Abstract
Stroke is the leading cause of disability and death worldwide. It severely affects patients’ quality of life and imposes a huge burden on the society in general. The diagnosis of stroke relies predominantly on the use of neuroimaging. The identification of stroke using electroencephalogram (EEG) in the clinical assessment of stroke has been underutilized. An EEG feature fusion based light gradient-boosting machine (LightGBM) model was proposed to achieve a fast diagnosis of non-stroke, ischemic stroke, and hemorrhagic stroke. This study aims to capture the essential difference between non-stroke, ischemic stroke, and hemorrhagic stroke. An optimal fusion feature set originated from approximate entropy and fuzzy entropy of EEG signal was constructed. To verify the effectiveness of the EEG fusion feature, the Tree-structured Parzen Estimator optimized LightGBM classifier (TPELGBM) was used for the classification. The ZJU4H EEG dataset used for analysis in this study was obtained from the Fourth Affiliated Hospital of Zhejiang University, China. The proposed ApFu-TPELGBM model exhibited excellent classification results, which achieved a precision of 0.9676, recall of 0.9669, and f1-score of 0.9672. To our knowledge, it was the most accurate classifier for EEG-based stroke diagnosis so far. The ApFu-TPELGBM model can determine the stroke type anywhere EEG signals can be collected, even before the patient is admitted to a hospital. Rapid and accurate diagnosis of stroke using EEG signals may become a promising approach in the clinical assessment of stroke.
Similar content being viewed by others
Introduction
Stroke is an acute cerebrovascular disease caused by the occlusion or rupture of cerebral blood vessels, and it has a high rate of disability and fatality1,2,3. Although mortality from stroke has declined with improvements in healthcare, the absolute number of people disabled due to stroke has globally increased across all age groups4,5. Stroke can be classified as hemorrhagic and ischemic based on cerebral blood circulation disorders. These stroke types have their corresponding treatment methods6,7,8. During stroke treatment, time is the most crucial factor, which is directly related to the therapeutic effect. International guidelines for stroke treatment recommend a door-to-needle time (DNT) within 60 min9. Furthermore, studies have shown that the timeliness of stroke diagnosis and treatment has a substantial impact on the prognosis; delaying the treatment reduces the probability of a good prognosis10,11,12. Stroke leads to reduced or even interrupted blood flow to the brain, causing severe brain tissue damage13,14,15. Therefore, rapid diagnosis is essential for stroke treatment.
Currently, the gold standard for clinical stroke diagnosis is imaging examination, such as computed tomography (CT), magnetic resonance imaging (MRI), etc. An Electroencephalogram (EEG) is only used as a diagnostic aid. EEG is a non-invasive test used to assess brain function by recording electrical activity on the scalp. It can be used to detect and record the electrical activity of neurons in the brain, as well as blood supply to blood vessels16. Compared with CT, MRI, and other methods, EEG has the advantages of portability, minimal interference, low cost, and low usage environmental requirements.
With the continuous development of neurology and medicine, EEG has been frequently studied as a routine examination method in clinical practice17,18. At present, EEG has increasingly become a major method for detecting several brain diseases, including brain tumors, epilepsy, and intracranial lesions19. However, EEG research on stroke is primarily focused on predicting poststroke epilepsy, cognitive decline, and functional outcomes; EEG has not received considerable attention concerning the clinical diagnosis of stroke. Until 2018, Maxim et al. have shown that EEG changes can be observed several seconds after decreased or blocked cerebral blood flow; these changes precede morphological changes in brain tissues (e.g., cell edema and necrosis)20. In 2021, Laura et al. proposed a strategy for detecting major pre-hospital vascular occlusive stroke using EEG, and they claimed that a reliable pre-hospital diagnostic method should meet the requirements of high diagnostic accuracy, fast speed, simple operation, small size, and low cost21. Subsequently, Laura et al. proposed an emergency-room EEG-based detection method for major vascular occlusive stroke and achieved a diagnostic accuracy of 0.83 using EEG δ-α ratio features22. Nevertheless, the analyses of stroke EEG signals in these studies are not comprehensive enough, and the accuracies of such studies are not yet up to the standard for clinical use.
In recent years, machine learning has been widely used in the field of EEG analysis. With machine learning methods, EEG signals can be used as indicators of difficult-to-detect conditions. To date, the common machine learning models include support vector machine (SVM), decision tree (DT), and random forest method (RF), etc23,24,25. Ke et al.26 proposed the light gradient-boosting machine (LightGBM) in 2017. The LightGBM model has many advantages in that the calculation speed is fast and can be applied to handle big data and provide better accuracy27,28. Due to these advantages, the LightGBM model is widely used by data scientists and has been applied in many fields29,30,31.
This paper introduces machine learning method for the diagnosis of stroke using EEG signals. We proposed a novel classification model (ApFu-TPELGBM) based on EEG feature fusion algorithm and LightGBM classifier. To our knowledge, this model is the first to introduce machine learning method for the diagnosis of stroke using EEG signals. In this study, we processed the raw EEG signals by filtering, bad channel elimination, and segmentation at first. Then the approximate entropy and fuzzy entropy of EEG signal were extracted and fused as a new feature. Finally, classification models are established by LightGBM to detect ischemic stroke patients, hemorrhagic stroke patients, and non-stroke patients at the individual level. Furthermore, the tree-structured Parzen estimator (TPE) algorithm was employed to optimize the hyperparameters of the ApFu-LightGBM model to further improve the classification performance of the model. The primary contributions of this research study can be listed as follows:
-
(1)
A novel ApFu-TPELGBM classification model based on our EEG feature fusion algorithm and LightGBM was established, which can achieve accurate and rapid stroke diagnosis in multiple scenarios, such as in ambulances, patients’ homes, or even outdoors. This model enriches the diagnosis methods of stroke and provides better treatment for patients.
-
(2)
A feature fusion algorithm combining approximate entropy and fuzzy entropy was proposed, which can profoundly distinguish the complexity and uncertainty features of EEG signals. This feature fusion algorithm was compared with the baselines through experiments, and its outstanding performance in stroke diagnosis was verified.
-
(3)
The TPE algorithm was used to optimize the LightGBM classifier, namely TPELGBM classifier, which displayed excellent classification accuracy on ZJU4H datasets, and its classifying performance was superior to the baselines, such as DT, SVM, RF, and LightGBM models. The results demonstrated that the proposed ApFu-TPELGBM model can provide reliable stroke diagnosis for clinicians, which has significant practical application value.
Materials and methods
Dataset
In this study, the ZJU4H dataset was used to test the proposed diagnosis model. EEG Data were obtained from the Fourth Affiliated Hospital of Zhejiang University, China, which included patients with stroke and suspected stroke. This research project has been approved by the Ethics Committee of the Fourth Affiliated Hospital of Zhejiang University School of Medicine. All test results appearing in the original medical records (including personal data, laboratory documents, etc.) will be kept completely confidential to the extent permitted by law. All subjects have signed informed consent for clinical research in this study. The data acquisition and screening process is depicted in Fig. 1. In all the suspected stroke patients (n = 73), patients with poor-quality EEG data (n = 1) and patients with incomplete information (n = 2) were excluded. Out of the 70 patients with high-quality EEG data, 42 were diagnosed with ischemic stroke, comprising 31 males and 11 females with ages ranging from 42 to 87 years. Additionally, there were 9 patients with hemorrhagic stroke, including 4 males and 5 females aged between 39 and 74 years. The remaining 19 patients had no history of stroke, with 12 males and 7 females aged from 49 to 73 years. A detailed summary of the patient demographics is presented in Table 1. It is noteworthy that the ischemic stroke group is significantly larger than the other two groups, and on average, the ischemic stroke patients tend to be older than those with hemorrhagic stroke.
In this study, XLTEK-EEG32U was used to collect EEG data. Specifically, the EEG acquisition system used herein comprised 32 channels, including AUX1, AUX2, AUX3, AUX4, AUX5, AUX6, AUX7, AUX8, PG1, PG2, A1, A2, C3, C4, CZ, F3, F4, F7, F8, FZ, FP1, FP2, FPZ, O1, O2, P3, P4, PZ, T3, T4, T5 and T6. Among them, C3, C4, CZ, F3, F4, F7, F8, FZ, FP1, FP2, FPZ, O1, O2, P3, P4, PZ, T3, T4, T5 and T6 were the 20 channels located in the brain by the international standard 10–20 EEG-electrode location and naming system. Moreover, the EEG sampling rate was 256 Hz and the EEG data of each patient were collected for approximately 5 min.
Methods
A machine learning model based on EEG signal feature fusion is introduced to distinguish among the EEG segments of hemorrhagic stroke, ischemic stroke and non-stroke. As depicted in Fig. 2, the model comprises three parts. Firstly, the original EEG signals are pre-processed, followed by which the obtained continuous EEG signals are divided into segments. Secondly, the approximate entropy (ApEn) and fuzzy entropy (FuEn) of EEG are calculated and fused as features. Finally, the LightGBM model is employed for classification. Specifically, the fused entropy features are divided into training and test sets. The training set is used as input to train the LightGBM model, and the tree-structured Parzen estimator (TPE) algorithm is employed to optimize the hyperparameters to obtain optimal hyperparameters. The test set is then input into the well-trained TPELGBM model for classification, and the diagnosis results are obtained.
Preprocessing
The main objective of EEG preprocessing is to reduce or remove mixed noise and interference in EEG signals. The preprocessing proposed in this study includes filtering, bad track elimination, and segmentation.
According to the frequency band characteristics of EEG signals in stroke patients20, we chose a bandpass filter that retained the 1–35 Hz portion of the signal. As the dataset contained missing channels, 18 of the 20 channels shared in the entire dataset were selected, including C3, C4, CZ, F3, F4, F7, F8, FZ, FP1, FP2, O1, O2, P3, P4, T3, T4, T5, and T6. Owing to its length, the original EEG signal was segmented in periods to obtain 18-dimensional data segments. The 1-s and 3-s EEG segments were evaluated in this study.
Feature extraction and fusion
In the field of information theory, entropy can be used to measure the uncertainty of an unknown state of a system. The more complex the system, the lower its predictability and the greater its entropy32,33. Considering EEG signals of a patient as a system, the entropy of this system can measure the disorder in the EEG signals. To date, this method has been successfully employed in different fields of EEG signal processing34,35,36.
The EEG signal itself is a non-equilibrium signal, meanwhile, there is noise interference when the EEG signal is collected in stroke patients. Therefore, we extract the approximate entropy (ApEn), sample entropy (SaEn) and fuzzy entropy (FuEn) of EEG as features to characterize the EEG signals commendable. These entropies have strong anti-noise and anti-interference abilities and are more suitable for clinical practice. The application of ApEn, SaEn, and FuEn can provide diverse insights into the data. By combining any two of these measures, we can attain a more holistic representation of the data’s traits. This approach not only enhances the descriptive power of the analysis but also enhances our ability to comprehend the intricacies of the data. The fusion of these unique metrics enriches our dataset, potentially revealing hidden patterns or characteristics that a singular evaluative strategy might have missed.
ApEn, SaEn and FuEn can be employed to describe the chaotic degree of a signal. ApEn is a nonlinear dynamic parameter used for quantifying the regularity and unpredictability of time-series fluctuations. It uses a non-negative number to represent the complexity of a time series, indicating the possibility of the occurrence of new information in the time series37. The ApEn of the signal \({X^N}=x(1),x(2), \ldots x(N)\) is calculated as follows:
Firstly, define the integers m and r, with m representing the length of the comparison vector and r representing the measure of similarity.
Reconstruct the original sequence for the m-dimensional vector as follows:
Where,
Then, calculate the Euclidean distance \(d\left[ {{X^m}(i),\;{X^m}(j)} \right]\) between any vector \({X^m}(i)\) and the rest of the vectors as follows:
Next, determine the number Nm(i) that satisfies d[Xm(i), Xm(j)] < r for each vector Xm(i), Subsequently, calculate the ratio Cim(r) between the number Nm(i) and the total number of vectors N-m+1 as follows :
Then, calculate the logarithm of Cim(r), then calculate their average value Φm(r) for all i as follows:
Further, increase the dimension to m + 1, and repeat calculating using Eqs. (1)–(5) to obtain Φm+1(r).
SaEn is another measure of the complexity of a time series and can be calculated based on ApEn. Compared with ApEn, SaEn does not include a comparison with its data segment when calculating the approximation (Eq. (7)), thus, the calculation gap is smaller38.
The SaEn can be derived from the following equation:
FuEn is obtained by introducing a fuzzy membership function based on SaEn39. When calculating the Nm(i)(in Eq. (4)), both ApEn and SaEn measure the similarity of vectors being compared using the Heaviside function, which can be represented as follows:
Here, x represents the difference between the distance between vectors and r(d[Xm(i), Xm(j)]-r). The Heaviside function leads to a two-state binary classifier, where the vectors are either close or not close. As a result, the mutation of calculation results is relatively large, the entropy value lacks continuity, and it is very sensitive to the value of threshold value r, and a slight change in r may lead to the mutation of results. Notably, FuEn uses the exponential function to blur the similarity-measurement formula (Eq. (10)). The continuity of the exponential function smoothly changes FuEn with a change in the parameters of the function.
Finally, the FuEn is calculated as follows:
We calculated the approximate entropy vector \(ApEn=\left[ {ApE{n_1},ApE{n_2}, \ldots ApE{n_{18}}} \right]\), the sample entropy vector \(SaEn=\left[ {SaE{n_1},SaE{n_2}, \ldots SaE{n_{18}}} \right]\) and the fuzzy entropy vector \(FuEn=\left[ {FuE{n_1},FuE{n_2}, \ldots FuE{n_{18}}} \right]\) of 18 channels for each segment. Then, the 18-dimensional ApEn feature vector and and the 18-dimensional FuEn feature vector were concatenated to obtain the 36-dimensional fusion feature vector as:
Similarly, the ApSa fushion feature and SaFu fushion feature can be calculated as:
The obtained new feature vector served as an input to the classifier.
Classification
The proposed model is based on the LightGBM classifier, which is a distributed gradient-boosting framework based on the DT model26. To further verify the performance of the TPELGBM classifier, it is compared with DT, SVM, and RF models in terms of precision, recall, and f1-score. The comparison results show that the TPELGBM can establish a more accurate stroke diagnosis model. In addition, to verify the effect of the TPE algorithm, we also compare the LightGBM classifier and the TPELGBM classifier. The experimental results show that adding the TPE algorithm optimizes the LightGBM classifier. Specific experimental results are described in detail in Sect. 3.
LightGBM classifier
The LightGBM classifier generates decision trees using the leaf-wise splitting method and searches for feature segmentation points using histogram-based algorithms. First, it uses the leaf-wise splitting method, which generates more complex trees than the horizontal splitting method, which can achieve higher accuracy. Second, a histogram-based algorithm is employed to convert the continuous eigenvalues to discrete values, and the histogram difference is used to accelerate the calculation of the brother-node histogram to speed up the training process and achieve less memory consumption. In addition, it supports parallel learning, including feature and data parallel learning, which brings high computation speed26. In summary, the LightGBM classifier includes the following algorithms:
Histogram-based DT Algorithm: The algorithm divides each feature in the dataset into a number of discrete intervals and builds a histogram by computing the class distribution of each interval. Then, feature selection and decision tree construction are carried out using the histogram information. It can improve the efficiency of feature selection and decision tree construction.
Leaf-wise Leaf Growth Strategy with Depth Limitation: The LightGBM adopts a leaf-wise growth strategy. From all current leaves at a time, the strategy finds the one with the largest splitting gain and splits, and so on. Therefore, compared with level-wise splitting, leaf-wise splitting has the following advantages: in case of the same number of splits, leaf-wise splitting can reduce more errors and obtain higher accuracy. Furthermore, the LightGBM adds a maximum depth limit, along with leaf-wise splitting, to prevent overfitting while ensuring high efficiency.
Exclusive Feature Bundling: The algorithm can bind several mutually exclusive features to form a new feature to capture more useful information and reduce the number of features. Redundant information and noise can be reduced to further improve model performance and efficiency. Gradient-based One-Side Sampling: This algorithm can reduce the number of data instances with only a low gradient so that only the remaining data with a high gradient can be used when calculating information gain. Compared with XGBoost, it saves a considerable amount of time and space overhead by traversing all eigenvalues.
Tree-structured Parzen estimator
Tree-structured Parzen Estimator (TPE) is a type of Bayesian optimization algorithm based on the tree structure and is used to solve the black-box global optimization problem40,41. TPE uses Gaussian Mixture Model (GMM) to learn hyperparameter model.
The Gaussian process-based approach directly simulates p(y|θ), which is the conditional probability that the hyperparameters is θ when the model loss is y.
Firstly, select a loss threshold y* based on the available data. And p(y|θ) can be expressed as:
The improvement in the model after choosing a new set of hyperparameters is given by:
TPE uses expected improvement (EI) as a collection function. Subsequently, the expectation of the improvement is:
l(θ) is probability density formed by the set of loss values less than y*, and g(θ) is the probability density formed by the remaining set. By constructing:
and
we can obtain
The TPE algorithm evaluates the hyperparameters using $g(θ)/l(θ) to find the hyperparameters θ* with maximum EI, the flowchart chart of this algorithm is depicted in Fig. 3.
Experimental results and evaluations
To explore the characteristics of EEG signals of stroke patients and verify our proposed diagnosis model, this study primarily conducted research and analysis from the following three aspects. Through different characteristics of stroke diagnosis experiments, the ApFu characteristics of stroke EEG characteristics were verified. Then the superiority of the proposed model was evaluated by comparing it with different lengths of EEG and different classifiers. Finally, the TPELGBM classifier was compared with the original LightGBM classifier. The advantages of the proposed model were further explained. The evaluation metrics used in this study were accuracy, precision, recall, and f1-score. In addition, we also draw the ROC curves and confusion matrices to evaluate the performance of the model. To ensure the stability of the model’s generalization capability, all our experiments were conducted using five-fold cross-validation.
Experiment settings
The proposed model is based on the feature fusion framework presented in Sect. 2. All training and testing procedures are executed on an RTX TITAN graphics processing unit with 32G random access memory using Python 3.8, sklearn 1.2.2, Mne 1.5.1 and Optuna 3.1.1.
In the experiment, the EEG of patients with ischemic stroke, hemorrhagic stroke and non-stroke was labeled 0, 1, 2 for machine learning model classification. And all 1-s length segmented EEG data sets are divided into training sets and test sets according to 7:3 ratio and 6:4 ratio. Figure 4 show the training performance of the 7:3 ratio and 6:4 ratio of approximate entropy and fuzzy entropy fusion feature data set under the TPELGBM classifier, where the blue line represents the accuracy versus iterations during the training process, and the orange line represents that during test process. It is evident that the dataset divided in a 6:4 ratio does not perform as well in training as the one divided in a 7:3 ratio, but it exhibits superior results in testing. Thus, it was ultimately decided to allocate the dataset with 70% for the training set and 30% for the testing set. Notably, the accuracy value converges to the upper limit. Therefore, no overfitting or underfitting is observed in the evaluated models.
Analysis of different feature vectors
In order to derive the optimal feature, six features of 1-s EEG segment are input to TPELGBM classifier, and they are ApEn, SaEn, FuEn, ApSa (fusion feature of approximate entropy and sample entropy), ApFu (fusion feature of approximate entropy and fuzzy entropy) and SaFu (fusion feature of sample entropy and fuzzy entropy). We then calculate the corresponding accuracy, precision, recall, and f1-score as evaluation indicators. Table 2 presents the results of the TPELGBM classifier based on various feature vectors. We can see that in the single-feature scenario, FuEn exhibits the best performance, and its accuracy, precision, recall and f1-score are 0.9395, 0.9312, 0.8975 and 0.9122, respectively. Obviously, the fusion entropy features are better than the single-entropy feature in terms of EEG-based stroke diagnosis. Meanwhile, among all fusion entropy features, the accuracy of the ApFu entropy feature reaches the highest value 0.9689, with precision, recall and f1-score of 0.9676, 0.9669 and 0.9672 respectively. For all 1-s segments, the ApFu entropy feature exhibits the best performance.
To show the superiority of the ApFu fusion feature more directly, we construct the confusion matrices of models with different features (Fig. 5). In Fig. 5, where class 0 represents ischemic stroke patients, class 1 represents hemorrhagic stroke patients and class 2 represents patients without stroke. In the confusion matrix, darker grids indicate more samples in that category, While the diagonal shows the number of samples accurately classified by the model. As we can see, the fusion features have better performance than single features, and the ApFu fusion feature perform best among all the features. In addition, from the color of the grid in the confusion matrix, we can see that hemorrhagic stroke EEG has far fewer samples (class 1) than the other two, which increases the difficulty of classification. Compared with other features, ApFu fusion feature has the best recognition of hemorrhagic stroke patients.
Analysis of different segmentation with different classifiers
To the best of our knowledge, there is no public research on utilizing machine learning methods to diagnose stroke with EEG signals for now. It was difficult for us to compare our approach with existing algorithms. However, we tested different classifiers and different EEG segmentations to demonstrate the superiority of our model.
Confusion matrix of prediction results of TPELGBM classifier with different features in test data. (a) The confusion matrix of approximate entropy classifier. (b) The confusion matrix of sample entropy classifier. (c) The confusion matrix of fuzzy entropy classifier. (d) The confusion matrix of approximate entropy and sample entropy fusion feature (ApSa) classifier. (e) The confusion matrix of approximate entropy and fuzzy entropy fusion feature (ApFu) classifier. (f) The confusion matrix of sample entropy and fuzzy entropy fusion feature (FuSa) classifier.
In order to compare the performance of different EEG segment lengths and classifiers on the experimental results, we conduct a comparative study. Under the condition that the experimental parameters and settings remain unchanged, different length segments and classifiers are used in the experiment. As shown in Table 3, we compared the classification performance of 1-s and 3-s segments in four different classifiers, which are DT, SVM, RF and TPELGBM.
For all four classifiers, performances of 1-s segmentation are better than those of 3-s segmentation. Taking the Precision as an example, the 1-s segment is 0.3123 higher than the 3-s segment for DT classifier, whereas the 1-s segment is 0.0389 higher than the 3-s segment for SVM classifier. Meanwhile, the 1-s segment is 0.0293 higher than the 3-s segment for RF classifier and the 1-s segment is 0.0384 higher than the 3-s segment for TPELGBM classifier. This phenomenon can be attributed to the fact that the computation of Fuzzy Entropy (FuEn) is not contingent upon the duration of EEG signal segments. Our proposed ApFu fusion feature inherits this beneficial characteristic. Additionally, dividing the EEG signal into 1-second segments, as opposed to 3-second segments, yields a larger number of samples, which in turn enhances the training efficacy and classification accuracy of the model. Among all experiments with 1-second segments, TPELGBM demonstrates superior classification performance, with each of its evaluation metrics markedly surpassing those of the other three classifiers. LightGBM, which is favored for its use of Gradient-based One-Side Sampling (GOSS) and Exclusive Feature Bundling (EFB), effectively minimizes redundant computations and bolsters training efficiency. Moreover, LightGBM provides a broad spectrum of parameter settings, allowing users to meticulously calibrate the model to specific challenges, thereby achieving optimal results. As a result, the TPELGBM model, fine-tuned by the TPE algorithm for hyperparameter optimization, stands out in classification tasks, with its evaluation metrics notably surpassing those of the other three classifiers.
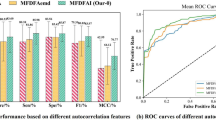
In order to show the superiority of TPELGBM classifier more clearly, we draw the ROC curves of DT, SVM, RF and TPELGBM classifiers in (Fig. 6). The corresponding AUC is also calculated and demonstrated. Different colors represent the classification performance of the model for different categories. To be specific, class 0 indicates ischemic stroke, class 1 indicates hemorrhagic stroke, and class 2 indicates non-stroke. Obviously, TPELGBM with average AUC of 0.9947 performs better than the other three classifiers for stroke diagnosis.
Ablation experiment of TPE optimizer
In order to verify the optimization effect of TPE module on LightGBM classifier, we conducted ablation experiments to compare the performance of LightGBM classifier, TPELGBM classifier and GSLGBM classifier (LGBM with grid search hyperparameter optimization). According to the previous experiments, the ApFu entropy feature of 1-s segments exhibits the best performance. Therefore, only the data of 1-s segments are tested for the TPE optimized LightGBM classifier and GS (grid search) optimized LightGBM classifier. To avoid experiment contingency, we perform five-fold cross-validation when performing TPE hyperparameter optimization and GS hyperparameter optimization. After the hyperparameter optimization, the classification performance of LightGBM is further improved. As shown in Fig. 7, after the pre-training of the TPE module and GS module, the precision, recall, and f1-score of the model are all improved for stroke diagnosis. Clearly, the TPE module significantly outperforms the GS method in enhancing the model’s performance. Specifically, the TPELGBM classifier achieved significant improvements in performance metrics, with accuracy, precision, recall, and F1-score ratings of 0.9689, 0.9676, 0.9669, and 0.9672, respectively. These figures represent enhancements of 0.0259, 0.0505, and 0.0394 over the LightGBM classifier’s respective scores. Furthermore, the TPELGBM classifier’s results surpass those of the GSLGBM classifier by margins of 0.004, 0.0072, and 0.0056 in precision, recall, and F1-score, respectively, underscoring its superior performance in these critical evaluation criteria. Meanwhile, the TPELGBM classifier also brings bright novelty in AUC (The average AUC = 0.9963). Compared to the default settings, the TPE algorithm successfully optimized the hyperparameters of the LightGBM model, as shown in Table 4. In particular, the finely adjusted learning rate has a significant impact on model performance, making the model more cautious in adjusting weights during the training process, thereby helping the model to converge more effectively and reduce the risk of overfitting. The increased number of trees (15000) enhances the model’s learning ability, enabling it to capture complex patterns in the data. These adjustments significantly improved the model’s classification accuracy, demonstrating the effectiveness of hyperparameter optimization in enhancing the performance of LightGBM.
Discussion
The utilization of EEG signals for stroke diagnosis is a burgeoning field that has yet to fully meet the intricate demands and rigorous criteria of clinical practice. To address this issue, we have developed the TPELGBM model, which aims to achieve rapid and accurate diagnosis and classification of stroke based on EEG signals. This model utilizes the approximate entropy and fuzzy entropy of EEG signals as key features, which are extremely sensitive to capturing changes in the complexity of EEG signals, aiding in the differentiation of various types of stroke patients, including ischemic and hemorrhagic strokes, as well as non-stroke individuals. The LGBM model has demonstrated the ability to classify these features efficiently and accurately. Furthermore, by meticulously optimizing the hyperparameters of LGBM using the TPE algorithm, we have further mitigated the risk of overfitting and enhanced the model’s generalization capabilities.
In order to enhance the model’s accuracy in stroke diagnosis and explore the factors affecting diagnostic accuracy, we conducted a series of experiments to identify the optimal model parameters. By adjusting the ratio of the training set to the test set in the dataset division, we further optimized the model performance. As shown in Fig. 4, the model trained with a 7:3 dataset split ratio demonstrated superior generalization capabilities. Furthermore, the data in Table 3 indicates that the ApFu features of 1-second EEG signal segments are significantly better in distinguishing stroke types than those of 3-second segments. This advantage is attributed to the fact that the calculation of FuEn is not restricted by the length of the EEG signal, a characteristic inherited by the ApFu features. Therefore, the model shows better classification performance when dealing with the larger data volume of 1-second segment features. After meticulous parameter adjustment and hyperparameter optimization, the model achieved accuracy, precision, recall, and F1 scores of 0.9689, 0.9676, 0.9669, and 0.9672, respectively. As shown in Table 5, compared to existing methods, our model has significantly improved the accuracy of stroke diagnosis. By assessing the complexity of EEG signals, the proposed ApFu feature can sensitively detect changes in the frequency bands of EEG signals in stroke patients. Furthermore, the TPELGBM model demonstrates efficient discriminative power for these features. These results indicate that the method has great potential for clinical application and may provide more comprehensive medical services to patients.
The application of EEG signals in stroke diagnosis offers advantages such as high accuracy, speed, ease of operation, portability, and low cost, demonstrating significant potential. However, deploying our model in actual clinical scenarios presents challenges. Principally, to affirm the model’s clinical relevance and expedite stroke diagnosis, the EEG data collection process must be swift and precise. We are exploring methods to simplify the EEG data collection process to meet the time-sensitive demands of clinical settings. This includes improving data collection protocols and potentially integrating our method with existing clinical workflows. Furthermore, the pre-hospital stroke diagnosis mandates stringent standards for EEG data acquisition. For this purpose, it is crucial to provide medical staff with extensive training in EEG data collection across diverse scenarios. While our preliminary results are encouraging, our current dataset is limited in scale. To ensure the robustness and generalization of our method across different patient populations, a broader dataset is needed to support its application in clinical environments. Given the current dataset’s lack of breadth, we are actively involved in ongoing efforts to collaborate with other research groups to access more diverse datasets. We are also continuously monitoring the publication of new datasets in our field that may become suitable for validating our method. We are hopeful that these efforts will yield results in the near future. Furthermore, we are contemplating the inclusion of patient age and gender as factors that could influence stroke diagnosis, which will be a focus of future work. We believe that these steps will help bridge the gap between our current research and the potential clinical application of our method. We are committed to overcoming these challenges and advancing our work to make a meaningful contribution to the diagnosis and treatment of stroke.
Conclusion
In this paper, we introduce a novel ApFu-TPELGBM model for classifying stroke patients using an EEG signal. We proposed a ApFu feature fusion method, which can profoundly distinguish the complexity and uncertainty features of EEG signals. In addition, we optimize LightGBM classifier by TPE algorithm to improve the classification performance of the model. Experimental results showed that the ApFu-TPELGBM model could exploit the deep characteristics of EEG signals and perform excellently on the ZJU4H dataset, which achieved accuracy of 0.9689, precision of 0.9676, recall of 0.9669 and f1-score of 0.9672. Our research indicates that the model enables clinicians to determine whether a stroke is present and what type of stroke it is. Meanwhile, the EEG-based ApFu-TPELGBM model meets the requirements of high diagnostic accuracy, fast speed, simple operation, small size, and low cost, which makes it can be used in more scenarios, especially in resource-limited settings such as pre-hospital. Furthermore, this model will change the current stroke treatment procedure and improve the treatment outcomes of thousands of stroke patients.
Data availability
Date used in the present study will be available from the corresponding author upon reasonable request.
References
Syland, M. H. et al. Wake-up stroke and unknown-onset stroke; occurrence and characteristics from the nationwide norwegian stroke register. Eur. Stroke J. 7(2), 143–150 (2022).
Hung, S. H. et al. Pre-stroke physical activity and admission stroke severity: A systematic review. Int. J. Stroke. 16(9), 1009–1018 (2021).
Saini, V., Guada, L. & Yavagal, D. R. Global epidemiology of stroke and access to acute ischemic stroke interventions. Neurology 97(Supplement 2), S6–S16 (2021).
Ntaios, G. Embolic stroke of undetermined source Jacc review topic of the week. J. Am. Coll. Cardiol. 3, 75 (2020).
Iadecola, C., Buckwalter, M. S. & Anrather, J. Immune responses to stroke: mechanisms, modulation, and therapeutic potential. J. Clin. Invest. 130(6), 2777–2788 (2020).
Bersano, A. et al. Heritable and non-heritable uncommon causes of stroke. J. Neurol. 268, 2780–2807 (2021).
Flach, C., Muruet, W., Wolfe, C. D. A., Bhalla, A. & Douiri, A. Risk and secondary prevention of stroke recurrence: A population-base cohort study. Stroke 51 8 (2020).
Rücker, V. et al. Twenty-year time trends in long-term case-fatality and recurrence rates after ischemic stroke stratified by etiology. Stroke 51(9), STROKEAHA120029972 (2020).
Singletary, E. M., Zideman, D. A., Bendall, J. C., Berry, D. C. & Lang, E. 2020 international consensus on first aid science with treatment recommendations. Circulation 142 (2020).
Bentes, C. et al. Quantitative eeg and functional outcome following acute ischemic stroke. Clin. Neurophysiol. 129(8), 1680–1687 (2018).
Phipps, M. S. & Cronin, C. A. Management of acute ischemic stroke. BMJ 368, 6983 (2020).
Mendelson, S. J. & Prabhakaran, S. Diagnosis and management of transient ischemic attack and acute ischemic stroke: A review’’. JAMA J. Am. Med. Assoc. 325(11), 1088 (2021).
Diener, H. C. et al. Review and update of the concept of embolic stroke of undetermined source,‘’ nature reviews. Neurology no. 8, 18 (2022).
Zhao, Y., Zhang, X., Chen, X. & Wei, Y. Neuronal injuries in cerebral infarction and ischemic stroke: From mechanisms to treatment (review). Int. J. Mol. Med. 49(2), 1–14 (2021).
Dong, Y. et al. Chinese stroke association guidelines for clinical management of cerebrovascular disorders: Executive summary and 2019 update of clinical management of spontaneous subarachnoid haemorrhage. Stroke Vasc. Neurol. (2019).
Can, W., Fenglian, L. I., Fengyun, H. U., Xueying, Z. & Wenhui, J. Classification of stroke eeg signals based on feature fusion. Comput. Eng. Appl. (2019).
Shreve, L., Kaur, A., Vo, C., Wu, J. & Cramer, S. C. Electroencephalography measures are useful for identifying large acute ischemic stroke in the emergency department. J. Stroke Cerebrovasc. Dis. 28(8), 2280–2286 (2019).
Vatinno, A. A. et al. The prognostic utility of electroencephalography in stroke recovery: A systematic review and meta-analysis. Neurorehabil. Neural Repair 36(4–5), 255–268 (2022).
Supriya, S., Siuly, S., Wang, H. & Zhang, Y. Epilepsy detection from eeg using complex network techniques: A review. IEEE Rev. Biomed. Eng. (2021).
Mulder, M. J. et al. Time to endovascular treatment and outcome in acute ischemic stroke: Mr clean registry results. Circulation 138(3), 232–240 (2018).
van Meenen, L. C. et al. Detection of large vessel occlusion stroke in the prehospital setting: Electroencephalography as a potential triage instrument. Stroke 52(7), e347–e355 (2021).
Van Meenen, L. C. C. et al. Detection of large vessel occlusion stroke with electroencephalography in the emergency room: first results of the electra-stroke study’’. J. Neurol. 269(4), 2030–2038 (2022).
Hosseini, M. P., Hosseini, A. & Ahi, K. A review on machine learning for Eeg signal processing in bioengineering. IEEE Rev. Biomed. Eng. 14, 204–218 (2021).
Zhang, L. et al. Applications of machine learning methods in drug toxicity prediction. Curr. Top. Med. Chem. 18, 987–997 (2018).
Maffulli, N. et al. Artificial intelligence and machine learning in orthopedic surgery: a systematic review protocol. BioMed Central (2020).
Meng, Q. Lightgbm: A highly efficient gradient boosting decision tree. Neural Inf. Process. Syst. (2017).
Pan, H. et al. The lightgbm-based classification algorithm for chinese characters speech imagery bci system. Cogn. Neurodyn. 1–12 (2022).
Yan, J. et al. Lightgbm: accelerated genomically designed crop breeding through ensemble learning. Genome Biol. 22, 1–24 (2021).
Liang, J. et al. Estimation of stellar atmospheric parameters with light gradient boosting machine algorithm and principal component analysis. Astron. J. 163((4), 153 (2022).
Li, D. & Peng, J. He,Aero-engine exhaust gas temperature predic- Tion based on Lightgbm optimized by improved Bat algorithm. Natl. Libr. Serbia. 25(2), 845–858 (2021).
Lin, L., Zhang, J., Zhang, N., Shi, J. & Chen, C. Optimized light- Gbm power fingerprint identification based on entropy features. Entropy 24(11), 1558 (2022).
Khoshnevis, S. A. & Sankar, R. Applications of higher order statistics in electroencephalography signal processing: A comprehensive survey. IEEE Rev. Biomed. Eng. 13, 169–183 (2020).
Peri, Z. H., Deli, V., Stamenkovi, Z. & Pokrajac, D. D. Advanced signal processing and adaptive learning methods. Comput. Intell. Neurosci. (2019).
Zhu, S. et al. Engineering entropy-driven based multiple signal amplification strategy for visualized assay of Mirna by naked eye. Talanta 235, 122810 (2021).
Kosciessa, J. Q., Kloosterman, N. A. & Garrett, D. D. Standard mul- Tiscale entropy reflects neural dynamics at mismatched Temporal scales: what’s signal irregularity got to do with it? PLoS Comput. Biol. 16(5), e1007885 (2020).
Hou, F., Zhang, L., Qin, B., Gaggioni, G. & Vandewalle, G. Changes in Eeg permutation entropy in the evening and in the transition from wake to sleep. Sleep 44, 6 (2020).
Pang, Q., Liu, X., Sun, B. & Ling, Q. Approximate entropy based fault localization and fault type recognition for non-solidly earthed network. Meas. Sci. Rev. 12(6), 309–313 (2012).
Alcaraz, R. & José, J. R. A review on sample entropy applications for the non-invasive analysis of atrial fibrillation electrocardiograms. Biomed. Signal Process. Control 5(1), 1–14 (2010).
Samantha, S., Pedro, E. & Daniel, A. Fuzzy entropy analysis of the electroencephalogram in patients with Alzheimer’s disease: is the method superior to sample entropy. Entropy 20(1), 21 (2018).
Hong, Y. & Zhang, X. Questionnaire and Lgbm model for assessing health literacy levels of Mongolians in China. BMC Public Health 22(1), 1–11 (2022).
Behera, R. N., Baral, P., Saha, S. & Dash, S. Emotion based classification of human voice using an optimized machine learning approach emotion based classification of human voice using an optimized machine learning approach. Int. J. Control Theory Appl. 10, 18 (2017).
Tong, W. et al. MSE-VGG: A novel deep learning approach based on EEG for rapid ischemic stroke detection. Sensors 24(13), 4234 (2024).
Funding
This study was supported by the Key Research and Development Project of Zhejiang Province (2022C03043, 2023C03080, 2020C03071), the key projects of major health science and technology plan of Zhejiang Province (WKJ-ZJ-2129) and the National Natural Science Foundation of China (61601409).
Author information
Authors and Affiliations
Contributions
W.T., F.C. and J.Z. contributed to the conception of the study; W.T. prepared the experimental task and performed the experiment; W.T., F.C. and W.S. contributed significantly to analysis and manuscript preparation; W.T. and F.C. performed the data analyses and wrote the manuscript; L.Z. and J.W. helped perform the analysis with constructive discussions. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
This research project has been approved by the Ethics Committee of the Fourth Affiliated Hospital of Zhejiang University School of Medicine (IRB approval code: K2021135, Approved date: 2021/12/17). This study was conducted with adherence to the highest ethical standards and in compliance with the guidelines and regulations of Fourth Affiliated Hospital of Zhejiang University School of Medicine. All test results as recorded in the original medical records, including personal data and laboratory documents, will be maintained with utmost confidentiality in accordance with the law.
Informed consent
All subjects have signed informed consent for clinical research in this study.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tong, W., Zhang, J., Chen, F. et al. A novel stroke classification model based on EEG feature fusion. Sci Rep 15, 14287 (2025). https://doi.org/10.1038/s41598-025-92807-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-92807-x