Abstract
Previous research has demonstrated a close connection between the development of osteosarcoma (OS) and variations in the abundance of specific gut microbiota (GM). Generally speaking, GM play a role in human health mostly through metabolites. However, the causal relationship between GM, plasma metabolites, and OS remains unclear. Hence, in our study, we aim to clarify this relationship between GM, plasma metabolites, and OS, by employing a Mendelian randomization (MR) approach. In this study, pooled data were derived from the public genome-wide association study (GWAS) in GM (GCST90032172 to GCST90032644), plasma metabolites (GCST90199621 to GCST90204603) and OS (finngen_R10_C3_OSTEOSARCOMA_EXALLC). The two-sample and two-step MR methods were used for the current analysis: (1) genetic causality between GM and plasma metabolites on OS; (2) mediation effects of plasma metabolites. For evaluating the previously described causal relationship, the inverse variance weighted (IVW) method was primarily used, with complementary approaches including the weighted median, MR-Egger, weighted mode, and simple mode. Moreover, the MR-Egger intercept test and the mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) were employed to assess the horizontal multiplicity. The reliability of causality is verified by “leave-one-out” sensitivity analysis and the Cochran’s Q test for heterogeneity. The STROBE-MR checklist for the reporting of MR studies was used in this study. First, according to the IVW results, 13 types of GM, specifically, were identified to have a potential causal relationship with OS. After FDR correction, Phocea was defined as a strain with a clear causal relationship with OS (FDR-adjusted p < 0.05). Second, a total of 48 plasma metabolites were identified to have a potential causal relationship with OS, including 30 currently known metabolites, 7 metabolites not yet studied, and 11 metabolite ratios. Finally, we further explored whether plasma metabolites mediate the causal relationship between Phocea and OS. And as a result, two plasma metabolites, Eugenol sulfate levels (mediated proportion: 7.74% (14.2%, 1.3%)) and N-acetylphenylalanine levels (mediated proportion: 3.52% (6.18%, 0.867%)), that may mediate the causal link between Phocea and OS were identified. All of the above results were subjected to sensitivity analysis. The causal relationship between GM, plasma metabolites, and OS was revealed in this MR study. Importantly, this study also demonstrated the mediating role of plasma metabolites levels of Eugenol sulfate levels and N-acetylphenylalanine levels in modulating the causal relationship between Phocea and OS. Of course, further research needs to be conducted to verify the above findings.
Similar content being viewed by others
Introduction
The most common primary malignant tumor of bone, osteosarcoma (OS), is characterized by a high degree of malignancy, aggressiveness, rapid progression, and extremely high mortality, mainly affecting children, adolescents, and young adults1. OS is also regarded as a serious threat to human health worldwide. More often than in the spine, pelvis, and sacrum, OS manifests as a single lesion in the metaphysis of long tubular bones2. Localized pain and swelling, as well as occasionally joint dysfunction, are the primary clinical symptoms of OS. Notably, nearly 10–20% of patients experienced detectable metastatic disease prior to actual OS identification, with the most common location being the lungs (85%), followed by the bones (8–10%)3. The prognosis for patients with OS has significantly improved with surgery and chemotherapy. Nevertheless, the prognosis for metastatic or recurrent OS remains unsatisfactory. Metastatic disease represents a warning sign of a poor prognosis for OS. Studies have indicated that the 5-year survival rate of individuals with OS is less than 20% once they encounter metastases4. For the most part, the causes of OS are still unknown5. While it has been indicated that specific chromosomes experience complicated alterations in OS, the research is not well-developed6. Therefore, the identification of genetic variants associated with OS will facilitate the development of new strategies for the early screening, diagnosis, and treatment of OS.
10 to 100 trillion microorganisms, mostly bacteria along with viruses, protozoa, and fungus, constitute the human gut microbiota (GM). A wide range of bacteria in the gastrointestinal system play important roles in many aspects of host health, including metabolism, immunity, nutrition, etc7. In recent years, the regulation of the GM and its impact on human health is a current research hotspot. An essential component of the body, the GM evolved with the host. A growing body of research indicates that although some microbiota members are conserved, their abundance in the gastrointestinal system varies depending on the individual’s health8. Several immune-related diseases, including many types of tumors9, autoimmune diseases10, and inflammation11, have been related to variations in the abundance of GM. Chen et al. have identified significant differences in oral microbial communities between OS patients and healthy controls12. They analysis revealed a significant reduction in fourteen genera, including Rothia, Halomonas, Rhodococcus, and Granulicatella, while concurrently observing an enrichment of Alloprevotella, Prevotella, Selenomonas, and Campylobacter in OS patients12. Species of GM were identified that could adjust immune checkpoint responses by generating different metabolites. A study has demonstrated that in colorectal cancer, F. nucleatum-derived succinic acid inhibits the cGAS-interferon-β pathway, thereby suppressing the anti-tumor response by limiting the traffic of CD8 T cells to the tumor microenvironment (TME) in vivo13. And the usage of the antibiotic metronidazole decreases serum levels of succinic acid and re-sensitizes tumors in the body to immunotherapy.
Moreover, present studies are also focused on investigating the function of metabolites originating from the GM in an array of tumor disorders attempting to propose relevant therapeutic strategies to affect tumor progression. A recent study has revealed the presence of an altered abundance of the amino acid metabolism-related genera Alloprevotella, Rikenellaceae_RC9_gut_group, and Muribaculum in murine models of OS14. The study demonstrates that the GM can influence OS progression by regulating the metabolites they derive. The mechanism by microbial metabolites affects the prognosis of tumor patients is mainly due to the following: immunotherapy, when combined with microbial metabolites, can effectively stimulate the immune system, eradicate tumors, and overcome drug resistance15. And microbial metabolites have been demonstrated to decrease severe side effects and increase the effectiveness of chemotherapy and radiation treatments16. However, some GM metabolites can exert different functions that vary with environment (e.g., type of cancer). When employing a variety of treatments for cancer by altering the state of the GM and its derived metabolites, including fecal bacterial transplantation and phage transplantation, an individualized and comprehensive approach is required based on the patient’s condition. Therefore, it is particularly important to clarify the causal relationship between the GM, metabolites, and OS. Mendelian randomization (MR), which employ genetic variants to create instrumental variables (IVs) for exposure and incorporate data gathered from genome-wide association studies (GWAS), offer a novel approach to assess the causal relationship between exposures and outcomes17. In this study, a mediation MR analysis was performed to assess the causal relationship between GM, metabolites, and OS using the GWAS summary data.
Methods
Study design
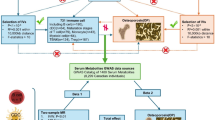
In this study, a mediation (two-step approach) MR study to deeply investigate the mediating effect of plasma metabolites as mediators between GM and OS. The research protocols adhered to the STROBE-MR checklist18. In order to ensure the efficacy of potential causal effects, MR analysis requires that three core assumptions should be satisfied:1 Assumption 1, genetic variants are strongly correlated with exposure (relevance assumption);2 Assumption 2, genetic variants are unaffected by confounding factors (independence assumption); and3 Assumption 3, genetic variants only affect the outcome as a result of exposure (exclusivity assumption)19,20 (Fig. 1).
Data sources
The GWAS catalog contains the full summary data for genetic variants of GM (GCST90032172 to GCST90032644) tested in 5,959 European individuals. The results show that 473 different taxa, which include 11 phyla, 18 classes, 24 orders, 62 families, 145 genera, and 213 species, were identified21.
The GWAS data for plasma metabolites were retrieved from the GWAS Catalog (GCST90199621 to GCST90204603). 1,091 blood metabolites and 309 metabolite ratios were included in the data, involving 8,299 samples and almost 150,000 Single nucleotide polymorphisms (SNPs)22.
GWAS summary statistics for OS were obtained from the FinnGen Consortium R10 release. The data can be obtained by visiting: https://storage.googleapis.com/finngen-public-data-r10/summary_stats/finngen_R10_C3_OSTEOSARCOMA_EXALLC.gz. The diagnostic criteria for OS are based on the international code of diseases.
Instrumental variables selection
To ensure the accuracy and reliability of our findings about the causal relationship between exposure and outcome, a rigorous quality assurance procedure was established for eliminating non-qualified IVs. Firstly, according to previous MR studies, potential IVs were identified as SNPs associated each exposure (P < 1.0 × 10− 5)23,24. Secondly, correlational analysis of relevant SNPs was carried out to remove inappropriate SNPs (R2 < 0.01 and clustering distance = 10,000 kb) which could lead to biased results resulting from linkage disequilibrium (LD). Thirdly, Palindromic SNPs were eliminated, and we only included SNPs with a minor allele frequency (MAF) > 0.01. Next, we calculated the F-statistic to assess the strength of IVs. IVs that exhibited an F-statistic less than < 10 were considered to be weak IVs and eliminated. F = R2(n-k-1)/k(1-R2); (R2, exposed variance explained by the selected IVs; n, sample size; k, number of IVs) (Fig. 1)
Statistical analysis
In this study, inverse variance weighted (IVW) was considered the dominant statistical method for assessing causality, with the weighted median, simple mode, weighted mode, and MR-Egger methods as the complementary methods25 (Fig. 1). To ensure the reliability of causality, the results of the remaining four complementary methods whose β-values were consistent with the β-values of IVW were included when the p-value of IVW was less than 0.05. In addition, false discovery rate (FDR) correction was conducted by applying the Bonferroni and benjaminian hochberg (BH) procedure. When the p-value for FDR was less than 0.05, we believe that there is a well-defined genetic causality between outcome and exposure. Potential horizontal pleiotropy was evaluated employing the MR-PRESSO method and the MR-Egger regression intercept analysis. The existence of horizontal pleiotropy could be verified if the intercept term was significant (p < 0.05). The results of horizontal pleiotropy that still existed after the removal of outlier SNPs by the Outlier-corrected MR-PRESSO tool will not be analyzed in subsequent analysis. In terms of heterogeneity, Cochran’s Q statistic of MR-IVW was employed as the main tool for detection. P-value greater than 0.05 indicates the absence of heterogeneity. To further ensure the reliability of our findings, we conducted a “leave-one-out” sensitivity analysis to find out if any specific SNPs significantly affected causality.
A statistical method, mediation analysis, is designed to investigate whether a variable mediates the relationship between two other variables. To investigate the mediating role of plasma metabolites in modulating the abundance of GM and OS causality, we performed a two-step MR analysis in the current research. First, we evaluated the effect of GM abundance on OS using MR analysis. Then, the other MR analysis was performed to explore the causal relationship between plasma metabolites and OS employing plasma metabolites as the exposure factor and OS as the outcome. Finally, the above obtained GM with a clear causal relationship with OS were used as exposures, and plasma metabolites with a clear causal relationship with OS were used as outcomes for MR analysis so as to obtain plasma metabolites with mediating effects. The mediator ratio is estimated using the formula: β12Ratio=(β1 × β2)/βAll26. (β1:MR effect of the GM on the metabolite, β2: MR effect of the metabolite on the OS, β1 × β2: “indirect” effect through the medium, and βAll: “total” effect of the GM on the OS.)
All statistical analyses were performed using R software (version 4.3.3, https://www.r-project.org/). The “TwoSampleMR”, “VariantAnnotation”, “gwasglue”, “data.table”, and the “MRPRESSO” package were used for all MR analysis.
Results
Genetic causality between gut microbiota and osteosarcoma
A total of 9,124 SNPs in the 473 taxa were identified in this study between GM and OS after a series of quality control steps. The detailed characteristics of the SNPs that are included in Supplementary Table S1.
According to the IVW results, 13 types of gut microbiota, specifically, Anaeromassilibacillus sp001305115 (OR = 6.20, 95% CI = 1.07–36.09, P = 0.042), Bacteroides thetaiotaomicron (OR = 0.14, 95% CI = 0.04–0.59, P = 0.007), Bifidobacterium adolescentis (OR = 2.08, 95% CI = 1.05–4.13, P = 0.036), CAG-884 sp000433875 (OR = 4.22, 95% CI = 1.14–15.69, P = 0.031), Dorea phocaeense (OR = 0.06, 95% CI = 0.01–0.47, P = 0.008), GCA-900,066,495 sp900066495 (OR = 17.88, 95% CI = 2.48-128.85, P = 0.004), Holdemania massiliensis (OR = 0.27, 95% CI = 0.08–0.91, P = 0.034), Hungatella sp900155545 (OR = 26.4, 95% CI = 1.07–651.80, P = 0.045), Johnsonella ignava (OR = 0.03, 95% CI = 0.01–0.66, P = 0.025), Mycoplasmataceae (OR = 68.97, 95% CI = 1.61-2961.18, P = 0.027), Phocea massiliensis (OR = 0.02, 95% CI = 0.01–0.18, P = 0.0004), Phocea (OR = 0.02, 95% CI = 0.01–0.14, P = 6.98E-05), and Syntrophorhabdia (OR = 0.01, 95% CI = 0.00-0.35, P = 0.019) were identified to have a potential causal relationship with OS (Table 1, Supplementary Figure S1, and Supplementary Table S2). After FDR correction, Phocea was defined as a strain with a clear causal relationship with OS (FDR-adjusted p < 0.05) (Table 1, Supplementary Table S3). Furthermore, scatter plots illustrating the distribution of effects for all SNPs revealed trends for the 5 different MR analysis methods (Supplementary Figure S2).
Genetic causality between metabolites and osteosarcoma
Similarly, 32,130 SNPs in 1,352 plasma metabolites were identified between metabolites and OS. The detailed characteristics of the included SNPs are shown in Supplementary Table S4.
According to the IVW results, 30 currently known metabolites, 7 metabolites not yet studied, and 11 metabolite ratios were identified to have a potential causal relationship with OS (Fig. 2, Supplementary Figure S3, and Supplementary Table S5). The causal relationship between metabolites and OS showed that 21 metabolites were genetically predicted to be associated with a reduced risk of OS and 27 were associated with an increased risk of OS. Moreover, scatter plots that depicted the distribution of effects for all SNPs also showed trends for each of the five MR methods of analysis. (Supplementary Figure S4).
Mediation analysis of Phocea and osteosarcoma
After investigating the genetic causality of GM, metabolites, and OS, we further explored whether metabolites mediate the causal relationship between GM and OS. In this study, Phocea remained significant after FDR correction and was treated as exposure. The 48 metabolites that were causally related to OS served as outcome. As a result, two plasma metabolites, Eugenol sulfate levels (mediated.
proportion: 7.74% (14.2%, 1.3%)) and N-acetylphenylalanine levels (mediated proportion: 3.52% (6.18%, 0.867%)), that may mediate the causal link between GM and OS were identified (Figs. 3 and 4; Table 2, Supplementary Figure S5, Supplementary Figure S6, and Supplementary Table S6). The findings provide light on the complicated connections that exist between metabolites, GM, and OS, as well as enhance our knowledge of the mechanisms related to the influence of GM on OS.
Sensitivity analysis
The F-statistics of the IVs for the causal relationship mentioned above were greater than 10, which eliminated the bias from weak IVs (Supplementary Table S1, Supplementary Table S4, and Supplementary Table S7). Cochran ‘s IVW Q-test indicated that no significant heterogeneity was observed in the IVs of the above results (P > 0.05) (Supplementary Table S8, Supplementary Table S9, and Supplementary Table S10). According to the findings of the MR-Egger regression intercept analysis, similarly, either of the IVs of the aforementioned results showed any discernible directional horizontal pleiotropy (all intercept P > 0.05) (Supplementary Table S11, Supplementary Table S12, and Supplementary Table S13,). Furthermore, we employed MR PRESSO to ensure global directional horizontal pleiotropy and increase the reliability of the study (Supplementary Table S14, Supplementary Table S15, and Supplementary Table S16). In addition, leave-one-out analysis were conducted to investigate the possible impact of specific SNPs on the observed correlations. When an SNP is eliminated individually, the overall error lines are all positioned on one side of the median line, implying that each SNP affects the results equally and excludes outlier SNPs from affecting the reliability of the results (Supplementary Figure S7, Supplementary Figure S8, and Supplementary Figure S9). Overall, the results of all these sensitivity analyses show how reliable our MR analysis is.
Discussion
In this study, a mediation MR study was performed to evaluate the causal relationship between GM, plasma metabolites, and OS. As a summary, first, the study revealed 13 GM with a potential causal relationship with OS, of which, after FDR correction, Phocea was defined as a strain with a clear causal relationship with OS. Second, our study also revealed 48 metabolites that have potential genetic causality with OS. Finally, we further analyzed the relationship between Phocea and the 48 metabolites obtained above, and identified that a was genetically causally related to Eugenol sulfate levels and N-acetylphenylalanine levels, respectively. In a way, this suggests that the causal relationship between Phocea and OS may be partly mediated by Eugenol sulfate levels and N-acetylphenylalanine levels in plasma. To the best of our knowledge, this is the first MR study, filling the gaps in this area, to investigate the potential causal relationship between GM, plasma metabolites, and OS.
It is widely acknowledged that GM play an essential part in human health and disease. As mentioned in the introduction, GM is closely related to host metabolism and immunity. In recent years, an increasing number of studies have focused on the contribution of GM to the development of various cancers. For example, an in vivo investigation found that OS model mice exhibited a higher alpha diversity of gut bacteria than controls. And the relative abundance of the Lachnospiraceae family changed more throughout time than that of controls27. In addition, altered gut microbial abundance may also affect the immune response to tumors. The study by Tian et al. assessed changes in the GM during OS growth and after chemotherapy with cisplatin (CDDP) or doxorubicin (DOX) in a mouse model and examined the association between OS progression and chemotherapy and the GM in mice9. Their findings revealed that certain chemotherapy medications affect the composition of the GM in mice, hence contributing to the formation of an anti-cancer immune response. The study also offers fresh perspectives on the progression and treatment of OS. In our study, a significant genetic causal relationship was identified between the abundance of Phocea and the development of OS. In other words, increased Phocea abundance may enhanced the incidence of OS. Currently, Phocea has been discovered to play a potential biological role in a number of disorders, including urothelial cancer28, Parkinson’s disease29, Type 1 Narcolepsy30, type 2 diabetes mellitus (T2DM)31, early vascular ageing32, and menopausal transition33. To some extent, our research contributes to the body of knowledge regarding the connection between Phocea and illness as well as provides insight on how OS develops.
Metabolites originating from GM may play a role in the development of cancer, in accordance to an increasing amount of research collected in recent years. To reveal the specific effects of plasma metabolites mediating the causal relationship between Phocea and OS, we included plasma metabolites in this study. In the present study, we established that two GM-derived metabolites (Eugenol sulfate levels and N-acetylphenylalanine levels) may play an important role in the development of OS. Previous studies have mostly revealed that metabolite abnormalities related to energy metabolism accompanied osteosarcoma throughout its development34. Because of their potential to benefit human health, phenolic phytochemicals have been investigated extensively by researchers. An example of a highly effective component found in cloves is Eugenol (4-allyl-1-hydroxy-2-methoxybenzene). As far as it is concerned, Eugenol has been found to play important roles in a wide range of human life activities, including neuroprotective and cardioprotective functions through different antioxidant mechanisms35,36, immunomodulatory functions against toxins37, inhibition of chronic inflammation38, and so on. Importantly, the researchers have discovered that Eugenol can act as a promising natural anticancer agent in a variety of tumors39,40. Eugenol sulfate levels, as a plasma metabolite related to Eugenol, was found in the present study to potentially mediate the causal role of Phocea and OS, enriching the field of related research.
An amino acid derivative consisting of phenylalanine and acetyl group, N-acetylphenylalanine, exerts a variety of biological functions and pharmacological effects in vivo41. N-acetylphenylalanine may be involved in several physiological processes such as protein synthesis, neurotransmitter synthesis, etc42. Furthermore, it has been employed as a substance in pharmaceutical studies and might contain biological effects like analgesic, antioxidant, and anti-inflammatory properties43. Previous studies have mostly shown N-acetylphenylalanine excretion in patients with inborn defects in amino acid metabolism including maple syrup urine disease, hereditary tyrosinemia, and phenylketonuria44. Research on how it is utilized in cancer, however, is limited number. But a recent untargeted metabolomics study identified a group of urinary biomarkers for the diagnosis of uroepithelial carcinoma of the bladder, and N-acetylphenylalanine became one of them. This work makes it possible for scientists to investigate N-acetylphenylalanine ‘s function in malignancies. In addition, a recent study showed that the abundance of specified intestinal genera associated with amino acid metabolism are altered in osteosarcoma mice, which is similar to the results of our study14.
Overall, this study exhibits several notable strengths. Firstly, through MR analysis, we identified a total of 13 GM and 48 plasma metabolites that may possess a genetically causal association with OS. Secondly, our study underwent FDR correction to delineate Phocea with a clear causal relationship to OS. Significantly, we conducted mediator analysis to explore the involvement of plasma metabolites in the causal relationship between Phocea and OS, elucidating the roles of Eugenol sulfate levels and N-acetylphenylalanine levels in Phocea regulation of OS. Undoubtedly, these findings shed new light on the etiology of OS and offer valuable insights into its diagnosis and treatment. However, our study is not without its limitations. Firstly, since the majority of participants in the data pooling were of European descent, the generalizability of our findings to other ethnic groups may be constrained. Secondly, the absence of individual-level data limited our ability to explore more intricate relationships and may have overlooked nonlinear associations between GM, metabolites, and OS. Thirdly, while MR analysis identified GM and metabolites potentially linked to OS development, it’s important to consider biological mechanisms when interpreting the findings. Statistical effect values alone cannot unveil biological mechanisms, underscoring the necessity for stronger biological evidence to support our study’s conclusions. Therefore, the principal limitation of this paper is the absence of experimental evaluation of the results of the online analysis in this study. This depends on further experimental studies by researchers to remedy the shortcomings.
Conclusion
In conclusion, the current study identified 13 GM and 48 plasma metabolites with potential causal associations with OS. Notably, it was discovered that Eugenol sulfate levels and N-acetylphenylalanine levels in plasma were helpful in modulating the causal relationship between Phocea and OS. Our finding offers fresh perspectives on the causes, identification, and management of OS.
Data availability
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Abbreviations
- OS:
-
Osteosarcoma
- GM:
-
Gut microbiota
- MR:
-
Mendelian randomization
- IVW:
-
Inverse variance weighted
- MR-PRESSO:
-
Mendelian randomization pleiotropy residual sum and outlier
- TME:
-
Tumor microenvironment
- IVs:
-
Instrumental variables
- GWAS:
-
Genome-wide association studies
- SNPs:
-
Single nucleotide polymorphisms
- LD:
-
Linkage disequilibrium
- BH:
-
Benjaminian Hochberg
- MAF:
-
Minor allele frequency
- FDR:
-
False discovery rate
- CDDP:
-
Chemotherapy with cisplatin
- DOX:
-
Doxorubicin
- T2DM:
-
Type 2 diabetes mellitus
References
Chakrabarti, S., Kamgar, M. & Mahipal, A. Systemic therapy of metastatic pancreatic adenocarcinoma: current status, challenges, and opportunities. Cancers. 14(11). (2022).
Zhao, X., Wu, Q., Gong, X., Liu, J. & Ma, Y. Osteosarcoma: a review of current and future therapeutic approaches. Biomed. Eng. Online. 20(1), 24 (2021).
Estrada-Villaseor, E. et al. Association of metastasis with clinicopathological data in Mexican patients with osteosarcoma, giant cell tumor of bone and chondrosarcoma. Asian Pac. J. Cancer Prev. 16(17), 7689–7694 (2015).
Shoaib, Z., Fan, T. M. & Irudayaraj, J. M. K. Osteosarcoma mechanobiology and therapeutic targets. Br. J. Pharmacol. 179(2), 201–217 (2022).
Mazziotta, C. et al. Regulatory mechanisms of circular RNAs during human mesenchymal stem cell osteogenic differentiation. Theranostics 14(1), 143–158 (2024).
Entz-Werle, N. et al. Genetic alterations in primary osteosarcoma from 54 children and adolescents by targeted allelotyping. Br. J. Cancer. 88(12), 1925–1931 (2003).
Wilson, I. D. & Nicholson, J. K. Gut microbiome interactions with drug metabolism, efficacy, and toxicity. Transl Res. 179, 204–222 (2017).
Borkent, J., Ioannou, M., Laman, J. D., Haarman, B. C. M. & Sommer, I. E. C. Role of the gut microbiome in three major psychiatric disorders. Psychol. Med. 52(7), 1222–1242 (2022).
Tian, Z. et al. Cisplatin and doxorubicin chemotherapy alters gut microbiota in a murine osteosarcoma model. Aging (Albany NY). 16(2), 1336–1351 (2024).
Xu, Q. et al. Causal relationship between gut microbiota and autoimmune diseases: A two-sample Mendelian randomization study. Front. Immunol. 12, 746998 (2021).
Cai, J., Sun, L. & Gonzalez, F. J. Gut microbiota-derived bile acids in intestinal immunity, inflammation, and tumorigenesis. Cell. Host Microbe. 30(3), 289–300 (2022).
Chen, Y. et al. Oral microbiota distinguishes patients with osteosarcoma from healthy controls. Front. Cell. Infect. Microbiol. 14, 1383878 (2024).
Jiang, S. S. et al. Fusobacterium nucleatum-derived succinic acid induces tumor resistance to immunotherapy in colorectal cancer. Cell. Host Microbe. 31(5), 781–97e9 (2023).
Li, Y. et al. Characterization of the gut microbiota and fecal metabolome in the osteosarcoma mouse model. Aging (Albany NY). 16(13), 10841–10859 (2024).
Yang, Q. et al. A review of gut microbiota-derived metabolites in tumor progression and cancer therapy. Adv. Sci. (Weinh). 10(15), e2207366 (2023).
Luu, M., Schütz, B., Lauth, M. & Visekruna, A. The impact of gut Microbiota-Derived metabolites on the tumor immune microenvironment. Cancers. 15(5). (2023).
Burgess, S., Timpson, N. J., Ebrahim, S. & Davey Smith, G. Mendelian randomization: where are we now and where are we going? Int. J. Epidemiol. 44(2), 379–388 (2015).
Skrivankova, V. W. et al. Strengthening the reporting of observational studies in epidemiology using Mendelian randomization: the STROBE-MR statement. Jama 326(16), 1614–1621 (2021).
Davies, N. M., Holmes, M. V. & Davey Smith, G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. Bmj 362, k601 (2018).
Smith, G. D. & Ebrahim, S. Mendelian randomization’: can genetic epidemiology contribute to Understanding environmental determinants of disease? Int. J. Epidemiol. 32(1), 1–22 (2003).
Qin, Y. et al. Combined effects of host genetics and diet on human gut microbiota and incident disease in a single population cohort. Nat. Genet. 54(2), 134–142 (2022).
Chen, Y. et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet. 55(1), 44–53 (2023).
Liu, X. et al. Mendelian randomization analyses support causal relationships between blood metabolites and the gut microbiome. Nat. Genet. 54(1), 52–61 (2022).
Li, J. et al. Association between gut microbiota and spinal stenosis: a two-sample Mendelian randomization study. Front. Immunol. 15, 1360132 (2024).
Fu, Y. et al. Causal effects of gut microbiome on autoimmune liver disease: a two-sample Mendelian randomization study. BMC Med. Genomics. 16(1), 232 (2023).
Chen, L. et al. Systematic Mendelian randomization using the human plasma proteome to discover potential therapeutic targets for stroke. Nat. Commun. 13(1), 6143 (2022).
Le, D., Chambers, M. M., Mercado, K. & Gutowski, C. J. Characterization of the gut microbiome in an osteosarcoma mouse model. J. Orthop. Res. 41(12), 2730–2739 (2023).
Miyake, M. et al. Probiotics enhances anti-tumor immune response induced by gemcitabine plus cisplatin chemotherapy for urothelial cancer. Cancer Sci. 114(3), 1118–1130 (2023).
Jang, J. H. et al. Acupuncture inhibits neuroinflammation and gut microbial dysbiosis in a mouse model of Parkinson’s disease. Brain Behav. Immun. 89, 641–655 (2020).
Zhang, R. et al. Gut microbiota in patients with type 1 narcolepsy. Nat. Sci. Sleep. 13, 2007–2018 (2021).
Wang, Y. et al. Phocea, Pseudoflavonifractor and Lactobacillus intestinalis: three potential biomarkers of gut microbiota that affect progression and complications of obesity-induced type 2 diabetes mellitus. Diabetes Metab. Syndr. Obes. 13, 835–850 (2020).
Salvado, R. et al. Gut microbiota and its relationship with early vascular ageing in a Spanish population (MIVAS study). Eur. J. Clin. Invest. :e14228. (2024).
Dai, R. et al. Gut microbiota and metabolites in estrus cycle and their changes in a menopausal transition rat model with typical neuroendocrine aging. Front. Endocrinol. (Lausanne). 14, 1282694 (2023).
Quintero Escobar, M. et al. Insights in osteosarcoma by proton nuclear magnetic resonance serum metabonomics. Front. Oncol. 10, 506959 (2020).
Jung, M. J. et al. Eugenol relieves the pathological manifestations of Alzheimer’s disease in 5×FAD mice. Phytomedicine 118, 154930 (2023).
Kumar, A. & Sharma, B. Cardioprotective effect of eugenol against Cd-induced inflammation, oxidative stress, and dyslipidemia in male rats: an in vivo and molecular docking study. Biol. Trace Elem. Res. (2024).
Dibazar, S. P., Fateh, S. & Daneshmandi, S. Immunomodulatory effects of clove (Syzygium aromaticum) constituents on macrophages: in vitro evaluations of aqueous and ethanolic components. J. Immunotoxicol. 12(2), 124–131 (2015).
Truzzi, F. et al. Spermidine-eugenol supplement preserved Inflammation-Challenged intestinal cells by stimulating autophagy. Int. J. Mol. Sci. ;24(4). (2023).
Esmeeta, A. et al. Plant-derived bioactive compounds in colon cancer treatment: an updated review. Biomed. Pharmacother. 153, 113384 (2022).
Abdullah, M. L., Al-Shabanah, O., Hassan, Z. K. & Hafez, M. M. Eugenol-induced autophagy and apoptosis in breast cancer cells via PI3K/AKT/FOXO3a pathway Inhibition. Int. J. Mol. Sci. 22(17). (2021).
Okajima, K., Inoue, M. & Morino, Y. Studies on the mechanism for renal elimination of N-acetylphenylalanine: its pathophysiologic significance in phenylketonuria. J. Lab. Clin. Med. 105(1), 132–138 (1985).
Maxwell, B. D. et al. The synthesis of [(14) C]4-acetylphenylalanine, effect on cell viability, and assessment of protein incorporation in male rat hepatocytes. J. Label. Comp. Radiopharm. 60(8), 352–356 (2017).
Veronesi, F. et al. Chondroprotective activity of N-acetyl phenylalanine glucosamine derivative on knee joint structure and inflammation in a murine model of osteoarthritis. Osteoarthr. Cartil. 25(4), 589–599 (2017).
Jellum, E., Horn, L., Thoresen, O., Kvittingen, E. A. & Stokke, O. Urinary excretion of N-acetyl amino acids in patients with some inborn errors of amino acid metabolism. Scand. J. Clin. Lab. Invest. Suppl. 184, 21–26 (1986).
Acknowledgements
The authors appreciate the researchers and participants for providing the gut microbiota, plasma metabolites, and osteosarcoma GWAS summary statistics.
Author information
Authors and Affiliations
Contributions
Xiuyu Qin and Zhuming Fan contributed to data collection, analysis, and interpretation; Xiuyu Qin and Shaopeng Qiao contributed to manuscript preparation and editing; Jian Li contributed to revision of the manuscript; Jia Lv was involved in supervision. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Consent for publication
All authors have seen and approved the final version of the manuscript being submitted.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Qin, X., Fan, Z., Qiao, S. et al. Genetically predicted plasma metabolites mediate the causal relationship between gut microbiota and osteosarcoma. Sci Rep 15, 7277 (2025). https://doi.org/10.1038/s41598-025-91869-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-91869-1