Abstract
Age-related macular degeneration (AMD) and Parkinson’s disease (PD) are prevalent and debilitating conditions that lead to irreversible blindness and dyskinesia, respectively. Emerging evidences imply that retinal abnormalities may serve as early indicators for monitoring PD. This study endeavors to explore the complex interactions and focus on their shared molecular and pathological mechanisms. We employed a comprehensive approach by integrating single-cell RNA sequencing (scRNA-seq) datasets, obtained from dry AMD retinas and PD brain tissues, along with Weighted Gene Co-expression Network Analysis (WGCNA)-related computational analysis. Gene Set Enrichment Analysis (GSEA) was conducted to analyze PD-related genes within retinal ganglion cells in dry AMD. Cell-cell chat was utilized to predict intercellular communication and signaling pathways. Module eigengenes (MEs) were calculated to identify specific gene modules. Dysregulation of PALLD, FYN and ZMZ1 may lead to cell structural abnormalities, impaired mitochondrial functions, and increased susceptibility to neuroinflammation, contributing to the AMD and PD progression. Additionally, this study highlighted an astrocyte–oligodendrocyte precursor cell (OPCs) signaling axis mediated by Neuregulin (NRG), which is hypothesized to influence neuroinflammatory processes characteristic of dry AMD and PD pathogenesis. Notably, ME-salmon module associated with gene dysregulation exhibited a strong positive correlation with the ME-blue module, linked to neurodegenerative impairment, and the ME-yellow module, related to mitochondrial dysfunction. The comprehensive investigation on astrocytes-OPCs signaling axis, and the NRG signaling pathway advances our understanding of the intricate biological processes underpinning AMD and PD. This research underscores the critical importance about exploring glial-related cell interactions, providing valuable insights into potential novel therapeutic approaches for these complex diseases.
Similar content being viewed by others
Introduction
Age-related macular degeneration (AMD) is a dominating contributor to blindness among the elderly, with 288 million individuals projected to be affected by 20401. AMD primarily impacts the macula, the central area of the retina responsible for high acuity and color vision2. The disease is categorized into two types: dry (non-neovascular) and wet (neovascular) AMD. Dry AMD is the more prevalent type, accounting for approximately 85–90%, characterized by geographic atrophy3. While wet AMD can lead to rapid and severe vision loss, it can be effectively intervened with anti-vascular endothelial growth factor (anti-VEGF) therapy. In contrast, there is currently no effective prevention or treatment for dry AMD, highlighting the urgency of exploring more novel therapeutic strategies4. Parkinson’s disease (PD) is an incurable and progressive neurodegenerative disorder, targeting the dopaminergic neurons in the substantia nigra. With a prevalence of approximately 2–3% among individuals over 60 years old, while the global rates of disability and mortality are rising swiftly, launching a rigorous challenge for families and public health5. PD may appear typical motor symptoms such as bradykinesia, tremors, rigidity, impaired balance, as well as mood and cognitive disorders (non-motor symptoms)6. Although there is no disease-modifying therapy available, several therapies are currently used to manage PD symptoms and improve quality of life. These include L-dopa (levodopa), which remains the gold standard for motor symptom control, as well as non-pharmacological interventions such as transcranial stimulation to aid in symptom management.
The retina is frequently regarded as an extension of the central nervous system (CNS), with analogous anatomical and functional to the brain, offering a significant vantage point for investigating diverse CNS pathologies. Recent studies have shed light on the potential association between AMD and PD. A retrospective cohort study involving 20,848 patients had indicated that individuals with AMD may face an elevated risk of developing PD compared to those without AMD7. Another clinical study revealed a higher prevalence of AMD among patients with PD than in the general population, where 15,800 out of 172,726 PD patients being subsequently diagnosed with AMD during follow-up8. Interestingly, the presence of amyloid beta plaques and elevated cholesterol levels observed in the retina of individuals with AMD, indicating a syndrome that accelerates brain aging9. Hence, several hypotheses have been proposed. One possibility is that both disorders share common genetic susceptibilities or environmental risk factors (hyperglycemia and hyperlipidemia) that contribute to their development. Additionally, they share common mechanisms in pathogenesis, including oxidative stress, inflammation, cell senescence and impaired autophagy10,11. Neuroinflammation is a complex process involving immune cells activation and inflammatory molecules release. The potential relationship between AMD and PD through neuroinflammation is an area of active research. Common signaling pathways and molecules involved in neuroinflammation, such as nuclear factor-kappa B (NF-κB) and toll-like receptors (TLRs), have been implicated in both AMD and PD pathogenesis12,13. Furthermore, several common features between AMD and another neurodegenerative disease, Alzheimer’s disease (AD) have already been identified in the early stages, especially on microglial cells and upregulated pro-inflammatory microRNAs (miRNAs)14,15. However, the precise pathophysiological mechanisms underlying the neuroinflammation association between AMD and PD remain to be fully elucidated.
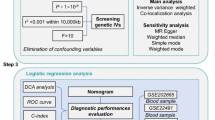
In order to address the intricate and enigmatical questions, we employed a comprehensive approach by integrating single-cell RNA sequencing (scRNA-seq) datasets, obtained from dry AMD retinas and PD brain tissues, along with Weighted Gene Co-expression Network Analysis (WGCNA)-related computational analysis (as shown in the Fig. 1). Traditionally, the pathological changes in retinal pigment epithelial (RPE) cells and photoreceptors have been paid more attention on AMD-related studies. Meanwhile, researches about PD have concentrated on the degeneration of dopaminergic neurons. Our work elucidated a common astrocyte and oligodendrocyte precursor cell (OPC) expression signature in human retina and brain tissue from AMD and PD patients respectively. We are the first to identify an astrocyte-to-OPC signaling axis mediated by Neuregulin (NRG), which may be a common driver for neuroinflammation characteristics of dry AMD and PD pathogenesis. Moreover, we found a close relationship between neuroinflammation and mitochondrial dysfunction through the regulation of the hub genes PALLD, FYN and ZMIZ1. These results highlighted the potential of targeting common neuroinflammatory pathways in both AMD and PD, offering new insights into the shared molecular mechanisms and their implications for future therapeutic strategies.
Results
Mapping the cellular landscape of dry AMD
After quality control procedures, 23,642 high-quality genes from GSE221042 were obtained for further biological analysis. As shown in Fig. 2A, we performed t-SNE for cell clustering and identified 12 distinct cell clusters: neuronal cell types (retinal ganglion cells, rod photoreceptors, horizontal cells, bipolar cells, cone photoreceptors), and non-neuronal cell types (Müller glia, OPCs, astrocytes). Furthermore, a heatmap was constructed to exhibit marker genes in each cell cluster (Fig. 2B, Supplementary Table S1), including retinal ganglion cells (RGCs) (NEFL, MAP1B, NEFM), rod photoreceptors (PDE6A, PDE6G, RHO), horizontal cells (GRIP1, GRIA4, LSAMP), bipolar cells (GRIK1, TRPM1, PTPRM), Müller glia (CTNNA2, TF, KAZN), cone photoreceptors (KCNB2, PEX5L, ARR3), OPCs (TNR, GRIA3, NAV3), astrocytes (SLC11A1, DGKG, GFAP). In addition, it was observed that RGCs and cone photoreceptors accounted for more than 50% of the retinal cells. The discovery of OPCs in the dataset provided a new insight into retinal cellular heterogeneity and worth further investigation. On the other hand, OPCs and astrocytes constituted a smaller proportion, comprising approximately 2% (Fig. 2C, Supplementary Table S2). These results shed light on the cellular heterogeneity and composition of the retina, providing a foundation for further investigations into the functional roles and interactions of these cell types in the context of vision and retinal physiology.
Link between retinal neurons in dry AMD and neurodegenerative disease
We retrieved gene expression data from two RGCs clusters, containing a total of 687 genes. Through the application of GSEA and KEGG pathway datasets, we revealed significant enrichment scores in pathways associated with various neurodegenerative diseases, including PD, Huntington’s disease (HD) and Alzheimer’s disease (AD) (Fig. 3A, Supplementary Table S3). The PD pathway was significantly enriched in both RGCs clusters (P = 0.002; P = 0.027) (Fig. 3B). Among these genes in the PD pathway, 9 genes were overlapped, which included MT-ATP6, TUBA1B, MT-ND4, MT-CO1, GNAS, VDAC1, MT-CO3, CALM1, MT-ND1 (Fig. 3C). It was also observed that there were 7 and 11 genes that overlapped in the two clusters related to HD and AD, respectively. These findings underscored the potential involvement of ganglion cell gene expression in the pathogenesis of these debilitating disorders, particularly highlighting the strong association between AMD and PD.
Detailed cell atlas and gene expression in PD
To establish an extended reference single-cell atlas and identify similarities between AMD and PD, we screened a total of 30,172 genes associated with PD (GSE161045). Following normalization and dimensionality reduction, these genes were classified into 9 various cell types (Fig. 4A, Supplementary Table S4): two clusters as oligodendrocytes, one clusters as astrocytes, two clusters as microglia, two clusters as neurons, one cluster as endothelial cells, one cluster as OPCs. Upon analyzing the cell types across the datasets, OPCs and astrocytes exhibited overlapping characteristics. Given their common traits and potential involvement in the pathogenesis of AMD and PD, we focused on OPCs/astrocytes axis. As shown in Fig. 4B, astrocytes were enriched in SLC1A2, GPM6A, ADGRV1. OPCs exhibited high expression levels in DSCAM, PCDH15, VCAN and neurons were characterized by RBFOX1, KCNIP4, ROBO2, SYT1. The cell population analysis revealed that astrocytes and OPCs accounted for approximately 10% within the total cells (Fig. 4C, Supplementary Table S5). Although these two cell types represent a relatively minor portion of the overall cell population, they are essential for understanding the pathogenesis of both AMD and PD.
Unveiling common hub genes in astrocytes and oligodendrocyte precursor cells across both datasets
To identify common cell types and further elucidate the molecular mechanisms involved in the pathogenesis of AMD and PD, we conducted a comprehensive analysis of DEGs in key cell types across both datasets. Astrocytes and OPCs were consistently present through cell clustering. As shown in Figs. 5A, 516 and 644 DEGs were severally identified in astrocytes and OPCs in the AMD dataset. Furthermore, we identified 728 DEGs for astrocytes and 720 DEGs for OPCs in the PD dataset (Supplementary Table S6). Venn diagrams revealed 22 genes that were highly expressed in both astrocytes and OPCs across the two datasets. These genes include FYN, ASAP1, PALLD, PITPNC1, TNRC6A, LSAMP, NTRK2, DCLK2, NAV3, NEBL, PTPRG, CSGALNACT1, NFIA, NAV2, LHFPL6, ATP2B4, NPAS3, RPS6KA2, ZMIZ1, KIAA1211, PTPRZ1 and TRIO. The dot plots from AMD dataset demonstrated that PALLD, LHFPL6, RPS6KA2 and ZMIZ1 were more specifically expressed in astrocytes and OPCs (Fig. 5B). In addition, NTRK2, PALLD, ATP2B4 and ZMIZ1 played dominant roles in astrocytes and OPCs in the PD dataset (Fig. 5C). Hence, PALLD and ZMIZ1 emerged as major predictive genes that correlated AMD and PD, which were also involved in the neural function regulation. These findings provided valuable insights into the shared pathophysiological processes underlying these neurodegenerative diseases and may offer novel targets for therapeutic intervention.
Deciphering the complexity of NRG signaling pathway in orchestrating astrocytes and OPCs in dry AMD
Intercellular communication networks play a crucial role in coordinating cellular functions through signaling interactions. CellChat, a tool for predicting cellular interactions, helps elucidate how cells affect or are affected by other cells. Analyzing the scRNA-seq data from AMD samples revealed numerous strong interactions between astrocytes and OPCs within the NRG signaling pathway, compared to the other 12 signaling pathways (Fig. 6A). As shown in Fig. 6B, the most cell populations in the AMD dataset received NRG signaling inputs, indicating the widespread influence of NRG signaling across the cellular landscape. In addition, our analysis demonstrated that paracrine signaling predominantly mediated intercellular interactions within NRG pathway, while astrocytes and Müller glia clusters exhibited significant autocrine signaling (Fig. 6C). Moreover, centrality analysis of the NRG signaling network (NRG1-ERBB4) indicated that bipolar cells, OPCs, and horizontal cells were chiefly served as senders, exerting their influence on astrocytes (Fig. 6D). In terms of gene expression about ligand-receptor, NRG1 was highly expressed in OPCs, playing a crucial role in their function and signaling processes, while ERBB4 was predominantly involved in astrocyte-related activities (Fig. 6E). Overall, our analysis underscored the complexity and specificity of intercellular communication within the AMD microenvironment.
Decoding the role of NRG signaling pathway in coordinating astrocytes and OPCs in PD
In the Fig. 7A, a strong interaction between astrocytes and OPCs within the NRG signaling pathway was evident. Furthermore, NRG signaling were identified as a source for the majority of cell clusters in the PD dataset (Fig. 7B). Notably, astrocytes were the primary driving force behind NRG signaling directed towards OPCs in the context of PD. Figure 7C provided insights into the nature of signaling interactions within the NRG pathway. Both astrocytes and OPCs exhibited significant paracrine and autocrine signaling capabilities, indicating their dual roles in communicating with neighboring cells and self-regulating their own functions. Astrocytes, in particular, emerged as major contributors within this network, acting as senders, receptors, mediators and influencers, thereby controlling communication within the NRG pathway and exerting substantial influence on OPCs (Fig. 7D). Additionally, our results revealed that both NRG1 and ERBB4 were involved in functional processes and signaling pathways related to astrocytes and OPCs (Fig. 7E). Collectively, the prominent roles of astrocytes and OPCs in the NRG signaling pathway highlighted their potential contributions to the disease’s pathogenesis. Understanding these complex intercellular communications could pave the way for identifying novel therapeutic targets aimed at modulating NRG signaling pathways.
Assembling weighted gene co-expression groups
To further identify crucial gene modules and key driver genes associated with these diseases and gain a deeper understanding of the molecular mechanisms, we performed WGCNA on transcriptome sequencing data from dry AMD (GSE29801). As shown in Fig. 8A, the soft-thresholding (14) was determined by the criteria of Sacle independence and Mean connectivity. The co-expressed gene clustering tree were visualized in Fig. 8B. By setting the abline to 0.2, the initial 17 co-expression modules were ultimately clustered into 10 distinct modules (Fig. 8C). Additionally, we calculated Pearson’s r and p-values associated with the module eigengenes (ME) to quantify the correlations between different modules. Figure 8D demonstrated that the ME-salmon module exhibited a strong positive correlation with the ME-blue and ME-yellow modules (p = 0.02, R² = 0.64; p = 0.04, R² = 0.72). The three modules were independent of each other, which was confirmed by 3D scatter plot (Fig. 8E). Furthermore, the intracellular genes were unlikely to be intermediate genes in Venn diagram (Fig. 8F). These findings indicated a robust regulatory relationship between specific gene modules, which could better realize the molecular underpinnings of dry AMD.
Core mechanisms and key genes among AMD development
Hence, ME-salmon, ME-yellow and ME-blue may modulate specific functions in dry AMD. Functional analysis of the gene modules revealed that the ME-salmon module was significantly enriched in the GO -- “0006397: mRNA processing (p = 2.83*10− 4)”; “0000398: mRNA splicing, via spliceosome (p = 9.83*10− 4)”; “0007601: visual perception (p = 0.001)” (Fig. 9A, Supplementary Table S7). KEGG-term “hsa03082: ATP-dependent chromatin remodeling (p = 0.026)”, which was exhibited in Fig. 9D. These processes are crucial for the proper transcription, modification and structural arrangement of genetic material, ultimately ensuring accurate gene expression. Given that AMD is a neurodegenerative disease of the retina and mounting evidence suggested that dysfunctional mRNA processing and metabolism may be key contributors to cellular senescence. Thus, ME-salmon may be defined as “Gene Dysregulation”. In Fig. 9B and E, ME- yellow were predominantly concentrated in GO “0030308: negative regulation of cell growth (p = 0.014)”, “0030182: neuron differentiation (p = 0.019)”, “0043065: positive regulation of apoptotic process (p = 0.036)”. KEGG-term “hsa05012: Parkinson disease (p = 0.038)”, “hsa05022: Pathways of neurodegeneration - multiple diseases (p = 0.063)”. These characteristics could contribute to the progressive degeneration and retinal cells dysfunction, ultimately leading to the characteristic features of AMD, such as the loss of central vision and retinal structural changes. Thus, ME-yellow may represent “Neurodegenerative impairment”. Additionally, ME- blue module exhibited prominent enrichment in GO “0042776: mitochondrial ATP synthesis coupled proton transport (p = 2.74*10− 20)”, “0032543: mitochondrial translation (p = 8.61*10− 19)”, “0006120: mitochondrial electron transport, NADH to ubiquinone (p = 3.80*10− 15)” (Fig. 9C). KEGG-term “hsa00190: Oxidative phosphorylation (p = 2.89*10− 25)”, “hsa05012: Parkinson disease (p = 6.69*10− 22)” (Fig. 9F). Excessive reactive oxygen species produced in mitochondria leads to oxidative stress, which are thought to be key factors in triggering RPE degeneration. Thus, ME-blue may classify as “Mitochondrial Dysfunction”. In summary, the study of “genetic dysregulation”, “neurodegenerative damage” and “mitochondrial dysfunction”, which are the key pathological mechanisms of dry AMD, might provide new therapeutic targets.
To identify key genes mediating these critical biological processes, we integrated the top 100 highly correlated genes from the ME-salmon, ME-yellow and ME-blue modules with the core targets identified through single-cell RNA-seq analyses (Fig. 10A-C, Supplementary Table S8). This integrative approach led to the identification of three hub genes: PALLD, FYN, and ZMIZ1. These genes serve as central nodes in the gene co-expression network and mediate essential biological processes. Therefore, PALLD, FYN and ZMIZ1may represent promising targets for future therapeutic strategies aimed at modulating the key pathological mechanisms of AMD, such as gene dysregulation, neurodegenerative impairment, and mitochondrial dysfunction.
Discussion
AMD is a complex neurodegenerative visual disorder characterized by the macula progressive deterioration, leading to central vision loss. PD is another neurodegenerative disease primarily associated with motor symptoms. Studies have indicated that these non-motor symptoms are not exclusively late-stage occurrences, potentially heralding the onset of PD or related degenerative changes, often preceding these conditions by several years. Among this, a less conspicuous yet crucial aspect that is frequently overlooked by PD patients themselves and may remain unnoticed. Alterations in olfactory function have emerged as potential early biomarkers for various neurodegenerative diseases16. Olfactory dysfunction (OD) is commonly observed in patients with early-stage neurodegeneration, with studies reporting that up to 90% of individuals with early-stage PD and 85% of those with early-stage AD exhibit olfactory impairments17. However, emerging evidence suggests that it can also involve visual impairments and retinal abnormalities, such as changes in retinal thickness, ganglion cell layer loss, and alterations in retinal vascularization18. Optical coherence tomography (OCT) has proven to be a valuable tool for visualizing and monitoring retinal changes in both AMD and PD, offering an early and non-invasive method for assessing disease progression19,20. The similarities in the pathophysiology of AMD and PD have sparked interest in exploring the underlying disease progression pathways. Therefore, we combined scRNA-seq and transcriptome sequencing data, as well as develop a machine learning pipeline to reveal the potential crosstalk and communication involved in the pathogenesis of these diseases. Investigating commonalities in molecular pathways, such as oxidative stress, neuroinflammation, mitochondrial dysfunction, and protein misfolding, may provide valuable insights into the mechanisms driving disease progression in both conditions.
RGCs reside within the retinal ganglion cell layer (GCL) that are particularly sensitive to neurodegenerative injury attributed to mitochondrial dysfunction, impaired axonal transport, oxidative stress and energy depletion. The progressive loss of RGCs and their axons can result in visual field defects and contribute to the advancement of AMD. In our study, we found that RGCs in AMD have an association with neurodegenerative diseases like PD, HD and AD. Notably, previous studies indicated that the contralateral eye, corresponding to the side of the body most affected in PD patients, exhibits more pronounced RGC loss21. HD is characterized by chorea movements and cognitive impairments in young adults. HD also demonstrates thinning of the retinal nerve fiber layer (RNFL) in the temporal region, which correlates with disease duration. In 1986, Hinton provided the initial compelling evidence linking AD to optic neuropathy22. Subsequently, a meta-analysis involving 887 AD patients and 864 controls examined peripapillary RNFL thickness, revealing significantly thinner RNFL measurements in AD patients compared to controls23.
In this study, we concentrated on the underlying mechanisms between astrocytes and OPCs among AMD and PD. Astrocytes, a subtype of glial cells, play a crucial role in the formation and maintenance of the blood-brain barrier (BBB) and blood-retinal barrier (BRB). These barriers protect the delicate neural tissue of the brain and retina by regulating the molecules exchange between the blood and the neural environment. In PD, the involvement of brain astrocytes in the pathophysiology of the disease has been extensively studied. Astrocytes in the substantia nigra, exhibit reactive changes and contribute to neuroinflammation through the release of inflammatory cytokines such as tumor necrosis factor-alpha (TNF-α), interleukin-1 beta (IL-1β) and interleukin-6 (IL-6)24. These inflammatory responses can exacerbate neuronal damage and contribute to the progression of PD. Furthermore, inhibition of NADPH oxidase 4 (NOX4) has been shown to reduce neuroinflammation in PD animal models by decreasing the expression of astrocytic lipocalin-225. Additionally, abnormal mitochondrial function has been implicated in PD pathogenesis, and targeting the Mfn2-cGAS signaling pathway to restore mitochondrial function delays astrocyte senescence in a mouse model of PD26. In contrast, the role of astrocytes has received less attention, with most studies focusing on the outer retina dysfunction in AMD. However, emerging evidence suggests that astrocytes may play a significant role in AMD pathology. Extensive retinal ischemia, a characteristic feature of advanced AMD, can lead to the loss of astrocytes and subsequent RGCs death27. Our study built on these findings by highlighting the potential for astrocytic changes to impact both neuroinflammation and retinal cell death in AMD and PD. OPCs are a unique and heterogeneous group of glial cells and responsible for forming and maintaining myelin sheaths, which insulate axons and facilitate rapid signal transmission. OPCs have been observed in the retina during the development of AMD, particularly in the RPE layer. Recent studies have indicated that OPCs may undergo reactive changes and contribute to the remodeling of the RPE-photoreceptor interface in response to AMD-related damage28. On the other hand, OPCs have been investigated for their role in promoting remyelination and restoring proper neuronal function in PD29. The high expression of LRRK2 gene in OPC is specifically recognized in PD30. However, research on the involvement of OPCs in AMD and PD is still in its early stages.
The potential relationship between astrocytes and OPCs has become a focal point. OPCs support axonal integrity by providing metabolic and nutritional support. Meanwhile, it was observed that the development of astrocyte endfoot coincided with OPCs migration31. A comprehensive cell-cell chat communication analysis was conducted to uncover the underlying signaling pathways mediating the communication between astrocytes and OPCs. We focused on ligand-receptor interactions (NRG1-ERBB4) and revealed that the neuregulin (NRG) signaling pathway emerged as the most critical route, exhibiting stronger interactions between astrocytes and OPCs. This intriguing result aligns with the pathogenesis of AMD and PD, suggesting that glial cell may act as key sources in modulating other cells behavior among these diseases. In neurodegenerative diseases like AMD and PD, the NRG1-ERBB4 signaling axis is tremendously important in maintaining neuronal health and function. NRG1 is a member of the neuro-modulatory protein family that plays a crucial role in regulating various physiological processes, including regeneration, inflammation, fibrosis and metabolism32,33. ERBB4 is one of the receptor tyrosine kinases, which is intimately involved in essential cellular processes such as cell differentiation, proliferation and survival34. Notably, NRG1-ERBB4 signaling axis could regulate the release of gamma-aminobutyric acid (GABA), contributing to the homeostatic balance. Studies have shown that NRG1β1 may protect nigrostriatal dopaminergic neurons in the 6-hydroxydopamine (6-OHDA) PD mouse model35. In addition, inhibition of clathrin-mediated endocytosis has been found to prevent amyloid-β-induced axonal damage, with inhibitors of adaptor-associated kinase 1 (AAK1) playing a key role in regulating this process36. Although the specific role of the NRG1-ERBB4 signaling pathway in AMD has not been adequately explored, elevated levels of NRG1 may offer protection against AMD progression by modulating inflammatory responses and promoting cell survival.
Furthermore, our findings elaborated that the PALLD, ZMZ1, FYN genes as the key components of a common regulatory pathway between AMD and PD development, which was demonstrated the reliability within the aid of WGCNA. This new mechanism surpasses the existing understanding of the immune cell axis and presents a more comprehensive perspective. It emphasizes the complex balance of cellular processes, and paves the way for improved clinical management, early intervention, and the development of novel therapeutic strategies targeting shared pathological pathways. PALLD was located in ME-salmon, which was defined as “Gene Dysregulation”. PALLD is a key cytoskeletal protein involved in maintaining the structural integrity of cells and regulating cell shape, adhesion and motility37. Dysregulation of gene expression can have profound effects on cell function and contribute to the development of various diseases, including AMD and PD. Although there is limited direct research on the relationship between PALLD, AMD and PD, it is possible to explore potential links based on known functions and pathways. PALLD involved in key processes such as inflammation, oxidative stress response, angiogenesis and protein aggregation38, which indicate that it may significantly influence the pathogenesis of both AMD and PD. ME-blue may classify as “Mitochondrial Dysfunction”. ZMIZ1 is a transcriptional co-activator in ME-blue modules and it may lead to a syndromic neurodevelopmental disorder. ZMIZ1 regulated various genes expression involved in cell proliferation, differentiation and survival39. Furthermore, ME-yellow may represent “Neurodegenerative impairment”. FYN, a member of the SRC non-receptor tyrosine kinase family, is associated with various cancers, neurodegenerative and immune diseases. By inhibiting the GSK-3β/FYN dependent Nrf2 nuclear translocation, FYN may enhance cellular defense mechanisms and maintain cellular homeostasis in the retinal tissue affected by dry AMD40. Moreover, emerging evidence indicated that targeting the pathways mediated by FYN will be a novel therapeutic approach for PD treatment41. A previous report found that pre-administration of the FYN inhibitor saracatinib (AZD0530) may lead to a significant reduction in locomotor deficits in wild-type PD mice42. Further research is needed to thoroughly understand the underlying mechanisms and to translate these findings into clinical applications.
Methods
ScRNA-seq data acquisition and quality control
The scRNA-seq datasets (Homo spaiens) were obtained from the Gene Expression Omnibus (GEO) database, including GSE221042 (four retinas with dry AMD), GSE161045 (four brain tissues with PD). To exclude cells with potential technical biases or low quality, cells (nCount_RNA < 15000, 500 < nFeature_RNA < 2500, mitochondrial < 5%) were filtered for subsequent analyses, which were performed with the Seurat package (version 4.4.0) in R (version 4.3.2).
Cell clustering, annotation and comparison
To overcome batch effects and ensure data consistency, all samples were integrated with “harmony” function. Subsequently, SCT (Seurat’s Centered Log-Ratio) was utilized to normalize the all data, followed by principal component analysis (PCA) to filter out the 2000 most highly variable genes. Then t-SNE (t-Distributed Stochastic Neighbor Embedding) analysis was performed on the top 10 principal components. By employing Seurat package, the function FindAllMarkers (only.pos = TRUE, min.pct = 0.25, logfc.threshold = 0.25) was performed to identify marker genes. Major cell types were annotated by leveraging the “Cell Markers 2.0” database and literature-reported markers43,44. Finally, the total number and proportion of cells were calculated in the entire dataset and within each specific group.
Gene set enrichment analysis
Gene set enrichment analysis (GSEA) calculates enrichment scores for a given gene set, classified under various functional categories, in order to identify significant and consistent differences between various cell clusters. In this study, we performed fgsea (version 1.28.0) and the Benjamini-Hochberg (BH) method for multiple testing correction to control the false discovery rate (FDR). Notably, we conducted GSEA to analyze all genes across the identified cell clusters by using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database.
Differential gene expression and correlation analysis
The FindMarkers function was used to determine differentially expressed genes (DEGs) in disease conditions compared to controls, with the following parameters: only.pos = TRUE, min.pct = 0.25, and logfc.threshold = 0.25. For all comparisons, we used base = exp (1) (the default parameter for calculating fold change in DEG analysis implemented in Seurat v4.4.0) to maintain consistency throughout the manuscript. Genes exhibiting a fold change > 0.25 and an adjusted p-value < 0.05 were considered statistically significant. We further performed a comparative analysis of DEGs within the same cell clusters across two datasets. Venn diagrams and dot plots were utilized to visualize overlapping genes and distinct molecular signatures.
Cell–cell communication
To uncover the potential regulatory mechanisms between different cell types, “Cellchat” function (version 1.6.1) was used to analyze cell-to-cell communication. Next, we imported a receptor-ligand database called “CellChatDB.human” and focused on analyzing intercellular communication through “secretion signaling”. The communication probabilities of all ligand-receptor interactions associated with each signaling pathway calculated with circular plots. The core official functions were applied to performed the visualization.
Weighted gene co-expression network analysis (WGCNA)
GSE29801 dataset (Human Genome Microarray from 16 dry AMD) were also downloaded from GEO database. Gene with expression variances greater than the 75th percentile of the whole genome (18989 genes) were utilized to construct a hierarchical cluster tree. We detected outliers by the function cutreeStatic (cutHeight = 90000, minsize = 4). Then, soft thresholds were determined by optimizing the scale-free topology fitting metrics (aiming for a value > 0.8 or reaching a plateau) and modules clustering was performed regarding the dissimilarity of the topological overlap matrix (dissTOM). “MinModulesSize” parameter was set to 100 and clustering trees and module information were integrated according to differently colored branches. Finally, the module eigengenes (MEs) were calculated, and highly similar modules were merged based on their dissimilarity.
Construction of Co-Expression modules
The first principal component of the gene modules was analyzed by the function “signedKME ()” in the WGCNA software package45. The higher absolute value of MEs, the closer relationship between genes and their corresponding modules. The pairwise similarity of co-expression matrixes was determined by Pearson’s correlation coefficients. Followingly, highly correlated modules were identified by Pearson’s r and p-value (< 0.05). The interactions between the modules were visualized in multiple dimensions by three-dimensional ScatterPlot3D.
Functional enrichment analysis
Significant genes in the module were employed to reveal uncharted biological function among modules. With the aid of the DAVID (https://david.ncifcrf.gov/) database (count > 3, p-value < 0.05), we conducted Gene ontology (GO) function and KEGG analysis to define module characteristics.
Identification of signature genes
To identify potential hub genes, we calculated the soft-connectivity within the high association modules and selected the top 100 genes. Subsequently, a cross-sectional association analysis was performed with the core genes analyzed in the single-cell analysis (Sect. 2.4) and screened the key targets with the most co-effective regulatory power. The results were observed in an R-package Nightingale rose diagram.
Conclusions
In conclusion, this study provided valuable insights into the cellular and molecular dynamics underlying the AMD and PD pathogenesis (Fig. 11). We have specifically concentrated on the interactions between astrocytes and OPCs in the retinal environment. NRG signaling pathway has been identified as a crucial mediator of cell-cell communication, contributing to our understanding of disease mechanisms. Furthermore, significant correlations have been discovered between gene modules involved in “Gene dysregulation”, “Neurodegenerative impairment”, and “Mitochondrial dysfunction”, offering potential targets for more precise and effective therapeutic interventions. These findings pave the way for the development of tailored therapeutic regimens and hold promise for enhancing disease management in AMD and PD patients. Ongoing research in this field is expected to further advance our knowledge and improve patient outcomes.
Flow chart of the study. A comprehensive approach was employed by integrating single-cell RNA sequencing (scRNA-seq) datasets, along with Weighted Gene Co-expression Network Analysis (WGCNA)-related computational analysis. Gene Set Enrichment Analysis (GSEA) was conducted to analyze PD-related genes within retinal ganglion cells in dry AMD. Cell-cell chat was utilized to predict intercellular communication and signaling pathways. Module eigengenes (MEs) were calculated to identify specific gene modules.
Mapping the cellular landscape of dry AMD (GSE221042). (A) TSNE plots show different samples (left panel) and 12 different cell types in dry AMD (right panel). (B) A heatmap exhibit retinal cells clustering and annotation. Marker genes in OPCs and astrocytes were highlighted (dashed red square). (C) Bar plots show the distribution of cell type proportions in dry AMD.
KEGG enriched biological pathways by GSEA in two retinal ganglion cells clusters (GSE221042). (A) Left, representative enriched biological pathway in KEGG in the retinal ganglion cells (1). Right, GSEA enrichment plot of the PD pathway. (B) Left, representative enriched biological pathway in KEGG in the retinal ganglion cells (2). Right, GSEA enrichment plot of the PD pathway. (C) The Venn diagrams show common genes in the two retinal ganglion cells clusters. All displayed pathways had p-value < 0.05.
Detailed cell atlas and gene expression in PD (GSE161045). (A) TSNE plots show different samples (left panel) and 9 different cell types in PD (right panel). (B) A heatmap demonstrate top three marker genes in each cluster in PD. The marker genes in OPCs and astrocytes are highlighted (dashed red square). (C) Bar plots show the distribution of cell type proportions in PD.
Unveiling common core genes in OPCs and astrocytes across both datasets. (A) The Venn diagrams demonstrate 22 highly expressed genes in both OPCs and astrocytes within AMD and PD. (B) The dot plots show 22 identical target genes in dry AMD (GSE221042). PALLD, LHFPL6, RPS6KA2 and ZMIZ1 are most commonly expressed in astrocytes and OPCs. (C) The dot plots show 22 hub genes in PD (GSE161045). NTRK2, PALLD, ATP2B4 and ZMIZ1 have greatest expression in astrocytes and OPCs in the PD dataset.
Intercellular communication among OPCs and astrocytes in GSE221042. (A) Outgoing and incoming signal strength of each signaling pathway in each cell population in PDR. (B) Heatmap of the NRG signaling pathway–mediated intercellular communication intensity. (C) Circle plot of intercellular communication network associated with the NRG signaling pathway in dry AMD. The thickness of the line and color represents the communication signal strength. (D) Heatmap of the sender, receiver, mediator and influencer of NRG signaling pathway network. (E) Expression level of the NRG signaling ligand and receptor (NRG1-ERBB4) in each cell population.
Cell–cell communication inferred by NRG signaling pathway in PD (GSE161045). (A) Outgoing and incoming signal strength of each signaling pathway in each cell population in PD. (B) Heatmap of the NRG signaling pathway–mediated intercellular communication intensity. (C) Circle plot of intercellular communication network associated with the NRG signaling pathway in PD. (D) Heatmap of the sender, receiver, mediator and influencer of NRG signaling pathway network. (E) Expression level of the NRG signaling ligand and receptor (NRG1-ERBB4) in each cell clusters.
Pivotal modules calculated by Weighted Gene Co-Expression Network Analysis with transcriptome sequencing data. (A) Analysis of network topology for powers of soft threshold. (B) Highly-relevant gene modules in accordance with the value of Module Eigengene (cur-off height = 0.2). (C) Gene clustering dendrogram contains modules hierarchical clustering and heatmap. (D) Pearson’s r and p-value between modules (all p < 0.05, R2 = 0.64,0.72 between ME-salmon, ME-yellow and ME-blue). (E) Geometric interpretation of gene expression in 3D scatter plot. (F) The Venn diagram show the relevancy genes between ME-salmon, yellow and blue modules.
Integrative approach identifies core genes influencing AMD development. (A) Top 100 hub genes in ME-salmon modules ranked by Soft-connectivity. (B) Top 100 hub genes in ME-yellow modules ranked by Soft-connectivity. (C) Top 100 hub genes in ME-blue modules ranked by Soft-connectivity. The hub genes in the three modules are integrated with the core targets identified in single-cell analyses. PALLD, FYN and ZMIZ1 are filtered out as three key genes.
Cellular and molecular mechanisms involved in the pathogenesis of AMD and PD. We focused on the interactions between astrocytes and OPC, which were common cell types in retina and brain tissues, and highlighted the critical role of the NRG signaling pathway in mediating cell-cell communication. It also illustrates significant correlations between gene modules associated with “Gene dysregulation,” “Neurodegenerative impairment,” and “Mitochondrial dysfunction,” suggesting potential targets for more precise therapeutic interventions. These findings provide a foundation for developing tailored treatment strategies aimed at improving disease management in AMD and PD.
Data availability
All data generated or analysed during this study are included in this published article and its supplementary information files.
References
Fleckenstein, M., Schmitz-Valckenberg, S. & Chakravarthy, U. Age-Related Macular Degeneration: Rev. JAMA. 331(2), 147–157 (2024).
Lee, K. S., Lin, S., Copland, D. A., Dick, A. D. & Liu, J. Cellular senescence in the aging retina and developments of senotherapies for Age-Related macular degeneration. J. Neuroinflammation. 18 (1), 32 (2021).
Sun, M. et al. Role of flavonoids in Age-Related macular degeneration. Biomed. Pharmacother. 159, 114259 (2023).
Cinar, E., Tel, B. C. & Sahin, G. Neuroinflammation in Parkinson’s disease and its treatment opportunities. Balkan Med. J. 39 (5), 318–333 (2022).
Mao, Q., Qin, W. Z., Zhang, A. & Ye, N. Recent advances in dopaminergic strategies for the treatment of Parkinson’s disease. Acta Pharmacol. Sin. 41 (4), 471–482 (2020).
Zaman, V. et al. Cellular and molecular pathophysiology in the progression of Parkinson’s disease. Metab. Brain Dis. 36 (5), 815–827 (2021).
Chen, P. J. et al. Increased risk of Parkinson’s disease among patients with Age-Related macular degeneration. BMC Ophthalmol. 21 (1), 426 (2021).
Yoon, B., Sa, H. S. & Kim, H. J. Incidence and risk factors of Age-Related macular degeneration in patients with Parkinson’s disease: A Population-Based study. Front. Aging Neurosci. 16, 1331786 (2024).
Wang, L. & Mao, X. Role of retinal Amyloid-Beta in neurodegenerative diseases: overlapping mechanisms and emerging clinical applications. Int. J. Mol. Sci. 22(5), 2360–2384 (2021).
Jabbehdari, S. et al. Age-Related macular degeneration and neurodegenerative disorders: shared pathways in complex interactions. Surv. Ophthalmol. 69 (3), 303–310 (2024).
Abokyi, S., To, C. H., Lam, T. T. & Tse, D. Y. Central Role of Oxidative Stress in Age-Related Macular Degeneration: Evidence from a Review of the Molecular Mechanisms and Animal Models. Oxid. Med. Cell. Longev. 7901270 (2020).
Heidari, A., Yazdanpanah, N. & Rezaei, N. The role of Toll-Like receptors and neuroinflammation in Parkinson’s disease. J. Neuroinflammation. 19 (1), 135 (2022).
Singh, S. S. et al. Nf-Kappab-Mediated neuroinflammation in Parkinson’s disease and potential therapeutic effect of polyphenols. Neurotox. Res. 37 (3), 491–507 (2020).
Donato, L. et al. Bridging Retinal and Cerebral Neurodegeneration: A Focus on Crosslinks Between Alzheimer-Perusini’s Disease and Retinal Dystrophies. Biomedicines. 11, (2023).
Pogue, A. I. & Lukiw, W. J. Up-Regulated Pro-Inflammatory Micrornas (Mirnas) in Alzheimer’s disease (Ad) and Age-Related macular degeneration (Amd). Cell. Mol. Neurobiol. 38, 1021–1031 (2018).
Franco, R., Garrigos, C. & Lillo, J. The Olfactory Trail of Neurodegenerative Diseases. Cells. 13(7), 615–630 (2024).
Dan, X. et al. Olfactory dysfunction in aging and neurodegenerative diseases. Ageing Res. Rev. 70, 101416 (2021).
Savitt, J. & Aouchiche, R. Management of visual dysfunction in patients with Parkinson’s disease. J. Parkinsons Dis. 10 (s1), S49–S56 (2020).
Kwan, S., Atiya, A., Hussaindeen, J. R., Praveen, S. & Ambika, S. Ocular features of patients with Parkinson’s disease examined at a Neuro-Optometry clinic in a tertiary eye care center. Indian J. Ophthalmol. 70 (3), 958–961 (2022).
Mailankody, P., Lenka, A. & Pal, P. K. The role of optical coherence tomography in parkinsonism: A critical review. J. Neurol. Sci. 403, 67–74 (2019).
La Morgia, C., Di Vito, L., Carelli, V. & Carbonelli, M. Patterns of retinal ganglion cell damage in neurodegenerative disorders: parvocellular vs magnocellular degeneration in optical coherence tomography studies. Front. Neurol. 8, 710 (2017).
Hinton, D. R., Sadun, A. A., Blanks, J. C. & Miller, C. A. Optic-Nerve degeneration in Alzheimer’s disease. N Engl. J. Med. 315 (8), 485–487 (1986).
den Haan, J., Verbraak, F. D., Visser, P. J. & Bouwman, F. H. Retinal thickness in Alzheimer’s disease: A systematic review and Meta-Analysis. Amst. 6, 162–170 (2017).
Araujo, B. et al. Neuroinflammation and Parkinson’s Disease-From Neurodegeneration to Therapeutic Opportunities. Cells. 11(18), 2908–2947 (2022).
Lin, Z. et al. Nox4 Exacerbates Parkinson’s Disease Pathology by Promoting Neuronal Ferroptosis and Neuroinflammation. Neural Regen. Res. 20(7), 2038–2052 (2025).
M, W. et al. Metformin normalizes mitochondrial function to delay astrocyte senescence in a mouse model of Parkinson’s disease through Mfn2-Cgas signaling. J. Neuroinflammation. 21 (1), 81 (2024).
X, Y. et al. A novel function and mechanism of Ischemia-Induced retinal Astrocyte-Derived exosomes for Rgc apoptosis of ischemic retinopathy. Mol. Ther. Nucleic Acids. 35, 102209 (2024).
S, N. et al. Activation of Shh/Smo is sufficient to maintain oligodendrocyte precursor cells in an undifferentiated state and is not necessary for Myelin formation and (Re)Myelination. Glia. 72 (8), 1469–1483 (2024).
J, S. et al. Oxidative stress and impaired oligodendrocyte precursor cell differentiation in neurological disorders. CMLS. 78 (10), 4615–4637 (2021).
Agarwal, D. et al. A Single-Cell atlas of the human substantia Nigra reveals Cell-Specific pathways associated with neurological disorders. Nat. Commun. 11 (1), 4183 (2020).
Duncan, G. J. & Emery, B. End of the road: astrocyte endfeet regulate Opc migration and myelination. Neuron. 111, 139–141 (2023).
De Keulenaer, G. W. et al. Mechanisms of the multitasking endothelial protein Nrg-1 as a compensatory factor during chronic heart failure. Circ. Heart Fail. 12(10), 6288–6302 (2019).
Ma, Y. et al. Neuregulin-1 regulates the conversion of M1/M2 microglia phenotype via Erbb4-Dependent Inhibition of the Nf-Kappab pathway. Mol. Biol. Rep. 49 (5), 3975–3986 (2022).
Iwakura, Y. & Nawa, H. Erbb1-4-Dependent Egf/Neuregulin signals and their cross talk in the central nervous system: pathological implications in schizophrenia and Parkinson’s disease. Front. Cell. Neurosci. 7, 4 (2013).
Depboylu, C., Rosler, T. W., de Andrade, A., Oertel, W. H. & Hoglinger, G. U. Systemically administered Neuregulin-1Beta1 rescues nigral dopaminergic neurons via the Erbb4 receptor tyrosine kinase in Mptp mouse models of Parkinson’s disease. J. Neurochem. 133 (4), 590–597 (2015).
Abdel-Magid, A. F. Inhibitors of Adaptor-Associated kinase 1 (Aak1) May treat neuropathic pain, schizophrenia, Parkinson’s disease, and other disorders. ACS Med. Chem. Lett. 8 (6), 595–597 (2017).
Zhang, X. et al. Palladin is a novel Microtubule-Associated protein responsible for spindle orientation. Sci. Rep. 7 (1), 11806 (2017).
Mastrototaro, G. et al. Ablation of Palladin in Adult Heart Causes Dilated Cardiomyopathy Associated with Intercalated Disc Abnormalities. eLife. 12, (2023).
K, C. R., Tiemroth, A. S., Thurmon, A. N., Meadows, S. M. & Galazo, M. J. Zmiz1 is a novel regulator of brain development associated with autism and intellectual disability. Front. Psychiatry. 15, 1375492 (2024).
X, Z. et al. Melatonin protects photoreceptor cells against ferroptosis in dry Amd disorder by inhibiting Gsk-3B/Fyn-Dependent Nrf2 nuclear translocation. Biochim. Biophys. Acta Mol. Basis Dis. 1870 (2), 166969 (2024).
Angelopoulou, E., Paudel, Y. N., Julian, T., Shaikh, M. F. & Piperi, C. Pivotal role of Fyn kinase in Parkinson’s disease and Levodopa-Induced dyskinesia: A novel therapeutic target?? Mol. Neurobiol. 58 (4), 1372–1391 (2021).
Sanz-Blasco, S. et al. The kinase Fyn as a novel intermediate in L-Dopa-Induced dyskinesia in Parkinson’s disease. Mol. Neurobiol.55 (6), 5125–5136 (2018).
J, X. et al. Human striatal glia differentially contribute to Ad- and Pd-Specific neurodegeneration. Nat. Aging. 3, 346–365 (2023).
M, K. et al. Single-Cell analysis reveals inflammatory interactions driving macular degeneration. Nat. Commun.14, 2589 (2023).
Y, J. et al. Metabolic reprogramming and macrophage expansion define Acpa-Negative rheumatoid arthritis: insights from Single-Cell Rna sequencing. Front. Immunol. 15, 1512483 (2024).
Acknowledgements
This is a short text to acknowledge the contributions of GSE221042, GSE161045 and GSE29801 original data.
Funding
This work was supported by National Key Research and Development Program of China: (Grant number: 2019YFC1711605).
Author information
Authors and Affiliations
Contributions
SYZ was a major contributor in writing the manuscript and original draft preparation; CZ reviewed the manuscript; YJZ, YBF reviewed and offered project administration. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, S., Zhang, C., Zhang, Y. et al. Unraveling the role of neuregulin-mediated astrocytes-OPCs axis in the pathogenesis of age-related macular degeneration and Parkinson’s disease. Sci Rep 15, 7352 (2025). https://doi.org/10.1038/s41598-025-92103-8
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-92103-8