reproduction number for T2. This timeframe is characterized by an upward trend in daily incidence. Initially, the value starts at approximately 1.1. with minor fluctuations, generally maintaining this level. As the simulation progresses, it slowly decreases, eventually dropping below 1, indicating a decrease in daily incidences. In comparison, results from the W–T and Cori methods tend to fluctuate around 1.2 for most of the simulation period but decline towards the end. The observed differences may be due to the high incidence rates and significant fluctuations of infections within this timeframe. The W–T and Cori method’s 30-day window could contribute to its insensitivity to these fluctuations. Nonetheless, the initial increase in daily incidence rates and subsequent levelling off and the decline appears to be better captured using our framework than the W–T method. To provide further insight, we extended the daily incidence graph by an additional 15 days, clearly showing that the moving average of daily incidence decreases as the value of \(\mathscr {R}_e\) falls below 1 as obtained by our method.
Finally, Fig. 9 provides the simulation results for the reproduction number during T3. The value of the reproduction number, as calculated by our method, starts a little above 0.95, with only slight variations, it stays consistent throughout this period. The daily incidence cases show similar flat behaviour as seen by the 5-day moving average values. Meanwhile, the result from the W–T method starts at almost the same value as ours but then briefly falls below this mark before experiencing a sudden upswing. This interesting behaviour might stem from the comparative methods initially including the data from before T3 in its 30-day sliding window, where the daily cases dropped quickly. After 30 days, the \(\mathscr {R}_e\) values from both the W–T method and Cori method start to closely align with our \(\mathscr {R}_e\) values.
Discussion and conclusions
In this article, we presented a new analytical methodology for accurately forecasting the basic (\(\mathscr {R}_0\)) and effective (\(\mathscr {R}_e\)) spatiotemporal reproduction numbers at any point in an epidemic, based on available infection spread data at that point. Furthermore, the methodology was anchored on a PDE-based, compartmental model of epidemic spread that: (1) accounts for stochastic aspects essential to describing spread dynamics influenced inevitably by uncertain factors, including human behaviour and pathogen transmission characteristics, and (2) was validated using COVID-19 spread data from both Hamilton County, Ohio, and the entire state of Ohio, United States, in our previous work. As the first step in the research methodology, we developed a new computational framework for numerically computing reproduction numbers from actual data—the computational framework faithfully mirrors the fundamental mathematical derivations of reproduction numbers obtained from the theory of PDEs applied to epidemic compartmental models. In the second step, we followed our previously developed strategy of training the PDE model to learn its key parameters from available infection spread data corresponding to a period of 30 days based on an optimization scheme, based upon which the model forecasts the spread for the next 15 days. The results from the first two steps then together facilitated our third step, wherein we numerically computed the predictions of the model for the reproduction numbers themselves. These predictions were validated against spread data from Hamilton County, Ohio, and further reinforced by a secondary set of validations against independent results obtained independently from applying the traditional Wallinga–Teunis (W–T) and Cori methods to identical data sets. Moreover, this exercise in the third step was repeated for infection spread datasets corresponding to three distinct phases witnessed by Hamilton County during COVID-19.
That the predictions of the effective reproduction number \(\mathscr {R}_e\) obtained from our framework—applied to the three distinct periods T1, T2, and T3—accurately trace the corresponding trends in actual infection spread data for each of those periods is a central result of this article. Indeed, this result establishes that this new approach to computing reproduction numbers rests on sound analytical foundations and inspires confidence about its predictive efficacy when supplied with real-time infection spread data as input. Furthermore, the framework predictions were observed to be accurate for T2 (a period marked by a significant rise in infections) and T1 and T3 ( both periods with decreasing numbers of infections). This is a testament to the robustness of the framework in predicting reproduction numbers during surges as well as recessions, both of which typically recur multiple times throughout an epidemic.
Since the predictions were also compared to reproduction numbers computed independently using the W–T and Cori methods, reiterating the fundamental contrast between these approaches appears to be in order here. Both the W–T and Cori methods are essentially retrospective or a posteriori methods and, as such, are not designed for forecasting reproduction numbers. Furthermore, significant errors that could potentially accrue should the W–T or Cori methods be wantonly employed for prediction have been illustrated as part of our discussion of the results from T2 and T3 in “Discussion and conclusions”. More to the point, comparison with actual data buttressed the credentials of the proposed approach for accurate forecasting.
Turning now to potential directions for further research, firstly, it will be interesting to investigate the fidelity of the computational framework across spatiotemporal scales. Moreover, the ability to predict \(\mathscr {R}_e\) from data collected at any level of granularity is a highlight that inspires confidence in the computational framework, particularly when employed for forecasting in smaller regions and for shorter durations.
Secondly, it will be interesting to apply the framework to compute reproduction numbers for other (periodic and otherwise) epidemics, such as influenza. While the specifics of the transmission dynamics can indeed be expected to change depending on the pathogen, we believe that the mechanistic nature of the underlying PDE model provides a foundation robust enough to effectively forecast \(\mathscr {R}_e\) in the case of other epidemics. In particular, we expect this robustness to result in reasonably reliable predictions even in the face of data moderately constrained owing to the presence of outliers. In other words, the proposed framework inherits all the inherent advantages of mechanistic models, which are, in general, less susceptible to incomplete data than exclusively data-driven approaches to forecasting.
A third interesting research direction will be to investigate the efficacy of the approach in predicting \(\mathscr {R}_e\) in the event of rapid surges in infection spread. Such surges could, for example, be triggered by superspreader events and were a major concern during the COVID-19 pandemic. We expect that the underlying PDE model will need to be analyzed more extensively to understand both the dynamic instabilities that will likely characterize the surges in infection spread as well as the post-surge relaxation dynamics that mark the return to equilibrium.
The framework presented in this article to compute reproduction numbers is based on the estimates of infection spread obtained from the underlying PDE model. Therefore, revisiting the computation of reproduction numbers guided by improvements to the PDE model constitutes an interesting fourth direction for future research. In particular, allowing the critical model parameters to be time-varying and estimating them using a dynamic optimizing process could enhance the accuracy of the PDE model. Motivated thus, we recently developed a time-varying infection parameter estimation model using Long Short-Term Memory (LSTM) networks and a data-driven approach. This iterative approach allows us to effectively capture the complex day-to-day dynamics of infection parameters while efficiently managing computational complexity. By leveraging the capabilities of LSTM networks, we can track infection dynamics over longer time frames and across larger regions with improved computational feasibility. This data-driven methodology not only enhances the accuracy of parameter estimation but also broadens the applicability of traditional PDE models in practical settings. The details are provided in our conference paper53, and we intend to investigate the implications of the results for the computation of reproduction numbers in future work.
Accurate forecasting is a critical precursor for the development and implementation of control interventions aimed at mitigating infection spread. Therefore, control-theoretic analyses of Non-Pharmaceutical interventions such as social distancing, as well as vaccinations, predicated on \(\mathscr {R}_e\) numbers obtained from the proposed framework present a fourth promising future research direction. Pursuing this line of research, we recently investigated a Nonlinear Model Predictive Control (NMPC) framework54 applied to the PDE model. This control method introduces an open loop control input into the system, the optimal value of which is computed using a receding horizon scheme. The computational results indicated a significant reduction in the number of infections upon implementation of the control input obtained from the NMPC framework. Investigating the expected reduction in reproduction numbers due to applying control to the PDE model represents yet another exciting direction for future research.
To conclude, reproduction numbers are fundamental quantitative metrics that characterize an epidemic spread, and the ability to accurately predict them from available data is a critical predicate for effective spread mitigation. The available data often being sparse—especially in the early stages of unprecedented epidemics such as COVID-19—predictive computations of reproduction numbers built upon mechanistic, mathematical models are essential, also as they tend to be more robust and reliable compared to exclusively data-driven methods. In this article, we presented such a predictive computational framework for forecasting reproduction numbers from available infection spread data and validated the same using COVID-19 data. We hope that the results reported in this article spur further research and, most importantly, open new pathways in forecasting reproduction numbers that will help significantly enhance our preparedness to swiftly conquer future epidemics.
Data Availability
Numerical code as well as corresponding datasets used for this study have been made available in our GitHub repository.
References
Delamater, P. L., Street, E. J., Leslie, T. F. & et al. Complexity of the basic reproduction number (\(r_0\)). Emerg. Infect. Dis. 25, 1–4. https://doi.org/10.3201/eid2501.171901 (2019).
Achaiah, N. C., Subbarajasetty, S. B. & Shetty, R. M. R0 and re of COVID-19: Can we predict when the pandemic outbreak will be contained?. Indian J. Crit. Care Med. 24, 1125–1127 (2020).
Seprianus, S. S., Nuraini. N. A computational model of epidemic process with three variants on a synthesized human interaction network. Sci. Rep. (2024).
Mok Jung, S., Endo, A., Akhmetzhanov, A. R. & Nishiura, H. Predicting the effective reproduction number of COVID-19: inference using human mobility, temperature, and risk awareness. Int. J. Infect. Dis. 113 (2021).
Ridenhour, B., Kowalik, J. M. & Shay, D. K. Unraveling r0: Considerations for public health applications. Am. J. Public Health 104, e32–e41. https://doi.org/10.2105/AJPH.2013.301704 (2014).
Diekmann, O., Heesterbeek, J. & Metz, J. On the definition and the computation of the basic reproduction ratio r0 in models for infectious diseases in heterogeneous populations. J. Math. Biol. 28, 365–382 (1990).
Jorge, D. C. P., Oliveira, J. F., Miranda, J. G. V., Andrade, R. F. S. & Pinho, S. T. R. Estimating the effective reproduction number for heterogeneous models using incidence data. R. Soc. Open Sci. 9, 220005 (2022).
Majumder, M. S. & Mandl, K. D. Early transmissibility assessment of a novel coronavirus in Wuhan, China. SSRN Electron. J. 3524675 (2020).
Balcan, D. et al. Modeling the spatial spread of infectious diseases: The global epidemic and mobility computational model. J. Comput. Sci. 1, 132–145. https://doi.org/10.1016/j.jocs.2010.07.002 (2010).
Kuddus, M. A. & Rahman, A. Analysis of COVID-19 using a modified SLIR model with nonlinear incidence. Results Phys. 27, 104478 (2021).
Brauer, F. A. Mathematical epidemiology: Past, present, and future. Infect. Dis. Model. (2017).
Mata, A. S. & Dourado, S. M. P. Mathematical modeling applied to epidemics: an overview. São Paulo J. Math. Sci. 15, 1025–1044 (2021).
Li, J. & Zou, X. Modeling spatial spread of infectious diseases with a fixed latent period in a spatially continuous domain. Bull. Math. Biol. 71, 2048–2079 (2009).
Brauer, F. Epidemic models with heterogeneous mixing and treatment. Bull. Math. Biol. 70, 1869–85. https://doi.org/10.1007/s11538-008-9326-1 (2008).
Mandal, M. et al. A model based study on the dynamics of covid-19: Prediction and control. Chaos Solitons Fractals 136, 109889 (2020).
Yang, W., Karspeck, A. & Shaman, J. Comparison of filtering methods for the modeling and retrospective forecasting of influenza epidemics. PLoS Comput. Biol. 10, 1–15. https://doi.org/10.1371/journal.pcbi.1003583 (2014).
Khan, A. et al. A numerical study of spatio-temporal covid-19 vaccine model via finite-difference operator-splitting and meshless techniques. Sci. Rep. (2023).
Deshpande, A., Kumar, M., & Ramakrishnan, S. Robot Swarm for Efficient Area Coverage Inspired by Ant Foraging: The Case of Adaptive Switching Between Brownian Motion and Lévy Flight, Dynamic Systems and Control Conference. https://doi.org/10.1115/DSCC2017-5229. V002T14A009. https://asmedigitalcollection.asme.org/DSCC/proceedings-pdf/DSCC2017/58288/V002T14A009/2376165/v002t14a009-dscc2017-5229.pdf.
Wang, W., Cai, Y., Wu, M., Wang, K. & Li, Z. Complex dynamics of a reaction-diffusion epidemic model. Nonlinear Anal. Real World Appl. 13, 2240–2258 (2012).
Wang, L. et al. A rigorous theoretical and numerical analysis of a nonlinear reaction–diffusion epidemic model pertaining dynamics of covid-19. Sci. Rep. (2024).
Gardiner, C. Handbook of Stochastic Methods (Springer, 1985).
Risken, H. & Haken, H. The Fokker–Planck Equation: Methods of Solution and Applications, 2nd edn (Springer, 1989).
Lavielle, M., Faron, M., Lefevre, J. H. & Zeitoun, J.-D. Predicting the propagation of COVID-19 at an international scale: extension of an SIR model. BMJ Open 11, e041472 (2021).
Yang, C. & Wang, J. Basic Reproduction Numbers for a Class of Reaction–Diffusion Epidemic Models. (Springer Science and Business Media LLC, 2020).
Wallinga, J. & Teunis, P. Different epidemic curves for severe acute respiratory syndrome reveal similar impacts of control measures. Am. J. Epidemiol. 160, 509–516. https://doi.org/10.1093/aje/kwh255 (2004).
Cori, A., Ferguson, N. M., Fraser, C. & Cauchemez, S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am. J. Epidemiol. 178, 1505–1512. https://doi.org/10.1093/aje/kwt133 (2013).
Kermack, W. O. & Mckendrick, A. G. A contribution to the mathematical theory of epidemics. In Proceedings of the Royal Society of London. Series A, Containing Papers of a Mathematical and Physical Character, vol. 115, no. 115, 700–721 (2022).
Dashtbali. M, M. M. A compartmental model that predicts the effect of social distancing and vaccination on controlling covid-19. Sci. Rep. (2021).
Zhuang, Q. & Wang, J. A spatial epidemic model with a moving boundary. Infect. Dis. Model. 6, 1046–1060 (2021).
Khyar, O. & Allali, K. Dynamic Analysis of SLIR Model Describing the Effectiveness of Quarantine Against the Spread of COVID-19, chap. Dynamic Analysis of SLIR Model Describing the Effectiveness of Quarantine Against the Spread of COVID-19 (Springer International Publishing, 2021).
Majid, F. et al. Non-pharmaceutical interventions as controls to mitigate the spread of epidemics: An analysis using a spatiotemporal pde model and covid-19 data. ISA Trans. 124, 215–224. https://doi.org/10.1016/j.isatra.2021.02.038 (2022).
van den Driessche, P. & Watmough, J. Reproduction numbers and sub-threshold endemic equilibria for compartmental models of disease transmission. Math. Biosci. 180, 29–48 (2002).
O, D., JA, H. & MG., R. The construction of next-generation matrices for compartmental epidemic models. J. R. Soc. Interface. 7(47), 873–885 (2009).
Allen, L. J. S., Bolker, B. M., Lou, Y. & Nevai, A. L. Asymptotic profiles of the steady states for an sis epidemic reaction–diffusion model. Discrete Contin. Dyn. Syst. 21, 1–20 (2008).
Wang, W. & Zhao, X.-Q. Basic reproduction numbers for reaction–diffusion epidemic models. SIAM J. Appl. Dyn. Syst. 11, 1652–1673. https://doi.org/10.1137/120872942 (2012).
Thieme, H. R. Spectral bound and reproduction number for infinite-dimensional population structure and time heterogeneity. SIAM J. Appl. Math. 70, 188–211. https://doi.org/10.1137/080732870 (2009).
Li, J., Blakeley, D. & Smith, R. J. The failure of r0. Comput. Math. Methods Med. 2011, 527610. https://doi.org/10.1155/2011/527610 (2011).
Gostic, K. M. et al. Practical considerations for measuring the effective reproductive number, rt. medRxiv 2020. PLoS Comput. Biol. 16(12), e1008409. https://doi.org/10.1101/2020.06.18.20134858 (2020).
Nishiura, H. & Chowell, G. The effective reproduction number as a prelude to statistical estimation of time-dependent epidemic trends. In Mathematical and Statistical Estimation Approaches in Epidemiology (eds Chowell, G. et al.) 103–121 (Springer, 2009).
Brauer, F. Compartmental Models in Epidemiology (Springer, 2008).
Bandoy, D. & Weimer, B. Analysis of sars-cov-2 genomic epidemiology reveals disease transmission coupled to variant emergence and allelic variation. Sci. Rep. 11. https://doi.org/10.1038/s41598-021-86265-4 (2021).
Smith, M., Trofimova, M., Weber, A. et al. Rapid incidence estimation from sars-cov-2 genomes reveals decreased case detection in Europe during summer 2020. Nat. Commun. 12. https://doi.org/10.1038/s41467-021-26267-y (2021).
Cori, A. Epiestim: Estimate time varying reproduction numbers from epidemic curves. https://CRAN.R-project.org/package=EpiEstim (2021). R package version 2.2-4.
Majid, F., Deshpande, A. M., Ramakrishnan, S., Ehrlich, S. & Kumar, M. Analysis of epidemic spread dynamics using a PDE model and covid-19 data from Hamilton county OH USA. IFAC-PapersOnLine 54, 322–327. https://doi.org/10.1016/j.ifacol.2021.11.194 (2021). (Modeling, Estimation and Control Conference MECC 2021).
Epidemiology Division of Hamilton County Public Health. Greg Kesterman (the Hamilton County Health Commissioner), David Carlson and Nathan Williams (2022).
He, X. et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. (2020).
Manson, S. et al. IPUMS National Historical Geographic Information System: Version 18.0 [dataset]. Minneapolis, MN: IPUMS (2023).
Team, Q. D. Qgis geographic information system. https://qgis.org (2023). (accessed 17 Nov 2024).
Yarsky, P. Using a genetic algorithm to fit parameters of a COVID-19 SEIR model for US states. Math. Comput. Simul. 185, 687–695 (2021).
Kristen, L. & Liu, F. Auto tuning sir model parameters using genetic algorithm. medRxiv https://doi.org/10.1101/2021.07.19.21260722 (2021).
Torlapati, J. & Clement, T. P. Using parallel genetic algorithms for estimating model parameters in complex reactive transport problems. Processes. 7, https://doi.org/10.3390/pr7100640 (2019).
CDC. CDC Museum COVID-19 Timeline (2023).
David, D. A., Street, L., Kumar, M. & Ramakrishnan, S. LSTM-Based Estimation of Time-Varying Parameters in a Spatiotemporal PDE Model for Prediction of Epidemic Spread. IFAC-PapersOnLine. 58(28), 468–473. https://doi.org/10.1016/j.ifacol.2025.01.090 (2024).
Street, L. et al. Nonlinear model predictive control for mitigating epidemic spread using a partial differential equation based compartmental dynamic model. IFAC-PapersOnLine. 58(28), 366–371. https://doi.org/10.1016/j.ifacol.2025.01.062 (2024).
Acknowledgements
This work was supported by the US National Science Foundation [Grant Numbers-2140405 (PI-Ramakrishnan), 2140420 (PI-Kumar), and 2140441 (PI-Ehrlich)]. We would like to express our gratitude to Greg Kesterman (the Hamilton County Health Commissioner), David Carlson, and Nathan Williams of The Epidemiology Division of Hamilton County Public Health, Ohio, USA for providing the essential data that served as a cornerstone for this study.
Author information
Authors and Affiliations
Contributions
D.A.D. developed the basic numerical algorithm and code, conducted analysis, and obtained results. D.A.D. also contributed to writing the manuscript. C.L. developed parts of the numerical algorithm and code, conducted portions of the analysis, and obtained results. L.S. contributed to the implementation and validation of the numerical analysis. S.E. contributed to the design of certain portions of the research, the analysis of the results, and the writing of portions of the manuscript. M.K. contributed to the conception and design of the research, the analysis, and manuscript writing and editing. MK also directed the research. S.R. contributed to the conception and design of the research, the analysis, and manuscript writing and editing. S.R. also directed the research. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
David, D.A., Liu, C., Street, L. et al. A novel approach to forecasting reproduction numbers of spatiotemporal stochastic epidemic spread using a PDE-based model and real-time infection data. Sci Rep 15, 9760 (2025). https://doi.org/10.1038/s41598-025-91811-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91811-5
This article is cited by
-
A new hybrid block collocation method for solving elliptic PDEs
Scientific Reports (2025)