Abstract
In medical image segmentation, traditional CNN-based models excel at extracting local features but have limitations in capturing global features. Conversely, Mamba, a novel network framework, effectively captures long-range feature dependencies and excels in processing linearly arranged image inputs, albeit at the cost of overlooking fine spatial relationships and local pixel interactions. This limitation highlights the need for hybrid approaches that combine the strengths of both architectures. To address this challenge, we propose CNN-Fusion-Mamba-based U-Net (CFM-UNet). The model integrates CNN-based Bottle2neck blocks for local feature extraction and Mamba-based visual state space blocks for global feature extraction. These parallel frameworks perform feature fusion through our designed SEF block, achieving complementary advantages. Experimental results demonstrate that CFM-UNet outperforms other advanced methods in segmenting medical image datasets, including liver organs, liver tumors, spine, and colon polyps, with notable generalization ability in liver organ segmentation. Our code is available at https://github.com/Jiacheng-Han/CFM-UNet.
Similar content being viewed by others
Introduction
Medical image segmentation aims to separate structures or tissues in medical images through computer algorithms. These segmentation results provide valuable information about anatomical regions required for detailed analysis, greatly helping physicians to characterize injuries, monitor disease progression, and specify further treatment options. As the demand for intelligent medical image analysis continues to grow, accurate and robust segmentation methods are becoming increasingly important1,2.
CNN-based medical image segmentation models3,4,5,6,7,8, represented by U-Net9, have achieved remarkable success in a variety of medical applications through accurate segmentation from MRIs, CT scans, PET/CT scans, and other medical imaging modalities, becoming mainstream methods. However, these methods have limitations in capturing remote relationships and global context, especially for objects with large inter-patient variations in texture, scale, and shape. Various strategies have emerged, such as Expanded Convolution3,4, Image Pyramid5,6, Prior-guided methods7,10, Multi-scale Fusion8,11, and Self-attention Mechanism12,13, all attempting to address these limitations in medical image segmentation. However, they still have weaknesses in extracting global contextual features, as shown in Fig. 1.
Recently, a new architecture based on the State Space Model (SSM) called Mamba14 has also been applied in medical image segmentation. VMamba15 constructs a visual state space (VSS) block based on this architecture, capable of capturing global contextual information of images with linear computational complexity, demonstrating powerful capabilities in establishing distant dependencies and understanding global context. However, existing Mamba-based visual modules15 capture global context by processing images as linear sequences along specific directions. While effective for capturing long-range dependencies, this approach inherently limits the ability to accurately capture local features. This limitation arises from the neglect of fine spatial relationships and local pixel interactions, which are crucial for detailed segmentation tasks. The insufficient capability to capture local features becomes particularly evident in scenarios with noisy or complex backgrounds. As shown in Fig. 1, the Mamba-based Swin-UMamba16 model is highly sensitive to such background complexities, often resulting in blurred boundaries and noticeable segmentation errors.
For liver organs with relatively fixed positions, CNN-based M2SNet outperforms Swin-UMamba. Conversely, for colon polyps with varying positions and shapes, Mamba-based Swin-UMamba performs better than M2SNet. This highlights the strengths of CNN-based methods in local detail and edge texture segmentation, while Mamba-based methods excel in capturing global features and contextual understanding.
Since Mamba-based medical image segmentation methods are still in their nascent stage, models emerging at this stage, such as VM-UNet17 and U-Mamba18, also extract and preserve local features of the image by introducing the VSS block into the U-shape framework using classical downsampling and skip connections, which are insufficient to improve the understanding of local details in the model itself. This limitation highlights the challenges faced by Mamba-based models in real-world applications, particularly in tasks requiring fine-grained segmentation. Therefore, the need arises for hybrid approaches that combine the strengths of Mamba and CNN architectures, enabling a more balanced extraction of both global and local features to improve segmentation performance. Inspired by the parallel network structure proposed by Peng et al.19, we migrate and construct a parallel network model of CNN and Mamba and apply it to medical image segmentation. Considering the performance characteristics and shortcomings of these two network structures, and noting the SE (Squeeze-and-Excitation) network’s20 versatility and adaptive feature weighting capability, which enhance feature expression while being simple and lightweight, we propose the CFM-UNet model. This model aims to improve the segmentation accuracy of medical images by leveraging the strengths of both CNN and Mamba without compromising their independent segmentation frameworks. It introduces the SEF (SE-based Feature Fusion) block to fuse features learned from both frameworks during the downsampling process and provide feedback. This approach enables complementary advantages between the two networks, thereby improving segmentation precision. Our contributions can be summarized as follows:
-
The encoder of CFM-UNet combines the strengths of CNN-based Bottle2neck21 and Mamba-based VSS15 frameworks, efficiently learning and capturing both local spatial contextual features and global features.
-
The SEF block feature fusion module does not require constructing a very deep network structure and can efficiently couple the two frameworks while effectively avoiding the problems of gradient vanishing and feature information loss.
-
The proposed CFM-UNet demonstrates excellent performance on the LiTS22 liver and liver tumor dataset, SPIDER23 spine dataset, and Kvasir-SEG24 colon polyp dataset, and also shows good generalization ability on the ATLAS25 liver dataset.
Related work
CNN-based medical image segmentation models
The fully convolutional network (FCN)26 was the first to introduce deep learning methods into image segmentation. The emergence of the U-Net9 further facilitated the rapid development of deep learning-based segmentation in medical imaging, establishing the dominance of the U-shape architecture in this field. Subsequently, various U-Net variants have been developed to improve segmentation accuracy. For instance, Attention-UNet27 incorporates the attention mechanism in the decoder, Res-UNet28 integrates residual learning into the network blocks, and U-Net++6 introduces a nested U-Net structure with a deep supervision mechanism. Despite the significant progress in CNN-based medical image segmentation, the inherent localization of convolutional operations and complex data access patterns continue to present challenges that require ongoing research.
ViT-based medical image segmentation models
The Visual Transformer (ViT)29, originally developed in the context of natural language processing, has emerged as a promising model for vision-based tasks. Models such as Swin-unet12, which are purely Transformer-based, represent notable advancements in this domain. To address the limitations of traditional CNNs, ViT is often integrated into existing frameworks or fused with CNN modules. For example, TransUNet13 adopts a hybrid CNN-Transformer-as-encoder approach, while TransFuse30 introduces pyramidal vision in PolypPVT31 and incorporates additional control mechanisms in MedT32 to enhance self-attention modules. TransAttUnet33 combines multilevel guided attention and multiscale jump connectivity. However, these models still require improvements in terms of inference speed and the performance of local feature extraction.
Mamba-based medical image segmentation models
Mamba, characterized by its linear computational complexity and ability to handle longer sequences, presents a challenge to Transformers in the field of medical image segmentation14. The VSS15 module demonstrates potential in addressing complex structures and patterns within medical images. Consequently, Mamba-based models have emerged as a viable alternative alongside CNN and ViT-based approaches. The VSS module performs “four-directional” 2D selective scans (top-left to bottom-right, top-right to bottom-left, bottom-left to top-right, and bottom-right to top-left) on the input image. It serializes the scanned results and applies the state space model for selective scanning. Subsequently, it restores and merges the results to obtain a comprehensive global view of the image. At this stage, exemplified by VM-UNet17, Mamba-based segmentation models are still in their early stages of development, but their inherent advantages are already apparent.
To leverage the potential of Mamba in medical image segmentation and address CNN’s limitations in capturing global features, we propose the CFM-UNet architecture. CFM-UNet adopts a parallel network branching approach for feature fusion, ensuring consistency and richness in 2D medical image segmentation. By harnessing the complementary strengths of both networks, CFM-UNet aims to further enhance segmentation accuracy.
Methods
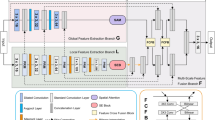
In this section, we first introduce the structure of CFM-UNet, depicted in Fig. 2. We then elaborate on the encoder and decoder components, along with the employed loss function in the training process.
Encoder
In the encoder, we designed a parallel framework comprising Local and Global feature blocks, leveraging the strengths of the Bottle2neck21 and VSS15 network structures, as shown in the blue and green components in Fig. 2. To exploit the adaptive feature weighting capabilities of the SE network20, which introduces channel attention mechanisms with minimal computational overhead, we developed the SEF block for feature fusion, as shown in the yellow component in Fig. 2. This module integrates features from both frameworks and incorporates them back into the networks. Its feedback mechanism is seamlessly integrated into the decoder, overcoming many of the shortcomings of other modules (such as addition fusion, multiplication fusion, etc.) in terms of handling feature importance or computational efficiency, and significantly enhancing the adaptive feature fusion effect.
Local branch
In the Local framework, the Bottle2neck module, based on the residual unit structure, serves as the primary method for local feature extraction. As shown in the blue module in Fig. 3, Bottle2neck divides the input into two independent information streams. One stream undergoes a \(1\times 1\) convolution, which is subsequently partitioned into several blocks based on channel number. Each block \(X_i\) undergoes a \(3\times 3\) convolution, and its result \(X_i\) is then added to the result of \(X_{i-1}\) from the previous convolution. This process generates outputs with varying quantities and receptive field sizes, thereby enhancing the capability to extract local features. Finally, after concatenating and merging all outputs via a \(1\times 1\) convolution layer, they are added back to the output of the other information stream.
Global branch
In the Global framework, the VSS block, derived from VMamba, functions as the primary module for global feature extraction. As shown in the green module in Fig. 3, the input to the VSS block undergoes processing through an initial linear embedding layer initially. Subsequently, it is split into two separate information streams. One stream passes through a \(3\times 3\) depth convolutional layer followed by SiLU activation and enters the 2D-Selective-Scan (SS2D) module. The normalized output from SS2D is then combined with the output from the other information stream, which also undergoes SiLU activation. The merged output constitutes the final result, where each output undergoes patch merging to increase channel numbers, reduce resolution, and normalize to produce the final output result.
SEF block for feature fusion
Although the sampling processes of these two feature extraction frameworks do not interfere with each other, they are not entirely independent. The SEF block integrates the downsampling outcomes from these two frameworks and feeds them back into their respective networks, thereby enhancing the model’s capability to extract global and local features simultaneously. In Fig. 2, the blue and green arrows represent information from the two frameworks entering the SEF block, while the yellow arrow indicates the result of feature fusion being fed back into these two networks.
The input image \(x \in {\mathbb {R}}^{H \times W \times C}\) is split into two information flows. One flows into the Local framework, where it undergoes a \(3\times 3\) deep convolutional layer followed by ReLU activation and max pooling, extracting shallow features to produce \(x_1 \in {\mathbb {R}}^{\frac{H}{4} \times \frac{W}{4} \times C'}\) (typically \(C' = 64\)). The other flow enters the Global framework, passing through a patch merging layer to obtain shallow features \(x_1' \in {\mathbb {R}}^{\frac{H}{4} \times \frac{W}{4} \times C'}\).
The results \(x_1\) and \(x_1'\) from these different feature extraction paths are concatenated and then fed through a \(3\times 3\) convolutional layer to restore the number of channels before entering the SEF block.
After regularization and sigmoid activation, the SE module evaluates the importance of each channel using global average pooling and two fully connected layers. It dynamically adjusts the weights of each channel to enhance important features and suppress irrelevant ones as follows:
Here, BN denotes batch normalization, \(W_1\) and \(W_2\) are the weights of the first and second fully connected layers respectively, and \(\sigma\) represents the Sigmoid activation function. Finally, the fused feature image \(x_1''\) is obtained. By adding the tensors \(x_1\) and \(x_1'\) to \(x_1''\), the subsequent deep feature extraction operations are continued.
The two frameworks each use their own feature extraction blocks, increasing the number of channels continuously according to \([2C', 4C', 8C', 16C']\), and reducing the feature dimensions. Each time, the obtained \(x_i\) and \(x_i'\) enter the SEF block for feature fusion to get \(x_i''\), which is then fed back to the two frameworks to continue downsampling. This multi-stage deep feature extraction and fusion allow global and local features to be maximally preserved and supplemented in both frameworks.
Decoder
The CFM-UNet encoder employs \(2\times 2\) deconvolutions to resize the image iteratively, as indicated by the gray arrow in Fig. 2. Each deconvolution output is integrated with \(x_i''\) using skip connections, combining deep and shallow features to preserve fine details. Following this, dual-layer \(3\times 3\) convolutions merge features with ReLU activation, progressively reducing channel numbers in the sequence [8C, 4C, 2C, C]. Ultimately, \(1\times 1\) convolutions decrease the channel count to 1, and interpolation restores the original image dimensions to produce the final segmentation output.
Loss function
To improve the segmentation accuracy towards the ground truth, we combine Dice loss and binary cross-entropy (BCE) using weighted factors in our loss function. The formulation of the loss function is given by:
where:
and \(\lambda _1\) and \(\lambda _2\) are the weights of the loss functions, with default values of 0.5 each. \(|X|\) and \(|Y|\) denote the sizes of the ground truth and prediction sets respectively, and \(y_i\) and \({\hat{y}}_i\) represent the true labels and predictions. This balanced strategy ensures a trade-off between maximizing overlap and enhancing classification accuracy, providing a robust starting point adaptable to various segmentation tasks. Such a configuration often yields satisfactory initial results and can be fine-tuned further based on task-specific performance needs.
Experiment and results
To validate our model’s segmentation performance on medical images, we selected datasets covering organs, tumors, bones, and tissues.
Data source
For human organs and tumor lesions, we used the LiTS22 dataset, which consists of 131 annotated CT scans for liver and liver tumor segmentation, presenting challenges such as class imbalance, varying tumor shapes, and scan variability. For human skeleton segmentation, we utilized the SPIDER23 dataset, which includes 3D MRI and CT scans annotated for vertebrae, intervertebral discs, and the spinal canal, designed for spinal anatomy segmentation but affected by variability in anatomy and scan quality. For human tissues, we chose the Kvasir-SEG24 colon polyp dataset, annotated for semantic segmentation of anatomical structures like polyps, mucosa, and lumen, with challenges such as image noise and class imbalance. To assess the model’s generalization ability, we conducted transfer testing on the ATLAS25 liver dataset, which features significant variability in tumor shape, size, and contrast, complicating accurate segmentation.
A smaller dataset scale we selected simulated realistic issues, including the high cost of acquiring medical image data, ethical restrictions involving patient privacy, and the difficulty of obtaining high-quality annotations. Meanwhile, we selected non-continuous slices from each patient when processing the CT datasets to enhance data processing efficiency, reduce redundancy caused by similar consecutive slices, and focus on key regions. Therefore, we randomly selected 1030 liver slices and 670 liver tumor slices from LiTS, 1000 complete spine slices from SPIDER, and all 1000 colon polyp endoscopic images from Kvasir-SEG. To further validate the model’s generalization ability, we also selected 800 liver slices from ATLAS for model transfer testing. For each segmented dataset mentioned above, we uniformly randomly divide the dataset into training and test sets using an 8:2 ratio. We evaluate the model performance using recommended metrics specific to each dataset. These metrics include the average cross-dice similarity coefficient (DSC), intersection-over-union (IoU), true positive rate (TPR), volumetric overlap error (VOE), relative volumetric difference (RVD), and average surface-to-surface distance (ASSD). For ease of comparison, we choose to display the absolute values.
Experimental environment
The experiments in this paper utilize the Mamba and PyTorch frameworks. The GPU employed is an RTX 4090, and Python version 3.10 is used. The model training epoch is set to 100, with a batch size of 4. The optimizer selected for training is Adam.
Experimental comparison
We compare our proposed CFM-UNet with 12 advanced 2D segmentation models, which can be roughly categorized into four groups. (1) general-purpose segmentation models6,9,27,28: the U-Net (Ronneberger et al., 2015), the ATT-UNet (Oktay et al., 2018), the Res-UNet (Xiao et al., 2018), and the U-Net++ (Zhou et al., 2018). (2) CNN-based medical image segmentation models8,34: the U-ReSNet (Estienne et al., 2019), M2SNet (Zhao et al., 2023). (3) Transformer-based medical image segmentation models13,35: TransUNet (Chen et al., 2021), META-Unet (Wu et al., 2023). (4) Mamba-based medical image segmentation Models16,36,37: Swin-UMamba (Liu et al., 2024), VM-UNet-V2 (Zhang et al., 2024), UltraLight VM-UNet (Wu et al., 2024). To be fair, none of the models used pre-training parameters in our comparison experiments. Each model trained on the training set was tested five times on the test set, and the average value for each metric was calculated.
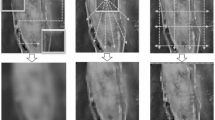
We selected two segmentation results from each dataset to represent the segmentation effect for visual analysis. The overall segmentation effect graph is depicted in Fig. 4. Subsequently, we will proceed with a detailed analysis of the model performance.
Liver and liver tumor segmentation
Liver segmentation
In sample a, aside from CFM-UNet, which accurately learns liver segmentation, the other models misclassify other organs as liver, as shown in Fig. 4. This highlights CFM-UNet’s effective utilization of the Mamba network, demonstrating a superior understanding of liver anatomy. In sample b, concerning detail handling, CFM-UNet and U-ResNet effectively utilize CNN’s capacity to capture local details, demonstrating precise localization in the lower right corner of the liver. In contrast, other models exhibit errors in this area.
Liver tumor segmentation
As demonstrated in Fig. 4, CFM-UNet, UNet++, U-ResNet, META-UNet, and UltraLight-VM-UNet, which employ more complex network structures, accurately segment liver tumors even in relatively small areas. In contrast, all other models exhibit noticeable noise, underscoring the effectiveness of deeper network architectures in comprehensively capturing liver tumor characteristics to a significant extent.
We preprocessed the LiTS dataset by slicing it into \(512 \times 512\) 2D images. Table 1 presents the quantization results of each model for segmenting this dataset.
The Ushape framework demonstrates outstanding performance in segmenting specific organs, emphasizing the importance of enhancing local accuracy control for improved accuracy. Our model utilizes a Local Branch to maximize local detail extraction capabilities, while the Global Branch leverages a feature fusion module to enhance learning of liver organ features. Given that liver tumors are typically small and scattered, demanding high model proficiency in local detail extraction, our approach harnesses the advantages of residual structures in both the lower convolution of the Local Branch and the feature fusion module. This strategy significantly enhances the model’s capacity to learn intricate local features of both the liver and liver tumors.
In comparison to other models, ours demonstrates robust segmentation capabilities across all three evaluation metrics, notably excelling in the core DICE metric. Although our model does not surpass others in all aspects of TPR and ASSD, the performance gap with the top model is minimal. Therefore, overall, our model exhibits excellent performance and remains highly competitive in the segmentation of both liver organs and liver tumors.
Spine segmentation
All models successfully segment the entire spine in Fig. 4. In sample g, CFM-UNet and M2SNet exhibit relatively less noise, while in sample h, CFM-UNet, META-UNet, and VM-UNet-V2 also show reduced noise. This suggests that CFM-UNet effectively leverages the Mamba’s characteristics to comprehensively understand each component of the spine. Additionally, it utilizes CNN capabilities to capture intricate details of the spine.
We preserved the entire spine structure in the SPIDER dataset and preprocessed it by slicing into \(512 \times 512\) 2D images. Table 2 presents the quantization results of each model for segmenting this dataset.
Retaining all spine components in the SPIDER dataset means that vertebrae, intervertebral discs, and the spinal canal may not appear simultaneously in a single slice, requiring the model to comprehend and learn the entire spine comprehensively. In our comparative experiments, our model achieved first place in four performance metrics, showcasing superior segmentation accuracy and robust feature learning capabilities.
Colon polyps segmentation
As demonstrated in Fig. 4, CFM-UNet, U-ResNet, M2SNet, META-UNet, VM-UNet-V2, and Swin-UMamba models achieve correct segmentation in sample e, while other models display noticeable segmentation errors. This highlights the specialized effectiveness of medical image segmentation methods over generalized approaches. The Mamba-based segmentation methods demonstrate superior capability in understanding the overall structure of the image. In sample f, only CFM-UNet correctly segments the colon polyps, showcasing its proficiency in leveraging the advantages of the Mamba network for accurate segmentation.
The Kvasir-SEG dataset comprises 2D images with varying pixel sizes, which we uniformly resize to \(512 \times 512\) dimensions as part of our preprocessing. Table 3 displays the quantization outcomes for each model in segmenting this dataset.
The segmentation of polyps in the Kvasir-SEG dataset is challenging due to varying sizes and unpredictable distribution caused by viewing angles. This variability imposes stringent demands on the model’s feature learning capabilities. A single network structure may provide incomplete polyp feature extraction, prompting the integration of our model with two network structures. This approach inherently advantages our model, leading to superior segmentation accuracy across all metrics compared to other models.
Significance analysis
To further demonstrate the superiority of our model approach on the aforementioned metrics in the experimental tasks, we employed the T-Test method to conduct significance testing between the top five models for each segmentation task and the CFM-UNet across the five repeated experiments on test set. We visualized the calculated p-values using a heatmap to intuitively assess the significance of CFM-UNet compared to other models on each metric.
If the p-value calculated using the T-Test is less than 0.05, the null hypothesis can be rejected, indicating a significant difference between the two models on that metric. Specifically, as shown in Fig. 5, the lighter the blue color in the blocks, the more apparent the difference between CFM-UNet and the corresponding model on the current metric for the task. From Fig. 5, we can observe in the “Experimental Comparison” section that the blocks for the metrics where CFM-UNet shows a clear advantage are light in color, with p-values much smaller than 0.05, indicating significant superiority. Although CFM-UNet does not perform at the top in terms of the TPR metric for the Spine Segmentation task, the corresponding block is darker, suggesting that the difference with the leading models is not significant. Moreover, since the ASSD quantification values of the leading models in the Spine Segmentation task do not show significant differences and the corresponding blocks are darker, it suggests that the performance in terms of boundary smoothness for spine segmentation is similar across these models.
Melt experiment
Number of parameters
Our model consists of two branches: one based on CNN and the other on Mamba. Therefore, determining the hyperparameter, which is the combination of the number of modules needed for each downsampling in these two branches, is crucial. For the parameters of Bottle2neck Blocks, we drew on the number of modules from three classic deep feature extraction networks, including Res2Net21, and attempted various combinations. Initially, we used the same number of modules as some common configurations, including the [2,2,2,2] structure from VM-UNet-V236.
We tested these parameter combinations on the LiTS liver dataset and evaluated the segmentation performance using the DICE metric. The quantitative results are presented in Table 4. Therefore, we have selected Bottle2neck Blocks configured as [3, 4, 6, 3] for the Global Branch and VSS blocks configured as [3, 3, 3, 3] for the Local Branch as the downsampling modules in our model.
Structure ablation
To assess the effectiveness of our parallel network structure, we conducted experiments by individually removing the Local Branch and Global Branch. Additionally, to evaluate the effectiveness of our SEF block, we conducted experiments where we removed the SEF block and instead employed additive fusion and multiplicative fusion methods.
All experiments were conducted on the LiTS liver dataset, and detailed metrics can be found in Table 5. The experiments demonstrate that removing any module from our model results in reduced accuracy. Furthermore, our SEF block, with its negative feedback mechanism, effectively integrates the strengths of both global and local networks, leading to comprehensive improvements in segmentation accuracy. In addition, selecting other CNN-based modules(dual convolutions and Bottleneck Blocks) as the core component of the Local Branch in CFM-UNet fails to surpass the Bottle2neck Blocks in fully leveraging CNN’s capability for local feature extraction.
Generalization ability
To verify the generalization ability of the model, we conducted generalization experiments through data augmentation and data migration without changing the training parameters of the model on the LiTS liver dataset.
Image rotation
The test image was rotated by \(0^\circ\), \(60^\circ\), \(120^\circ\), \(180^\circ\), \(240^\circ\), and \(300^\circ\), then resized to a pixel size of \(512 \times 512\) before reassessing segmentation accuracy.
Pixel resizing
The test image was resized to pixel dimensions of \(256 \times 256\), \(512 \times 512\), and \(1024 \times 1024\), followed by a reassessment of segmentation accuracy.
As shown in Table 6, the two key segmentation performance metrics, DICE and TPR, do not exhibit significant fluctuations overall. This indicates that data augmentation through image rotation and pixel adjustment has a minimal impact on the overall accuracy of the CFM-UNet.
Data migration
For the model migration test, we utilized the open-source liver dataset ATLAS. From this dataset, 800 images were randomly selected and uniformly resized to \(512\times 512\) pixels. Subsequently, the segmentation accuracy test was performed anew.
The visual representation in Fig. 6 shows that DICE coefficients results of CFM-UNet on ATLAS mostly fluctuate between 0.8 and 0.9, demonstrating robust and favorable quantitative performance.
As presented in Table 7, DICE and TPR metrics maintained at approximately 84%. In conclusion, CFM-UNet demonstrates robust generalization capabilities in both data augmentation and data migration experiments.
Conclusion and future work
We introduced the CFM-UNet model, which integrates two network architectures, CNN and Mamba, for image segmentation. In the Local Branch, we employed the CNN-based Bottle2neck module to extract local features, while in the Global Branch, the Mamba-based VSS module was utilized to capture global features. The SEF block, designed for feature fusion and feedback, enables leveraging the complementary strengths of both networks. Experimental results demonstrate that CFM-UNet achieves excellent performance on the LiTS, SPIDER, and Kvasir-SEG medical image datasets, and exhibits strong generalization capabilities on the ATLAS dataset.
In future experiments, we plan to further optimize CFM-UNet and explore its application in semi-supervised medical image tasks. This approach aims to effectively utilize unlabeled medical image data, thereby enhancing the model’s generalization ability and practical applicability.
Data availability
The datasets used in the study are publicly available at the following links: LiTS - https://competitions.codalab.org/competitions/17094. ATLAS - https://atlas-challenge.u-bourgogne.fr/dataset. Kvasir-SEG - https://datasets.simula.no/downloads/kvasir-seg.zip. SPIDER - https://spider.grand-challenge.org/data/.
References
Qureshi, I. et al. Medical image segmentation using deep semantic-based methods: A review of techniques, applications and emerging trends. Inf. Fusion 90, 316–352. https://doi.org/10.1016/j.inffus.2022.09.031 (2023).
Ramesh, K., Kumar, G. K., Swapna, K., Datta, D. & Rajest, S. S. A review of medical image segmentation algorithms. EAI Endors. Trans. Pervasive Health Technol. 7, e6–e6 (2021).
He, F. et al. Brain tumor image segmentation network based on dual attention mechanism. In International Conference on Intelligent Computing, 125–136 (Springer, 2023).
He, X., Qiao, Z., Huang, Y. & Hao, Q. Dilated-residual u-net for optical coherence tomography noise reduction and resolution improvement. In Optics in Health Care and Biomedical Optics XIII, Vol. 12770, 246–254 (SPIE, 2023).
Yuan, Y. et al. Dsca-pspnet: Dynamic spatial-channel attention pyramid scene parsing network for sugarcane field segmentation in satellite imagery. Front. Plant Sci. 14, 1324491 (2024).
Zhou, Z., Rahman Siddiquee, M. M., Tajbakhsh, N. & Liang, J. Unet++: A nested u-net architecture for medical image segmentation. In Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support: 4th International Workshop, DLMIA 2018, and 8th International Workshop, ML-CDS 2018, Held in Conjunction with MICCAI 2018, Granada, Spain, September 20, 2018, Proceedings 4, 3–11 (Springer, 2018).
Zhao, X. et al. Prior attention network for multi-lesion segmentation in medical images. IEEE Trans. Med. Imaging 41, 3812–3823 (2022).
Zhao, X. et al. M2snet: Multi-scale in multi-scale subtraction network for medical image segmentation. arXiv preprint arXiv:2303.10894 (2023).
Ronneberger, O., Fischer, P. & Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, October 5–9, 2015, Proceedings, Part III 18, 234–241 (Springer, 2015).
Guo, C. et al. Sa-unet: Spatial attention u-net for retinal vessel segmentation. In 2020 25th International Conference on Pattern Recognition (ICPR), 1236–1242 (IEEE, 2021).
Wang, Z., Zhang, H., Huang, Z., Lin, Z. & Wu, H. Multi-scale dense and attention mechanism for image semantic segmentation based on improved deeplabv3+. J. Electron. Imaging 31, 053006–053006 (2022).
Cao, H. et al. Swin-unet: Unet-like pure transformer for medical image segmentation. In European Conference on Computer Vision, 205–218 (Springer, 2022).
Chen, J. et al. Transunet: Transformers make strong encoders for medical image segmentation. arXiv preprint arXiv:2102.04306 (2021).
Gu, A. & Dao, T. Mamba: Linear-time sequence modeling with selective state spaces. arXiv preprint arXiv:2312.00752 (2023).
Liu, Y. et al. Vmamba: Visual state space model. arXiv preprint arXiv:2401.10166 (2024).
Liu, J. et al. Swin-umamba: Mamba-based unet with imagenet-based pretraining. arXiv preprint arXiv:2402.03302 (2024).
Ruan, J. & Xiang, S. Vm-unet: Vision mamba unet for medical image segmentation. arXiv preprint arXiv:2402.02491 (2024).
Ma, J., Li, F. & Wang, B. U-mamba: Enhancing long-range dependency for biomedical image segmentation. arXiv preprint arXiv:2401.04722 (2024).
Peng, Z. et al. Conformer: Local features coupling global representations for visual recognition. In Proceedings of the IEEE/CVF International Conference on Computer Vision, 367–376 (2021).
Hu, J., Shen, L. & Sun, G. Squeeze-and-excitation networks (2018).
Gao, S.-H. et al. Res2net: A new multi-scale backbone architecture. IEEE Trans. Pattern Anal. Mach. Intell. 43, 652–662 (2019).
Bilic, P. et al. The liver tumor segmentation benchmark (lits). Med. Image Anal. 84, 102680 (2023).
van der Graaf, J. W. et al. Lumbar spine segmentation in MR images: A dataset and a public benchmark. Sci. Data 11, 264 (2024).
Jha, D. et al. Kvasir-seg: A segmented polyp dataset. In MultiMedia Modeling: 26th International Conference, MMM 2020, Daejeon, South Korea, January 5–8, 2020, Proceedings, Part II 26, 451–462 (Springer, 2020).
Quinton, F. et al. A tumour and liver automatic segmentation (atlas) dataset on contrast-enhanced magnetic resonance imaging for hepatocellular carcinoma. Data 8, 79 (2023).
Long, J., Shelhamer, E. & Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, 3431–3440 (2015).
Oktay, O. et al. Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:1804.03999 (2018).
Diakogiannis, F. I., Waldner, F., Caccetta, P. & Wu, C. Resunet-a: A deep learning framework for semantic segmentation of remotely sensed data. ISPRS J. Photogramm. Remote. Sens. 162, 94–114 (2020).
Dosovitskiy, A. et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020).
Zhang, Y., Liu, H. & Hu, Q. Transfuse: Fusing transformers and cnns for medical image segmentation. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: 24th International Conference, Strasbourg, France, September 27–October 1, 2021, Proceedings, Part I 24, 14–24 (Springer, 2021).
Dong, B. et al. Polyp-pvt: Polyp segmentation with pyramid vision transformers. arXiv preprint arXiv:2108.06932 (2021).
Qi, Q., Lin, L., Zhang, R. & Xue, C. Medt: Using multimodal encoding-decoding network as in transformer for multimodal sentiment analysis. IEEE Access 10, 28750–28759 (2022).
Chen, B., Liu, Y., Zhang, Z., Lu, G. & Kong, A. W. K. Transattunet: Multi-level attention-guided u-net with transformer for medical image segmentation. IEEE Trans. Emerg. Top. Comput. Intell. 8(1), 55–68. https://doi.org/10.1109/TETCI.2023.3309626 (2023).
Estienne, T. et al. U-resnet: Ultimate coupling of registration and segmentation with deep nets. In Medical Image Computing and Computer Assisted Intervention–MICCAI 2019: 22nd International Conference, Shenzhen, China, October 13–17, 2019, Proceedings, Part III 22, 310–319 (Springer, 2019).
Wu, H., Zhao, Z. & Wang, Z. Meta-unet: Multi-scale efficient transformer attention unet for fast and high-accuracy polyp segmentation. IEEE Trans. Autom. Sci. Eng. 21(3), 4117–4128. https://doi.org/10.1109/TASE.2023.3292373 (2023).
Zhang, M., Yu, Y., Gu, L., Lin, T. & Tao, X. Vm-unet-v2 rethinking vision mamba unet for medical image segmentation. arXiv preprint arXiv:2403.09157 (2024).
Wu, R., Liu, Y., Liang, P. & Chang, Q. Ultralight vm-unet: Parallel vision mamba significantly reduces parameters for skin lesion segmentation. arXiv preprint arXiv:2403.20035 (2024).
Acknowledgements
Financial support for this project was provided by the Promoting the Classification and Development of Colleges and Universities-Student Innovation and Entrepreneurship Training Programme Project-School of Computer (5112410852).
Author information
Authors and Affiliations
Contributions
K.N.: Investigation, Conceptualization, validation, Resources, supervision. J.H.: Methodology, Software, Writing – original draft. J.C.: Formal analysis, Writing – original draft.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Niu, K., Han, J. & Cai, J. CFM-UNet: coupling local and global feature extraction networks for medical image segmentation. Sci Rep 15, 22236 (2025). https://doi.org/10.1038/s41598-025-92010-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-92010-y