Abstract
Alport syndrome is a common monogenic kidney disease resulting from pathogenic variants in COL4A3, COL4A4 or COL4A5 genes. The estimated global population prevalence is one in 106 individuals for autosomal dominant (AD) and one in 2,320 for sex-linked (XL) conditions. Here, we aimed to estimate the population prevalence of individuals carrying pathogenic variants that cause Alport syndrome in Singapore, and to stratify the prevalence by ancestry. We used population-scale genomic data of 9,051 unrelated subjects, comprising 5,443 (60.8%) Chinese, 1,922 (21.4%) Indian and 1,686 (17.8%) Malay individuals. The prevalence of individuals with pathogenic variants that cause AD and XL Alport syndrome are 1 in 165 and 1 in 2,262 respectively. Additionally, 0.8% of Chinese and 0.3% of Malay populations carry pathogenic Alport syndrome variants, with Chinese individuals being 2.7 times more affected than Malays (95% CI:1.147–6.437, P = 0.027). Interestingly, each pathogenic variant was associated with people of a single ancestry. The two most prevalent pathogenic variants, COL4A3: c.3856G > A (p.Gly1286Arg) (n = 8) and COL4A3: c.4793T > G (p.Leu1598Arg) (n = 4), were exclusively found in the Chinese population. In conclusion, AD Alport syndrome may be prevalent in Singapore, with higher frequencies among the Chinese. Furthermore, founder effects may exist within the ancestries.
Similar content being viewed by others
Introduction
Alport syndrome (AS) is a monogenic kidney disease due to defects in COL4A3, COL4A4 and COL4A5 genes which encode the α3, α4 and α5 chains of collagen IV respectively1. Autosomal dominant (AD) Alport syndrome is caused by a heterozygous variant in either COL4A3 or COL4A4, while autosomal recessive (AR) disease is caused by two pathogenic variants (in trans) in either COL4A3 or COL4A4. Sex-linked (XL) Alport syndrome occurs if there is a defect in the COL4A5 gene. Digenic Alport syndrome is due to defects in two of the three genes (COL4A3, COL4A4, COL4A5), either in cis or trans1.
The α3, α4 and α5 chains of collagen IV intertwine to form triple helices that assemble to a network in the basement membranes of the glomeruli, cochlea, retina and lens2. Genetic variants resulting in amino acid changes that distort this trimerization can affect the assembly of collagen IV networks, leading to abnormal basement membranes. Glycine, being the smallest amino acid and occurring at the first position of the Gly-Xaa-Yaa repeats in the intermediate domains, allows for tight packing of the collagen triple helix3. In Alport syndrome, replacing glycine with a larger amino acid can disturb the α3α4α5 helix formation and affect the collagen networks in the basement membranes4. This explains the possible multiorgan involvement in Alport syndrome, namely glomerular disease, sensorineural hearing loss, lens and retinal abnormalities1.
Common kidney phenotypes in Alport syndrome include persistent glomerular hematuria, proteinuria, and progressive kidney failure5. Histological findings include focal and segmental glomerulosclerosis (FSGS) and coincidental IgA nephropathy6,7. Kidney cysts can also occur8. The trajectory of kidney disease depends on the mode of inheritance as well as epigenetic and environmental factors, modifying genes and comorbidities. Individuals with AR disease and males with XL disease have the most severe phenotypes, while those with AD condition and females with XL disease have a milder disease trajectory9,10.
Traditionally known as a rare genetic disorder, the prevalence rates of Alport syndrome were previously reported to range from one in 5000 to one in 53,00011,12,13. However, a recent study based on the Genome Aggregation Database (gnomAD) of more than 122,000 subjects showed that one in 106 (0.9%) individuals have a heterozygous variant in either COL4A3 or COL4A4 gene that is predicted to be pathogenic and can cause AD Alport syndrome14. These variants were associated with hematuria in the United Kingdom 100,000 Genomes Project. Additionally, in that study, one in 2,320 individuals had a pathogenic variant predicted to cause XL Alport syndrome. These findings corroborated with the subsequent Geisinger MyCode/ DiscovEHR study which was based on an unselected health system-based cohort of 174,418 participants15. In the latter study, 1 in 433 (0.23%) individuals had a heterozygous COL4A3 variant that was predicted to be pathogenic. The true population prevalence of Alport syndrome may be higher than those suggested in these two studies because individuals with diagnosed disease were not included in the cohorts. Moreover, large structural genetic rearrangements and deep intronic splicing variants could not be detected with the genomic testing techniques used14.
These population cohort studies had an under-representation of Asians. Although Asians represent nearly 60% of the global population, Asian genomes comprise only 6.6% of the gnomAD data and 3% of population health studies16,17. Furthermore, Asian-centric genomic databases such as GenomeAsia 100k were only created recently and have not been fully recruited yet18. Minor allele frequencies (MAF) of Alport gene variants are known to vary among populations. Pathogenic COL4A5 variants are more common in European or African populations, while heterozygous pathogenic COL4A3 and COL4A4 variants are at least four times more common in East Asian ancestries compared with Ashkenazim and Finns14.
Due to historical migration patterns, the genetic ancestry of Singapore’s multi-ethnic population has strong correlations with Asian populations in different geographical locations19. Hence, the study of Singapore population genomics can reflect the human genetic diversity across Asia. In this study, we aimed to estimate the population prevalence of individuals carrying pathogenic variants that can cause Alport syndrome, using a population-scale genomic database of 10,323 Singaporeans20. In addition, we aimed to stratify the population prevalence by genetically-inferred ancestries to provide insights into the ancestry-specific variant landscape of Alport syndrome in Asia19.
Results
Genetic variants
The SG10K study dataset includes 9,770 genomes after quality control. Initial analysis revealed a total of 28,968 variants in the COL4A3, COL4A4 and COL4A5 genes (9,012 for COL4A3, 9,777 for COL4A4 and 10,179 for COL4A5). Most variants were intronic (97.3% for COL4A3, 94.2% for COL4A4, and 97.8% for COL4A5) (Figure S1).
These 28,968 unique variants consisted of single nucleotide variants (SNVs), indels, and others (Table S1). The SNVs were predominantly non-synonymous missense variants, followed by synonymous missense changes. Notably, there was one and four SNVs in COL4A3 and COL4A4 respectively that led to gain of premature stop codons; and one SNV in COL4A4 that resulted in loss of the stop codon. Moreover, four SNVs in COL4A3 and one SNV in COL4A4 were located at canonical splice sites (+/- 1 or 2). There were no SNVs that affected stop codons or canonical splice sites identified in COL4A5 (Table S1).
Indel variant types included frameshift or inframe insertions and deletions (Table S1). These include one variant in COL4A3 that resulted in a frameshift insertion, three variants in COL4A3 and four variants in COL4A4 that caused frameshift deletions, one variant in COL4A4 that resulted in an inframe insertion, and six variants that caused inframe deletions (four in COL4A3, one in COL4A4 and one in COL4A5). Two indel variants in COL4A3 gene were identified at canonical splice sites while such variants were absent in the COL4A4 and COL4A5 genes (Table S1).
All synonymous and intronic non-splicing variants were filtered out as part of the variant curation steps.
Pathogenic COL4A3, COL4A4 and COL4A5 variants
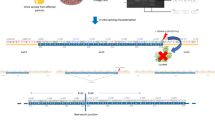
There were 44 pathogenic or likely pathogenic variants (Table 1 and S1) in all three genes. Of these, 19 (43%) were COL4A3, 22 (50%) were COL4A4 and 3 (7%) were COL4A5 (Fig. 1).
COL4A3, COL4A4 and COL4A5 variants curation flowchart and pathogenic variants distribution. Whole-genome sequencing (WGS) was performed on a total of 10,323 healthy samples, of which, 9,770 genomes passed quality control. After removal of related family members up to second-degree, 9,051 individuals consisting of 61% Chinese, 21% Indian and 18% Malay remained. Variant curation revealed 44 (likely) pathogenic variants and 6 unique variants of uncertain significance with high pathogenicity scores. Among the 9,051 individuals tested, 55 individuals had heterozygous pathogenic variants in COL4A3 or COL4A4 suggesting AD Alport syndrome, while 4 individuals (3 females, 1 male) carried a single copy of pathogenic variant in COL4A5 suggesting XL Alport syndrome. Comparing across the three ethnic groups in Singapore, the prevalence of Alport syndrome pathogenic variants among the Chinese population was 2.7 times (95% CI: 1.1–6.4) higher than that in the Malay population (P = 0.027, Fisher’s exact test). There was no statistical difference in the prevalence rates between the Chinese and Indian populations, and between the Indian and Malay populations (P > 0.05).
Non-synonymous missense variants predominated, accounting for 52% (23 out of 44) of all pathogenic variants. Almost all (22 out of 23) non-synonymous missense pathogenic variants cause substitutions of glycine occurring at the first position of the Gly-Xaa-Yaa repeats in the intermediate domains. The remaining variant (COL4A3: c.4793T > G (p.Leu1598Arg)) led to the substitution of a hydrophobic leucine with a hydrophilic arginine.
All five identified SNVs that cause gain of a premature stop codon were pathogenic. Similarly, 7 out of 8 identified frameshift deletions and insertions were pathogenic. In contrast, only 2 out of 7 inframe insertions or deletions were pathogenic. Of the 11 splicing pathogenic variants, 7 occurred at canonical splice sites, 3 were non-synonymous missense variants, while the remaining one was a 9 bp deletion (Table S1).
Population prevalence of individuals who carry pathogenic variants for Alport syndrome
Variants predicted to be pathogenic in COL4A3, COL4A4 and COL4A5 genes were found in 60 out of 9,051 individuals, indicating an estimated overall population frequency of one in 150.
Of these 60 individuals, 55 (92%) had a single heterozygous pathogenic variant that can cause AD Alport syndrome. This included the COL4A3 gene for which 17 pathogenic variants were present in 27 individuals; and the COL4A4 gene for which 22 heterozygous pathogenic variants were present in 28 individuals. Hence, the estimated prevalence of individuals carrying variants that can cause AD Alport syndrome is one in 165 (55 out of 9,051 individuals) (Fig. 1), which is slightly less than previously reported prevalence of 1 in 106 based on the gnomAD database14.
We identified 4 individuals with pathogenic variants that can cause XL Alport syndrome (7% of 60 individuals). These four individuals, namely one Indian hemizygote and three heterozygous females (two Chinese, and one Malay) carried three distinct pathogenic variants in the COL4A5 gene. The estimated population frequency for individuals who carry variants which can cause XL Alport syndrome is therefore one in 2,262 (4 out of 9,051 individuals) (Fig. 1), similar to the previously reported prevalence of one in 2,320 (Fisher’s exact test, P= 0.7966)14.
Additionally, we identified one individual (Malay male) who carried two heterozygous variants in COL4A3, namely c.4253-4_4258delGAAGGACCAG (p.Gly1418_Pro1419del) and c.4258_4259insTTTTTTTTTT (p.Ala1420Valfs*93)). Although it could not be determined if the two variants were in cis or in trans, we presumptively assigned this subject as autosomal recessive Alport syndrome. No individuals with digenic forms of Alport syndrome were found.
Ethnic distribution of pathogenic COL4A3, COL4A4 and COL4A5 variants
When stratified by ancestry, the prevalence rates for Chinese, Indian and Malay populations were 0.808% (44 in 5,443), 0.572% (11 in 1,922) and 0.297% (5 in 1,686) respectively (Fig. 1). The prevalence rate among the Chinese population was 2.7 times (95% CI: 1.147–6.437) higher than for the Malay population (P = 0.027, Fisher’s exact test). Conversely, there was no significant difference between the Chinese and Indian populations, or between the Indian and Malay populations (P = 0.451 and 0.223, respectively, Fisher’s exact test).
It is notable that each pathogenic variant occurred exclusively within a specific ethnic group, apart from the variant COL4A4: c.1323_1340del18 (p.Pro444_Leu449delProGlyAlaProGlyLeu) which appeared twice in the Chinese population and once in the Indian population, and the variant COL4A5: c.2119G > C (p.Gly707Arg) which affected one Indian individual and one Malay individual. The two most prevalent pathogenic variants, COL4A3: c.3856G > A (p.Gly1286Arg) identified in eight individuals, and COL4A3: c.4793T > G (p.Leu1598Arg) identified in four individuals, were only found in the Chinese population.
Variants of uncertain significance with high pathogenicity scores
Five variants in COL4A3 and one variant in COL4A4 each have a total pathogenicity score of 5 (Table 2). Among these six VUS, three were missense SNVs that caused substitutions of glycine at position 1 of the Gly-Xaa-Yaa repeats while the other two were inframe deletions in COL4A3. The missense SNVs remained as VUS primarily because the evidence for in silico tool prediction (PP3) were only of moderate strength (REVEL scores ranging from 0.773 to 0.932)52. Additional criteria will be needed to reclassify these variants53,54. For example, further genotyping or phenotyping of the proband or family members; or emergence of newly published cases in the future may allow for addition of PS4, PP1, PP4 or PM5 criteria.
Minor allele frequencies of pathogenic COL4A3, COL4A4 and COL4A5 variants
The MAF for each pathogenic variant are shown in Table 1 and S2. The commonest pathogenic variant is COL4A3: c.3856G > A (p.Gly1286Arg) which affects eight Chinese individuals and are present in 13% of the individuals carrying Alport gene alterations (8 of 60 subjects). This variant has a MAF of 0.00045 (8 out of 17,752 alleles) in our cohort, and this is higher than those reported in the gnomAD database: 0.00025 (11 out of 44,882 alleles) in East Asians and 2.542 × 10−6 (3 out of 1,179,994 alleles) in non-Finnish Europeans. This variant is absent in other ancestral groups of SG10K project.
Discussion
We have found that in Singapore, the prevalence of individuals carrying pathogenic variants that can cause AD Alport syndrome to be one in 165, which is slightly less than previously reported prevalence of 1 in 106 based on the gnomAD database14. To our knowledge, this is the first study that analyzed ethnic-specific prevalence in an Asian population. Our data also suggested that in Singapore, individuals carrying pathogenic variants for Alport syndrome are more common among the Chinese compared to the Malay population.
Located in the middle of Southeast Asia, Singapore’s population stemmed from immigrants from China, India and other parts of Asia over the past two centuries61. Because of this, Singapore’s current population of five million comprises of three major ethnic groups: Chinese (74.2%), Malay (13.7%), Indian (8.9%) which are of East Asian, Southeast Asian and South Asian ancestries respectively19. Our findings provide some insights into the variant landscape of Alport syndrome in Asia and could suggest that Alport syndrome may be more prevalent among Asian populations of Chinese descent compared to those of Malay descent. Further studies are needed to confirm these. Interestingly, our results contradicted the local observation that the prevalence of chronic kidney disease is higher among the Malay population compared to the other ethnic groups62. This may be explained by lifestyle and other environmental factors, gene-gene interactions or polygenic causes.
Our study was based on the SG10K_Health population-scale genomic database20which provides a rich resource to survey the population prevalence of genetic conditions. While our predicted population prevalence appears to be comparable to other population-based studies on Alport syndrome performed in global and Caucasian populations14,15, it is notable that the variant curation criteria differ widely among these studies. This is especially pertinent in variants that result in substitution of position 1 glycine in genes that encode collagen type IV. While there have been recommendations that such variants should be considered a “hot spot” for mutations according to the ACMG/AMP criteria for pathogenicity63, these have not been clearly defined in the newer guidelines released by the Clinical Genome (ClinGen) expert panels54. The impact is not miniscule since nearly half of the non-synonymous missense pathogenic variants identified in our study result in substitution of position 1 glycine (22 out of 55 variants).
The criteria used in our study were relatively more stringent compared to other similar population-based studies. Specifically, we converted the ACMG/AMP evidences into calculated odds of pathogenicity64and determined the variant classification using a Bayesian framework65in accordance to the ClinGen expert panels guidelines54. In contrast, Gibson et al. classified all variants that resulted in the substitution of position 1 glycine as “predicted pathogenic”14 while Solanki et al. used Varsome (www.varsome.com) to predict pathogenicity and classified variants as pathogenic if they have been reported in ClinVar as likely pathogenic or pathogenic at least once, regardless of the level of review supporting the variant classification14. Because of these differences, four variants that result in position 1 glycine substitutions in COL4A3 or COL4A4 genes remained as VUS with high pathogenicity scores in our study. Yet, they would have been categorized as “predicted pathogenic” if the criteria described by Gibson et al.14 was applied. This difference in variant curation criteria may partly explain the slightly lower prevalence in our study compared to other population-based studies, and clearly illustrates a real challenge in genomics medicine today.
A key limitation in our study is the lack of access to clinical or family details of the subjects in SG10K database. This limitation is partly ameliorated by our criteria for variant curation which were based on publicly available data such as GnomAD and ClinVar databases; clinical details of published cases in the literature as well as variant-specific details such as REVEL scores and published functional studies. Variants were classified as pathogenic if they achieved the pathogenicity score cut-offs without phenotype or family details specific to the SG10K subject. Nevertheless, we recognize some variants may be mis-classified especially if there are undetected evidence in the SG10K subjects that may suggest a benign tendency, such as lack of segregation in affected family members. Separately, we have described the VUS with high pathogenicity scores. These may be upgraded to likely pathogenic with added evidence from the clinical or family details of the subject in SG10K. These VUS were not included in the determination of population prevalence. Subsequent Phase II and III of the National Precision Medicine initiative in Singapore will include 100,000 genomes, and access to data in the electronic medical records of the study subjects66. Future analysis using this larger, more comprehensive dataset will provide better insights into disease trajectory and the population prevalence of Alport syndrome in Singapore.
We have also described the segregation of most variants in the COL4A3–COL4A5 genes into specific ethnic groups. The two most prevalent pathogenic variants, COL4A3: c.3856G > A (p.Gly1286Arg) and COL4A3: c.4793T > G (p.Leu1598Arg), were exclusively present in the Chinese population, suggesting a possible founder effect. Notably, the variant COL4A3: c.3856G > A (p.Gly1286Arg) has a higher MAF among the Chinese in the SG10K cohort compared to those in the overall gnomAD cohort and its East Asian subgroup. In addition, there was a notable absence of COL4A5: c.1871G > A (p.Gly624Asp) in our cohort even though this variant represented almost half the pathogenic COL4A5 variants in the European population14. These exemplify the need for population-specific MAF databases for better interpretation of the genetic variants.
It may be argued that the prevalence of individuals with pathogenic variants may not equate the prevalence of the disease. This is because about 30% of individuals who harbor pathogenic variants in Alport syndrome do not manifest disease15,67 and hence the presence of pathogenic variants may not render the diagnosis of Alport syndrome. Additionally, age-dependent penetrance may occur as disease manifestations occur only later in life. Furthermore, the presumably non-penetrant individual may have an increased genetic predisposition to chronic kidney disease especially in the presence of other kidney insults like nephrotoxins, diabetes mellitus and hypertension. Health prevention strategies targeted at such an individual may therefore be justified. To this end, from a public health perspective, knowledge on the population prevalence of individuals carrying pathogenic variants of Alport syndrome can provide directions on health prevention strategies targeted to the genetically at-risk individuals.
The findings of this study provide insights on the potential disease burden in Singapore, and this is important for the nationwide clinical implementation of genetic testing in nephrology services. Genetic testing in glomerular disease has been shown to have high clinical utility in a systematic analysis68. Specifically in Alport syndrome which is the commonest monogenic glomerular disease68,69, 70% and 90% of male patients with XL Alport syndrome develop kidney failure by age 30 and 40 respectively without treatment70.
In those with AD Alport syndrome, 29% develop chronic kidney disease and 15% reached kidney failure by 53 years47. Early and aggressive treatment with renin-angiotensin-aldosterone system blockade may delay kidney failure by two decades or more71,72. The crux is that the earlier the treatment is started, the greater the impact on kidney survival71. This partly explains why genetic testing in glomerular diseases is cost-effective73. However, while clinical benefits are clear, genetic testing in nephrology has not been routinely performed in Singapore because of the limited availability of genetic testing, absence of clinical algorithms and lack of genetic counselling expertise. This results in lost opportunities to reduce the incidence of chronic kidney disease in Singapore, known to have one of the highest prevalence rates of kidney failure in the world74. A clinical implementation pilot project has been underway to provide genetic services within the nephrology service75. With the new understanding on the variant landscape and genetic burden of Alport syndrome in our study, it will direct the implementation efforts in various aspects such as resource allocation and genetic counselling.
There are several other limitations in this study. Firstly, the SG10K cohort comprises of a diverse group of individuals, including those in good health and others with known medical conditions. Participants with late-stage chronic kidney disease were likely not included and this may result in an under-estimation of the true population prevalence of Alport syndrome. Secondly, despite the ability of next-generation sequencing techniques to detect SNVs and most indels, it may not detect long-length duplications, deletions and insertions. Our analysis also focused only on exonic variants and intronic variants within 50 base pairs of the exon-intron boundaries, potentially neglecting deep intronic pathogenic variants, which may induce pathogenicity by creating or activating alternative splice sites76. These will lead to an under-estimation of the prevalence.
In conclusion, one in 150 individuals in the Singaporean population carry pathogenic variants that can lead to Alport syndrome. The prevalence rate among the Chinese population is 2.7 times higher than that of the Malay population. Subsequent studies as part of the Phase II of Singapore’s National Precision Medicine program should enhance the accuracy of our estimates.
Methods
Datasets
The SG10K Health Project20 consisted of cross-sectional population-scale whole genome sequencing of 10,323 Singaporeans. These included a mix of healthy and diseased individuals with adult-onset diseases such as diabetes mellitus and eye diseases (Table S4). They were not known to have a genetic kidney disease, but the prevalence of chronic kidney disease within this group was not known.
Ethics approval
This present study was conducted using data derived from the SG10K Health Project, established through collaboration with six local cohorts. These studies were approved by the Institutional Review Board of the respective institutions, including the National University Hospital, Singapore (CIRB/E/2019/2655 for “GUSTO”), Nanyang Technological University (IRB 2016-11-030 for “HELIOS”), National University of Singapore (CIRB 13–512 for “SEED”), SingHealth Centralised Institutional Review Board (2013/605/C for “MEC”; 2018/2717, 2018/2921, 2012/487/A, 2015/2279, 2018/2006, 2018/2594, and 2018/2570 for “PRISM”), and the National Health Group (TB-2020-001 and BTC-2020-001 for the Tan Tock Seng Hospital cohort) (Table S4). These studies were performed in accordance with the 1975 Declaration of Helsinki and its 2013 amendment. Informed consent was obtained from all participants20.
Genomic sequencing
Details of the methodology have been described previously20. In brief, whole-genome sequencing (WGS) of 15x or 30x coverage was performed on a total of 10,323 healthy samples. Following joint variant calling and quality control based on the GenomeAnalysisToolKit (GATK, v4.0.6.0) best practices workflow77,78, 9,770 genomes remained. The samples were further filtered through kinship analysis, resulting in 9,051 remaining genomes inferred to be unrelated to the second degree. Females comprised 57.3% of this analysed dataset. The age of participants ranged from birth to 85 years old (median 47 years). Due to cryptic admixture, discordance between self-reported and genetically-inferred ancestry have been previously described79. Therefore, we used genetically-inferred ancestry for all our analysis. With these, 60.8% of the final cohort were Chinese, 21.4% Indian and 17.8% Malay.
Genetic variants
COL4A3-COL4A5 variants were aligned using the Genome Reference Consortium Human Build 38 (GRCh38/hg38). The MANE transcripts for the collagen type IV α3 chain, α4 chain and α5 chain are ENST00000396578.8, ENST00000396625.5 and ENST00000328300.11, respectively. Variants were classified based on their location and variant alteration type. SNV types included missense synonymous and nonsynonymous SNV and SNVs that resulted in gain or loss of stop codons. Indel variant types included frameshift or inframe insertions or deletions, and those are located at splice sites.
Variant curation
Variants were classified into five categories: “Benign”, “Likely Benign”, “VUS”, “Likely Pathogenic” and “Pathogenic” based on the ACMG / AMP guidelines53,63. In this study, only pathogenic and likely pathogenic variants were used to derive prevalence rates.
Firstly, four criteria were initially applied to filter out variants: (1) variants with MAF > 1% in ExAC (Exome Aggregation Consortium) and GnomAD (Genome Aggregation Database) v3.1.3 databases, (2) intronic variants that were more than 50 base pairs from the exon-intron boundaries, (3) variants classified as “Benign” and “Likely Benign” in ClinVar or InterVar and (4) variants classified as “Benign” and “Likely Benign” by the semiautomated online tool VarSome80 (www.varsome.com).
We then applied the ACMG/AMP criteria based on population databases and in silico tools: (1) Absence in population databases or occurrence in extremely low frequencies of ≤ 0.00007 for AR and ≤ 0.00002 for AD (PM2). The strength of PM2 was not higher than “Supporting”52,81; and (2) REVEL scores for missense variants (Supporting: 0.644–0.773; Moderate: 0.773–0.932; Strong: > = 0.932)52 (PP3).
According to the consensus by the ClinGen Sequence Variant Interpretation Working Group82, relevant data of the published cases carrying the same variant can be used to assign appropriate ACMG criteria. In light of these, an extensive literature search, empowered by Mastermind Genomenon®, was conducted to retrieve previously reported cases related to the variants83. The information on the published cases were obtained for the following ACMG evidences: (1) typical Alport syndrome phenotypes, such as absence of immunohistochemistry staining for collagen IV α5 in kidney or skin biopsies in XL and AR disease63 (PP4); (2) co-segregation or lack of segregation with disease respectively in multiple affected family members (PP1 or BS4); (3) de novo (both maternity and paternity confirmed or unconfirmed respectively) in a patient with the disease and no family history (PS2 or PM6). The BP6 or PP5 criteria (variant being reported as benign or pathogenic respectively by reputable sources) were no longer used82.
We also reviewed the literature to identify the following evidences for each variant: (1) same amino acid change as a previously established pathogenic variant regardless of nucleotide change (PS1); (2) novel missense change at an amino acid residue where a different missense change determined to be pathogenic has been described (PM5); (3) well-established in vitro or in vivo functional studies supportive of a damaging or benign effect respectively on the gene or gene product (PS3 or BS3). These scores can be additive across different reported families.
The strengths of each of these ACMG/AMP evidences were then transformed into scores which were calculated odds of pathogenicity64: Benign_Strong, −4; Benign_Supporting, −1; Indeterminate, 0; Pathogenic_Supporting, + 1; Pathogenic_Moderate, + 2; Pathogenic_Strong, + 4; Pathogenic_Very Strong, + 8. The added scores were used to determine the variant classification using a Bayesian framework65: >=10, Pathogenic; 6–9, Likely pathogenic; 0–5, VUS; −1 to −3, Likely benign; <= −4, Benign.
The VUS that each had a total pathogenicity score of 5 were analyzed. These VUS may potentially be “upgraded” to ‘Likely Pathogenic’ with the addition of 1 more point. This additional point may be obtained from the ACMG/AMP evidence derived from the clinical or family details specific to the subject in SG10K, such as family segregation of the variant (PP1) or phenotype specific to the genetic condition (PP4).
Minor allele frequency and population prevalence
Minor allele frequency (MAF) was calculated as the percentage of number of copies of the minor allele compared with the total number of alleles. For accuracy, individuals with variant loci that did not meet sequencing quality control standards were excluded. Therefore, the total number of alleles used for calculating MAF was less than the total number of alleles in the individuals recruited in the whole cohort.
Population prevalence was determined by dividing the number of individuals carrying the pathogenic variants by the total number of individuals in the cohort (n = 9,051). Individuals carrying a single heterozygous pathogenic variant in either COL4A3 or COL4A4 can develop AD Alport syndrome while individuals carrying a single pathogenic variant in COL4A5 can develop XL Alport syndrome. Gender was not considered in the determination of prevalence for XL Alport syndrome because both males and females may develop the disease, albeit with differing severity.
We could determine the number of individuals who had co-occurrence of more than one pathogenic variant in these three genes. Individuals carrying two pathogenic variants in the same COL4A3 or COL4A4 gene were assumed to have autosomal recessive Alport syndrome, even though it was not possible to determine if the variants were occurring in cis or trans.
The population prevalences were also compared between the Chinese, Indian and Malay ethnicities in Singapore.
Statistical analysis
Data was analyzed using SPSS (v.29). The statistical difference between the allele frequencies in different populations was calculated using the two-sided Fisher’s exact test. A P value of less than 0.05 was considered significant.
Data availability
The SG10K datasheet is incorporated into the genome Aggregation Database (gnomAD v4.1.0) under the East Asian subgroup (https://gnomad.broadinstitute.org/). SG10K_Health datasets, including allele frequencies, protein-drug interactions, imputation and polygenic risk scores, are accessible via the SG10K_Health web portal at https://npm.a-star.edu.sg.
References
Kashtan, C. E. et al. Alport syndrome: A unified classification of genetic disorders of collagen IV alpha345: A position paper of the Alport syndrome classification working group. Kidney Int. 93, 1045–1051. https://doi.org/10.1016/j.kint.2017.12.018 (2018).
Gubler, M. C. Inherited diseases of the glomerular basement membrane. Nat. Clin. Pract. Nephrol. 4, 24–37. https://doi.org/10.1038/ncpneph0671 (2008).
Gibson, J. T. et al. Genotype–phenotype correlations for COL4A3–COL4A5 variants resulting in gly substitutions in Alport syndrome. Sci. Rep. 12, 2722. https://doi.org/10.1038/s41598-022-06525-9 (2022).
Yeo, J. et al. Adverse effects of Alport syndrome-related Gly missense mutations on collagen type IV: Insights from molecular simulations and experiments. Biomaterials 240, 119857. https://doi.org/10.1016/j.biomaterials.2020.119857 (2020).
Adone, A. & Anjankar, A. Alport Syndrome Compr. Rev. Cureus 15, e47129. https://doi.org/10.7759/cureus.47129 (2023).
Savige, J. & Harraka, P. Pathogenic variants in the genes affected in Alport syndrome (COL4A3-COL4A5) and their association with other kidney conditions: A review. Am. J. Kidney Dis. 78, 857–864. https://doi.org/10.1053/j.ajkd.2021.04.017 (2021).
Bhattacharyya, A., Huang, Y., Khan, S. H., Drachenberg, C. B. & Malone, L. C. Tale of two nephropathies; co-occurring Alport syndrome and IgA nephropathy, a case report. BMC Nephrol. 22, 358. https://doi.org/10.1186/s12882-021-02567-9 (2021).
Gulati, A. et al. Collagen IV gene mutations in adults with bilateral renal cysts and CKD. Kidney Int. Rep. 5, 103–108. https://doi.org/10.1016/j.ekir.2019.09.004 (2020).
Savige, J. et al. Thin basement membrane nephropathy. Kidney Int. 64, 1169–1178. https://doi.org/10.1046/j.1523-1755.2003.00234.x (2003).
Deltas, C., Savva, I., Voskarides, K., Papazachariou, L. & Pierides, A. Carriers of autosomal recessive Alport syndrome with thin basement membrane nephropathy presenting as focal segmental glomerulosclerosis in later life. Nephron 130, 271–280. https://doi.org/10.1159/000435789 (2015).
Persson, U., Hertz, J. M., Wieslander, J. & Segelmark, M. Alport syndrome in Southern Sweden. Clin. Nephrol. 64, 85–90. https://doi.org/10.5414/cnp64085 (2005).
Pajari, H., Kääriäinen, H., Muhonen, T. & Koskimies, O. Alport’s syndrome in 78 patients: Epidemiological and clinical study. Acta Paediatr. 85, 1300–1306. https://doi.org/10.1111/j.1651-2227.1996.tb13915.x (1996).
Hasstedt, S. J. & Atkin, C. L. X-linked inheritance of Alport syndrome: Family P revisited. Am. J. Hum. Genet. 35, 1241–1251 (1983).
Gibson, J. et al. Prevalence estimates of predicted pathogenic COL4A3-COL4A5 variants in a population sequencing database and their implications for Alport syndrome. J. Am. Soc. Nephrol. 32, 2273–2290. https://doi.org/10.1681/ASN.2020071065 (2021).
Solanki, K. V. et al. The phenotypic spectrum of COL4A3 heterozygotes. Kidney Int. Rep. 8, 2088–2099. https://doi.org/10.1016/j.ekir.2023.07.010 (2023).
Grzymski, J. J. et al. Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat. Med. 26, 1235–1239. https://doi.org/10.1038/s41591-020-0982-5 (2020).
Wang, Q. et al. Rare variant contribution to human disease in 281,104 UK biobank exomes. Nature 597, 527–532. https://doi.org/10.1038/s41586-021-03855-y (2021).
GenomeAsia, K. C. The genomeasia 100K project enables genetic discoveries across Asia. Nature 576, 106–111. https://doi.org/10.1038/s41586-019-1793-z (2019).
Consortium, H. P. A. S. et al. Mapping human genetic diversity in Asia. Science 326, 1541–1545. https://doi.org/10.1126/science.1177074 (2009).
Chan, S. H. et al. Analysis of clinically relevant variants from ancestrally diverse Asian genomes. Nat. Commun. 13, 6694. https://doi.org/10.1038/s41467-022-34116-9 (2022).
Chiereghin, C. et al. Alport syndrome cold cases: Missing mutations identified by exome sequencing and functional analysis. PLoS One 12, e0178630. https://doi.org/10.1371/journal.pone.0178630 (2017).
Webb, B. D. et al. A founder mutation in COL4A3 causes autosomal recessive Alport syndrome in the Ashkenazi Jewish population. Clin. Genet. 86, 155–160. https://doi.org/10.1111/cge.12247 (2014).
Zhang, Y. et al. Genotype-phenotype correlations and nephroprotective effects of RAAS Inhibition in patients with autosomal recessive Alport syndrome. Pediatr. Nephrol. 36, 2719–2730. https://doi.org/10.1007/s00467-021-05040-9 (2021).
Zhang, Y. et al. Genotype-phenotype correlations in 17 Chinese patients with autosomal recessive Alport syndrome. Am. J. Med. Genet. A. 158a, 2188–2193. https://doi.org/10.1002/ajmg.a.35528 (2012).
Morinière, V. et al. Improving mutation screening in Familial hematuric nephropathies through next generation sequencing. J. Am. Soc. Nephrol. 25, 2740–2751. https://doi.org/10.1681/asn.2013080912 (2014).
Liu, H. F., Li, Q. & Peng, Y. Q. Alport syndrome combined with lupus nephritis in a Chinese family: A case report. World J. Clin. Cases 9, 4721–4727. https://doi.org/10.12998/wjcc.v9.i18.4721 (2021).
He, L. et al. Targeted next-generation sequencing and parental genotyping in sporadic Chinese Han deaf patients. Clin. Genet. 93, 899–904. https://doi.org/10.1111/cge.13182 (2018).
Rosado, C., Bueno, E., Felipe, C., Valverde, S. & González-Sarmiento, R. Study of the true clinical progression of autosomal dominant Alport syndrome in a European population. Kidney Blood Press. Res. 40, 435–442. https://doi.org/10.1159/000368519 (2015).
Zacchia, M. et al. Nephroplex: a kidney-focused NGS panel highlights the challenges of PKD1 sequencing and identifies a founder BBS4 mutation. J. Nephrol. 34, 1855–1874. https://doi.org/10.1007/s40620-021-01048-4 (2021).
Cook, C., Friedrich, C. A., Baliga, R. & Novel COL4A3 mutations in African American siblings with autosomal recessive Alport syndrome. Am. J. Kidney Dis. 51, e25–28. https://doi.org/10.1053/j.ajkd.2007.09.028 (2008).
Nagel, M., Nagorka, S. & Gross, O. Novel COL4A5, COL4A4, and COL4A3 mutations in Alport syndrome. Hum. Mutat. 26, 60. https://doi.org/10.1002/humu.9349 (2005).
Deng, H., Zhang, Y., Ding, J. & Wang, F. Presumed COL4A3/COL4A4 missense/synonymous variants induce aberrant splicing. Front. Med. (Lausanne) 9, 838983. https://doi.org/10.3389/fmed.2022.838983 (2022).
Gao, M. et al. Diagnostic application of exome sequencing in Chinese children with suspected inherited kidney diseases. Front. Genet. 13, 933636. https://doi.org/10.3389/fgene.2022.933636 (2022).
Oka, M. et al. Natural history of genetically proven autosomal recessive Alport syndrome. Pediatr. Nephrol. 29, 1535–1544. https://doi.org/10.1007/s00467-014-2797-4 (2014).
Omori, A. et al. Disruption of the glomerular basement membrane associated with nutcracker syndrome and double inferior Vena Cava in Noonan syndrome: A case report. BMC Nephrol. 23, 65. https://doi.org/10.1186/s12882-022-02671-4 (2022).
Zhang, L., Sun, B. C., Zhao, B. G. & Ma, Q. S. An overview of the multi-pronged approach in the diagnosis of Alport syndrome for 22 children in Northeast China. BMC Nephrol. 21, 294. https://doi.org/10.1186/s12882-020-01962-y (2020).
Weber, S. et al. Identification of 47 novel mutations in patients with Alport syndrome and thin basement membrane nephropathy. Pediatr. Nephrol. 31, 941–955. https://doi.org/10.1007/s00467-015-3302-4 (2016).
Nozu, K. et al. Genetic background, recent advances in molecular biology, and development of novel therapy in Alport syndrome. Kidney Res. Clin. Pract. 39, 402–413. https://doi.org/10.23876/j.krcp.20.111 (2020).
Tokhmafshan, F. et al. A no-nonsense approach to hereditary kidney disease. Pediatr. Nephrol. 35, 2031–2042. https://doi.org/10.1007/s00467-019-04394-5 (2020).
Zhu, F., Li, W., Li, Z., Zhu, H. & Xiong, J. Identification of a novel COL4A4 variant in compound-heterozygous state in a patient with Alport syndrome and histological findings similar to focal segmental glomerulosclerosis (FSGS). Front. Genet. 9, 748. https://doi.org/10.3389/fgene.2018.00748 (2018).
Gao, Y. et al. Identification of COL4A4 variants in Chinese patients with Familial hematuria. Front. Genet. 13, 1064491. https://doi.org/10.3389/fgene.2022.1064491 (2022).
Liu, J. H. et al. Novel mutations in COL4A3, COL4A4, and COL4A5 in Chinese patients with Alport syndrome. PLoS One. 12, e0177685. https://doi.org/10.1371/journal.pone.0177685 (2017).
Papazachariou, L. et al. Frequent COL4 mutations in Familial microhematuria accompanied by later-onset Alport nephropathy due to focal segmental glomerulosclerosis. Clin. Genet. 92, 517–527. https://doi.org/10.1111/cge.13077 (2017).
Slajpah, M. et al. Sixteen novel mutations identified in COL4A3, COL4A4, and COL4A5 genes in Slovenian families with Alport syndrome and benign Familial hematuria. Kidney Int. 71, 1287–1295. https://doi.org/10.1038/sj.ki.5002221 (2007).
Longo, I. et al. Autosomal recessive Alport syndrome: An in-depth clinical and molecular analysis of five families. Nephrol. Dial Transplant. 21, 665–671. https://doi.org/10.1093/ndt/gfi312 (2006).
Storey, H., Savige, J., Sivakumar, V., Abbs, S. & Flinter, F. A. COL4A3/COL4A4 mutations and features in individuals with autosomal recessive Alport syndrome. J. Am. Soc. Nephrol. 24, 1945–1954. https://doi.org/10.1681/asn.2012100985 (2013).
Matthaiou, A., Poulli, T. & Deltas, C. Prevalence of clinical, pathological and molecular features of glomerular basement membrane nephropathy caused by COL4A3 or COL4A4 mutations: A systematic review. Clin. Kidney J. 13, 1025–1036. https://doi.org/10.1093/ckj/sfz176 (2020).
Xie, J. et al. COL4A3 mutations cause focal segmental glomerulosclerosis. J. Mol. Cell Biol. 6, 498–505. https://doi.org/10.1093/jmcb/mju040 (2014).
Lata, S. et al. Whole-Exome sequencing in adults with chronic kidney disease: A pilot study. Ann. Intern. Med. 168, 100–109. https://doi.org/10.7326/m17-1319 (2018).
Pinto, E. V. F. et al. Genomics integration into nephrology practice. Kidney Med. 3, 785–798. https://doi.org/10.1016/j.xkme.2021.04.014 (2021).
Xu, H. et al. Collagen IV and Podocyte-Related gene variants in patients with concurrent IgA nephropathy and thin basement membrane nephropathy. Nephron 147, 301–310. https://doi.org/10.1159/000526971 (2023).
Pejaver, V. et al. Calibration of computational tools for missense variant pathogenicity classification and ClinGen recommendations for PP3/BP4 criteria. Am. J. Hum. Genet. 109, 2163–2177. https://doi.org/10.1016/j.ajhg.2022.10.013 (2022).
Richards, S. et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American college of medical genetics and genomics and the association for molecular pathology. Genet. Med. 17, 405–424. https://doi.org/10.1038/gim.2015.30 (2015).
Ellard, S. et al. ACGS Best Practice Guidelines for Variant Classification in Rare DIsease 2020. (2020). www.acgs.uk.com
Yang, W. et al. Fetal hyperechoic kidney cohort study and a meta-analysis. Front. Genet. 14, 1237912. https://doi.org/10.3389/fgene.2023.1237912 (2023).
Alge, J. L. et al. Variants in genes coding for collagen type IV α-chains are frequent causes of persistent, isolated hematuria during childhood. Pediatr. Nephrol. 38, 687–695. https://doi.org/10.1007/s00467-022-05627-w (2023).
Zupan, A., Matjašič, A., Grubelnik, G., Tasić, V. & Momirovska, A. Mutations in collagen genes in the context of an isolated population. Genes (Basel) 11 https://doi.org/10.3390/genes11111377 (2020).
Yokota, K. et al. Female X-linked Alport syndrome with somatic mosaicism. Clin. Exp. Nephrol. 21, 877–883. https://doi.org/10.1007/s10157-016-1352-y (2017).
Bu, L. et al. Somatic mosaicism in a male patient with X-linked Alport syndrome. Kidney Int. Rep. 4, 1031–1035. https://doi.org/10.1016/j.ekir.2019.03.005 (2019).
Aoto, Y. et al. A case with somatic and germline mosaicism in COL4A5 detected by multiplex ligation-dependent probe amplification in X-linked Alport syndrome. CEN Case Rep. 9, 431–436. https://doi.org/10.1007/s13730-020-00503-8 (2020).
Swee-Hock, S. The Population of Singapore (Third Edition) (Institute of Southeast Asian Studies, 2012).
Sabanayagam, C. et al. Ethnic disparities in prevalence and impact of risk factors of chronic kidney disease. Nephrol. Dialysis Transplant.25, 2564–2570. https://doi.org/10.1093/ndt/gfq084 (2010).
Savige, J. et al. Consensus statement on standards and guidelines for the molecular diagnostics of Alport syndrome: Refining the ACMG criteria. Eur. J. Hum. Genet. 29, 1186–1197. https://doi.org/10.1038/s41431-021-00858-1 (2021).
Tavtigian, S. V., Harrison, S. M., Boucher, K. M. & Biesecker, L. G. Fitting a naturally scaled point system to the ACMG/AMP variant classification guidelines. Hum. Mutat. 41, 1734–1737. https://doi.org/10.1002/humu.24088 (2020).
Tavtigian, S. V. et al. Modeling the ACMG/AMP variant classification guidelines as a Bayesian classification framework. Genet. Med. 20, 1054–1060. https://doi.org/10.1038/gim.2017.210 (2018).
Wong, E. et al. The Singapore national precision medicine strategy. Nat. Genet. 55, 178–186. https://doi.org/10.1038/s41588-022-01274-x (2023).
Buzza, M., Wilson, D. & Savige, J. Segregation of hematuria in thin basement membrane disease with haplotypes at the loci for Alport syndrome. Kidney Int. 59, 1670–1676. https://doi.org/10.1046/j.1523-1755.2001.0590051670.x (2001).
Schott, C. et al. Utility of genetic testing in adults with chronic kidney disease: A systematic review and meta-analysis. Clin. J. Am. Soc. Nephrol. https://doi.org/10.2215/cjn.0000000000000564 (2024).
Groopman, E. E. et al. Diagnostic utility of exome sequencing for kidney disease. N Engl. J. Med. 380, 142–151. https://doi.org/10.1056/NEJMoa1806891 (2019).
Puapatanakul, P. & Miner, J. H. Alport syndrome and Alport kidney diseases - elucidating the disease spectrum. Curr. Opin. Nephrol. Hypertens. 33, 283–290. https://doi.org/10.1097/mnh.0000000000000983 (2024).
Zeng, M., Di, H., Liang, J. & Liu, Z. Effectiveness of renin-angiotensin-aldosterone system blockers in patients with Alport syndrome: A systematic review and meta-analysis. Nephrol. Dial Transplant. 38, 2485–2493. https://doi.org/10.1093/ndt/gfad105 (2023).
Kashtan, C. E. & Gross, O. Clinical practice recommendations for the diagnosis and management of Alport syndrome in children, adolescents, and young adults-an update for 2020. Pediatr. Nephrol. 36, 711–719. https://doi.org/10.1007/s00467-020-04819-6 (2021).
Wu, Y. et al. Genomic testing for suspected Monogenic kidney disease in children and adults: A health economic evaluation. Genet. Med. 25, 100942. https://doi.org/10.1016/j.gim.2023.100942 (2023).
United States Renal Data System. USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States (National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, 2023).
Lim, C. et al. Clinical implementation of Nephrologist-led genomic testing for glomerular diseases in Singapore: Rationale and protocol. Am. J. Nephrol. 1–20. https://doi.org/10.1159/000542942 (2024).
Boisson, M. et al. A wave of deep intronic mutations in X-linked Alport syndrome. Kidney Int. 104, 367–377. https://doi.org/10.1016/j.kint.2023.05.006 (2023).
Geraldine, A. & Van der Auwera, B. D. O. C. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra (1st Edition). (O’Reilly Media, Inc., (2020).
Ryan, P. et al. Scaling accurate genetic variant discovery to tens of thousands of samples. BioRxiv 201178 https://doi.org/10.1101/201178 (2018).
Mersha, T. B. & Abebe, T. Self-reported race/ethnicity in the age of genomic research: its potential impact on Understanding health disparities. Hum. Genomics. 9, 1. https://doi.org/10.1186/s40246-014-0023-x (2015).
Kopanos, C. et al. VarSome: the human genomic variant search engine. Bioinformatics 35, 1978–1980. https://doi.org/10.1093/bioinformatics/bty897 (2019).
Group, C. S. V. I. S. W. ClinGen Sequence Variant Interpretation Recommendation for PM2 - Version 1.0, (2020). https://clinicalgenome.org/site/assets/files/5182/pm2_-_svi_recommendation_-_approved_sept2020.pdf
Biesecker, L. G. & Harrison, S. M. ClinGen sequence variant interpretation working, G. The ACMG/AMP reputable source criteria for the interpretation of sequence variants. Genet. Med. 20, 1687–1688. https://doi.org/10.1038/gim.2018.42 (2018).
Chunn, L. M. et al. Mastermind: A comprehensive genomic association search engine for empirical evidence curation and genetic variant interpretation. Front. Genet. 11, 577152. https://doi.org/10.3389/fgene.2020.577152 (2020).
Acknowledgements
This study is supported by the National Precision Medicine Programme (NPM) PHASE II FUNDING to W.K.L (MOH-000588) and to the Clinical Implementation Pilots project “Health and Economic Impact of Next Generation Sequencing in Primary Glomerular Diseases in Singapore (PGD)” from Precision Health Research, Singapore (PRECISE). This study made use of data generated as part of the Singapore National Precision Medicine program funded by the Industry Alignment Fund (Pre-Positioning) (IAF-PP: H17/01/a0/007). This study made use of data / samples collected in the following cohorts in Singapore: (1) The Health for Life in Singapore (HELIOS) study at the Lee Kong Chian School of Medicine, Nanyang Technological University, Singapore (supported by grants from a Strategic Initiative at Lee Kong Chian School of Medicine, the Singapore Ministry of Health (MOH) under its Singapore Translational Research Investigator Award (NMRC/STaR/0028/2017) and the IAF-PP: H18/01/a0/016); (2) The Growing up in Singapore Towards Healthy Outcomes (GUSTO) study, which is jointly hosted by the National University Hospital (NUH), KK Women’s and Children’s Hospital (KKH), the National University of Singapore (NUS) and the Singapore Institute for Clinical Sciences (SICS), Agency for Science Technology and Research (A*STAR) (supported by the Singapore National Research Foundation under its Translational and Clinical Research (TCR) Flagship Programme and administered by the Singapore Ministry of Health’s National Medical Research Council (NMRC), Singapore - NMRC/TCR/004-NUS/2008; NMRC/TCR/012-NUHS/2014. Additional funding is provided by SICS and IAF-PP H17/01/a0/005); (3) The Singapore Epidemiology of Eye Diseases (SEED) cohort at Singapore Eye Research Institute (SERI) (supported by NMRC/CIRG/1417/2015; NMRC/CIRG/1488/2018; NMRC/OFLCG/004/2018); (4) The Multi-Ethnic Cohort (MEC) cohort (supported by NMRC grant 0838/2004; BMRC grant 03/1/27/18/216; 05/1/21/19/425; 11/1/21/19/678, Ministry of Health, Singapore, National University of Singapore and National University Health System, Singapore); (5) The SingHealth Duke-NUS Institute of Precision Medicine (PRISM) cohort (supported by NMRC/CG/M006/2017_NHCS; NMRC/STaR/0011/2012, NMRC/STaR/ 0026/2015, Lee Foundation and Tanoto Foundation); (6) The TTSH Personalised Medicine Normal Controls (TTSH) cohort funded (supported by NMRC/CG12AUG17 and CGAug16M012). The views expressed are those of the author(s) are not necessarily those of the National Precision Medicine investigators, or institutional partners. We thank all investigators, staff members and study participants who made the National Precision Medicine Project possible.
Author information
Authors and Affiliations
Contributions
L.S.T. and Z.Y.C. designed and executed the study, and drafted the manuscript. S.J. revised the manuscript and significantly contributed to variant curation. N.Y.J. conducted gene annotation and MAF calculation. C.H.L. and L.W.K. contributed to variant curation. K.C.T., N.J.L., C.G.C., Y.S.C., L.H.M. and Y.J. revised the manuscript and played critical roles in interpreting the results. N.K.H. conceived and designed the study, revised the manuscript, and was instrumental in interpreting the results. N.K.H. and Z.Y.C. share senior authorship of this publication. All authors approved the final version of the manuscript. The corresponding author confirms full access to the study data and assumes final responsibility for the decision to submit for publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lim, T.S.T., Koh, C.T., Savige, J. et al. Pathogenic variants in the Alport genes are prevalent in the Singapore multiethnic population with highest frequency in the Chinese. Sci Rep 15, 7691 (2025). https://doi.org/10.1038/s41598-025-92520-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-92520-9