Abstract
Cohesin and condensin, two related protein complexes, play essential roles in ensuring the accurate segregation of the genome into daughter cells during cell division. However, the interaction between cohesin and condensin in embryonic stem cells remains unclear, as does the specific function of the meiosis-specific cohesin complex. Cohesin maintains the cohesion of replicated sister chromatids until their separation at anaphase, whereas condensin facilitates the reorganization of chromosomes into a highly compact structure characteristic of mitosis. First, we found via ChIP-seq analysis that cohesins (SMC3, RAD21, and REC8) and condensin (SMC4) share DNA binding sites in close proximity and directly interact with the insulator protein CTCF. Second, siRNA-regulated SMC3 depletion led to nuclear accumulation of SMC4. Third, embryonic stem (ES) cells uniquely harbor cohesin complexes containing the meiotic kleisin subunit REC8. RAD21 knockdown increased the proportion of SMC3–REC8 complexes. Our findings indicate that cohesin and condensin make important contributions to the functions of the chromosomal organization, and that meiotic cohesin may be specifically required for the mitotic program in ES cells.
Similar content being viewed by others
Introduction
Embryonic stem (ES) cells rely on specific cell cycle machinery to sustain self-renewal and pluripotency1,2. These cells exhibit dynamic chromatin changes that are crucial for gene expression and chromosome condensation during differentiation3,4,5. Unlike differentiated cells, ES cells possess an enhanced self-renewal capacity, elevated expression of proteins related to DNA repair, rapid proliferation with an extended S-phase, and high tolerance to replication and DNA damage stress6,7. However, the mechanisms underlying the maintenance of genome integrity and the response to chromosomal abnormalities and replication stress in ES cells remain largely unexplored.
Cohesin, a ring-shaped protein complex, ensures proper chromosome segregation by holding sister chromatids together until their separation during anaphase8,9. This complex comprises SMC1, SMC3, STAG1, STAG2, and an α-kleisin subunit, which is represented by RAD21 in mitotic cells or REC8 and RAD21L in meiotic cells9,10. In eukaryotes, chromatids progressively separate along their arms during prophase and at the centromeres during anaphase, whereas in others, separation occurs in a single step during anaphase11,12. This process involves the cleavage of the α-kleisin subunit, opening of the cohesin ring, and its dissociation from chromosomes, enabling sister chromatid segregation9,13. This release mechanism supports a model in which cohesin encloses the chromatids within a single ring.
The condensin complex is a highly conserved protein assembly found across eukaryotes, from yeast to humans14,15. This complex comprises five subunits, including two core components, SMC2 and SMC4, which are members of the structural maintenance of chromosomes (SMC) family, a group of chromosomal ATPases16,17,18,19. SMC proteins are essential for higher-order chromosome dynamics, encompassing chromosome condensation, sister chromatid cohesion, and recombinational repair20. The remaining three subunits (CAP-D2, CAP-G, and CAP-H) of the condensin complex do not belong to the SMC family; however, they share structural motifs with proteins involved in cohesin16,21. Functionally, the condensin holo-complex induces positive supercoiling of DNA in an ATP hydrolysis-dependent manner in vitro. This activity is thought to facilitate the initial steps of mitotic chromosome assembly within cells18,21.
Recent studies indicate that the mitotic cohesin, meiotic cohesin, and condensin complexes play pivotal roles in establishing or maintaining topological domains and managing topological changes in chromosomes in ES cells as their binding sites are enriched at domain boundaries22,23,24. It has been proposed that mitotic cohesin, meiotic cohesin and condensin collaborate to facilitate long-range genomic interactions in ES cells25. Evidence supporting this hypothesis includes the extensive co-localization of the mitotic cohesin core factor SMC3 with mitotic or meiotic cohesin subunits (RAD21, REC8, and STAG1/STAG2/STAG3) across mammalian genomes, their shared involvement in mediating long-range interactions, and the role of cohesin in enhancing the chromatin insulation function of the CCCTC-binding factor (CTCF)26,27,28.
These findings imply that the mitotic cohesin, meiotic cohesin, and condensin complexes regulate chromosomal morphogenesis during cellular progression22,25,29. However, the relationship between cohesin and condensin complexes, which share DNA binding sites, remains poorly understood in ES cells.
The present study addresses the relationship between the cohesin and condensin complexes, which share DNA binding sites, and the interaction between the meiotic factor REC8 and the mitotic factor RAD21 in ES cells. First, we discovered that cohesin (SMC3) and condensin (SMC4) share common DNA binding sites with the insulator protein CTCF. Furthermore, the inhibition of cohesin factor (SMC3) expression suppressed the expression of the condensin core factor SMC4, leading to its nuclear accumulation. These findings provide insights into the mechanisms underlying aberrant chromosome compaction and SMC4 accumulation caused by cohesin depletion. Second, we observed that RAD21 depletion in ES cells increased the interaction ratio between the meiotic kleisin subunit REC8 and SMC3. Thus, our findings highlight the essential roles of condensin and meiotic cohesin complexes in mitotic ES cells.
Materials and methods
Cell lines
Mouse ES cells and mouse embryonic fibroblasts (MEFs) were prepared as previously described22. ES cells were seeded on 0.2% gelatin-coated 6-cm plates in Dulbecco’s Modified Eagle’s medium with high glucose (Thermo Fisher, Cat. 10569) supplemented with 10% horse serum (Thermo Fisher, Cat. 16050122), 2 mM L-glutamine (Thermo Fisher, Cat. 25030081), 0.1 mM MEM non-essential amino acids (Thermo Fisher, Cat. 11140050), 10 mM HEPES (Thermo Fisher, Cat. 15630080), 0.1 mM β-mercaptoethanol (Thermo Fisher, Cat. 21985023), 100 U/ml penicillin–streptomycin (Thermo Fisher, Cat. 15140122), and 100 U/mL mouse leukemia inhibitory factor (mLIF) (Merck Millipore, Cat. ESG1107). MEFs were cultured on six-well plates in Dulbecco’s Modified Eagle’s Medium high glucose (Thermo Fisher, Cat. 10569) supplemented with 10% fetal bovine serum (Thermo Fisher, Cat. 16000044), and 100 U/ml penicillin-streptomycin (Thermo Fisher, Cat. 15140122). Cells were cultured at 37 °C in a humidified incubator with 5% CO2.
Antibodies
The following primary antibodies were used in this study: anti-SMC3 (Abcam, Cat. ab9263), anti-REC8 (Abcam, Cat. ab192241), anti-RAD21 (Abcam, Cat. ab154769), anti-STAG1 (Abcam, Cat. 4457), anti-STAG2 (Santa Cruz, Cat. 81852), anti-OCT4 (Santa Cruz, Cat. 5279), anti-SMC4 (Abcam, Cat. 17958), anti-NCAPH2 (Abcam, Cat. ab221722), anti-CTCF (Cell signaling, Cat. #2899), anti-PCNA (Santa Cruz, Cat. 56), anti-GAPDH (Abcam, Cat. ab8245), anti-α-tubulin (Abcam, Cat. ab4074), Rhodamine anti-α-tubulin (Bio-Rad, Cat. 12004165) and anti-Lamin B1 (Santa Cruz, Cat.56144). The following secondary antibodies were used: goat anti-mouse IgG (Jackson IR, Cat. 115-035-003), goat anti-rabbit IgG (Jackson IR, Cat. 111-035-003), and anti-rabbit TRITC (Jackson IR, Cat. 111-025-003).
Immunoblot and Immunoprecipitation (IP)
mES cells were prepared as previously described22,30. For the IP assay, ES cells were cultured at a concentration of 1 × 106 cells/ml per 6 cm culture plate. Cells were washed by phosphate buffered saline (PBS) to remove horse serum and treated 0.25% Trypsin-EDTA for 1 to 2 min at 37 °C in an incubator. Subsequently, the cells were resuspended in fresh medium and centrifuged. Pellets were resuspended in lysis buffer (0.1 mM EDTA, 1% sodium deoxycholate, 0.15% NP-40, 120 mM NaCl, and 25 mM Tris-HCl; pH 7.6) for 10 min on ice. After centrifugation for 5 min; max RPM, Cell lysates were concentrated by the bicinchoninate acid (BCA) assay. Soluble proteins were reacted with primary antibodies (1–1.5 ug) overnight and mixed with 20 ul 50% agarose A/G beads for 3 h at 4 °C. Subsequently, the soluble proteins were centrifuged at 5000 rpm for 3 min and supernatants were removed. The agarose beads were washed three times by cleaning buffer (0.15% NP-40, 1 mM DTT, 100 mM KCl, 2 mM MgCl2, and 10 mM HEPES). After centrifugation, 2x SDS buffer was mixed with protein agarose A/G beads and boiled for 5 min at 95 °C on a heat block. Soluble proteins were analyzed using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Cell cycle synchronization
To analyzing of cohesin and condensin subunits expression pattern, ES cells were synchronized at G1-phase by reacting with 2 µM thymidine (Sigma, Cat. 89270) for 24 h, after which 1st thymidine was washed and added a fresh medium. Subsequently, 2nd thymidine was added in ES cells for 24 h. Cells were washed with 1x PBS and replaced with fresh medium to release cell cycle from the 2nd thymidine block. To acquire synchronization, duplicated cultures were collected after thymidine block release at 2 h intervals.
Immunostaining
ES cells were attached on poly-L-lysine-coated coverslips (Deckglaser, Cat. 0111520) and fixed with 4% paraformaldehyde for 10 min. Cells were treated with triton X-100 (0.1%) for 15 min to membrane permeabilization and washed with PBS-T (0.1% tween-20 in PBS) for three times. Cells were blocked with 2% BSA in PBS-T for 1 h and incubated for 2 h with primary antibody: anti-REC8 (Abcam, Cat. ab192241). Cells were washed with 1x PBS-T, and incubated for 30 min with the fluorescence secondary antibody: anti-TRITC (Jackson lR, Cat. 109-025-003). Images were captured and analyzed using the Eclipse Ti2 inverted microscope system (Nikon).
RNA interference
A commercially available predesigned siRNA specific to the target genes was used to knock-down endogenous expression in ES cells. The siRNA pool included the following sequences: #1 siSmc3: 5’-GAGGUUGGCUCAAGCUACA(dTdT)−3’, #1 siSmc3: 5’-CAGAUGAAGUCAGCACGAA(dTdT)−3’, #3 siSmc3: 5’- AGAGAAGUAGCUGGUACUA (dTdT)−3’, siRad21: 5’-GAGCUAGUGAUAACUCACU(dTdT)−3’, and siRec8: 5’-GAGAUCAGUCGAGGAGACU(dTdT)−3’. The siRNAs were transfected with RNAiMAX Lipofectamine (Thermo, Cat. 13778150) according to the manufacturer’s protocol. Transcription was suppressed by siRNAs, and the cells were incubated in serum free medium for 48 h.
RNA-Seq library preparation and sequencing
Total RNA was isolated using a RNeasy Mini Kit (Qiagen, 74004) according to the manufacturer’s protocol. RNA concentration was analyzed using a Nanodrop 2000 spectrophotometer (Thermo), and RNA quality was measured with an Agilent Bioanalyzer 2000 (Agilent Technologies). For each RNA sample, library construction was performed using the SENSE mRNA-Seq Library Prep Kit (Lexogen) according to the manufacturer’s instructions. Briefly, 2 mg of total RNA was prepared from each sample and incubated with magnetic beads coated with oligo dT, after which all RNAs (except for mRNAs) were removed with a washing solution provided in the kit. Library was produced by the random hybridization of heterodimers (starter/stopper) to the poly(A) RNA bound to the magnetic beads. These heterodimers contained Illumina-compatible linker sequences. Reverse transcription and ligation reaction was performed to extend the starter to the next hybridized heterodimer. High-throughput sequencing was performed as paired-end 100 sequencing using a HiSeq 2000 instrument (Illumina, USA).
Data analysis
The raw sequencing data were quality checked using FastQC. Adapter and low-quality reads (< Q20) were removed using the FASTX Trimmer and BBMap tools. The trimmed reads were then mapped to the reference genome using TopHat. Gene expression levels were estimated as FPKM (fragments per kb of transcript per million reads) values using Cufflinks. The FPKM values were normalized based on the quantile normalization method using edgeR in R. Data mining and graphic visualization were performed using the ExDEGA software (E-biogen).
Accession numbers
The RNA sequencing datasets generated and/or analyzed during the current study are available in the BioProject repository (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA338725), (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1055310), accession number: PRJNA338725, PRJNA1055310.
Statistical analysis
All data for this study were analyzed by the GraphPad Prism (version 9.0). Quantification of all western blot data were analyzed by the Image Lab 6.0 software (Bio-Rad). Statistically significant differences (ns; no significant, *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001) between several groups were determined via paired two-tailed t-test. For all figures, three biological replicates were performed. P-values (paired two-tailed t-test) were evaluated using GraphPad Prism 9.0 software. For Fig. 4F, the level of intensities was quantified by NIS advanced software (Nikon). The error bars represent the mean ± SD values of three independent experiments (at least 50 nuclei per condition were counted). P-values (paired two-tailed t-test) were evaluated using the GraphPad Prism 9.0 software.
Results
Knockdown of mitotic and meiotic cohesin components changes SMC4 expression levels
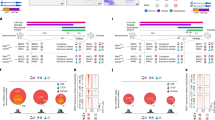
ES cells possess distinct characteristics, including pluripotency and unlimited self-renewal, and are characterized by unique gene expression patterns associated with genomic integrity, chromatin modifications and structures. Therefore, we first analyzed the protein expression patterns of the cohesin and condensin components in ES cells and mouse embryonic fibroblasts (MEFs) using immunoblotting (Fig. 1a and b). Mitotic cohesin components, including SMC3, RAD21, STAG1, STAG2, and the condensin core factor SMC4, were expressed in both ES cells and MEFs (Fig. 1b). Notably, the meiotic cohesin subunit REC8 were highly expressed in ES cells but were absent in MEFs. In addition, RNA sequencing analysis revealed that the transcription levels of mitotic and meiotic cohesin and condensin components were consistent with the protein expression levels observed through immunoblotting (Fig. 1c-e). These findings suggested that cohesin complexes containing meiotic REC8 may perform specific functions within the mitotic program of ES cells.
Expression of cohesin and condensin components in embryonic stem (ES) cells. (A) Cohesin and condensin complex structure. The cohesin complex comprises long coiled-coil structural maintenance of chromosomes (SMC) subunits, specifically SMC3 and SMC1α in mitotic cohesins, and SMC3 and SMC1β in meiotic cohesins8. These SMC subunits, together with α-kleisin subunits—RAD21 for mitosis and REC8 for meiosis—assemble into a characteristic V-shaped structure. The condensin complex is classified into two subtypes, Condensin I and Condensin II. Condensin I consists of subunits SMC2, SMC4, CAPH, CAPD2, and CAPG, whereas Condensin II is composed of subunits SMC2, SMC4, CAPH2, CAPD3, and CAPG216. (B) Expression analysis of cohesin and condensin components. Soluble proteins were extracted from asynchronous ES cells and mouse embryonic fibroblasts (MEFs). α-tubulin and OCT4 were used as a house keeping gene and stemness marker, respectively. (C) Comparison of expression levels of cohesin, condensin, and stemness-related genes between ES cells and MEFs, as assessed using RNA-sequencing. (D) Comparison of gene expression of cohesin and condensin. Expression levels from immunoblotting and RNA sequencing were analyzed to evaluate cohesin and condensin related gene expression. The expression levels measured using RNA-sequencing are represented as average values of two biological replicates. (E) Raw value of RNA-sequencing data. The error bar denotes the ± SD value from the biological replicates. (F) Comparison of cohesin components knockdown efficiency by siRNA pool against SMC3, RAD21, and REC8. (The original western blot image of Oct4 was cropped. Therefore, we have provided the full, uncropped image from the repeated experiments in the supplementary data). (G) Comparison of expression levels of cohesin, condensin, stemness, and proliferation-related genes between cells treated with siCtrl and cohesin component knockdown siRNA, assessed using RNA-sequencing data.
To investigate the specific roles of mitotic and meiotic cohesin components in ES cells, we knocked down the mitotic cohesin components SMC3 and RAD21 and the meiotic cohesin component REC8 by treating ES cells with pooled siRNAs targeting SMC3, RAD21, and REC8 (siSmc3, siRad21, and siRec8, respectively) in ES cells (Fig. 1f). Interestingly, transcript analysis by RNA-sequencing showed that knockdown of mitotic and meiotic cohesin factors suppressed the transcription level of the condensin core component SMC4, but did not affect the expression levels of other cohesin components (Fig. 1g). Furthermore, in ES cells, mitotic (RAD21) and meiotic (REC8) cohesin factors are commonly involved in regulating apoptotic processes, cell death, DNA repair, translation, and cell differentiation (Fig. S1).
The siRNAs effectively reduce the expression of SMC3, RAD21, and REC8 to similar levels. However, cells treated with siSmc3 exhibited a significantly suppressed level of SMC4 expression, which was reduced by more than 45%. Additionally, the inhibition of mitotic and meiotic cohesin component expression resulted in a more than 20% reduction in the transcription levels of NANOG and OCT4, a stemness-associated gene in ES cells (Fig. 1g). Kim and colleagues reported that significantly lower levels of SMC3, RAD21, and REC8 induced stem cell differentiation, compared to normal-state ES cells22. Taken together, our findings indicate that mitotic/meiotic cohesin components are required for maintaining the pluripotency of ES cells and condensin functions.
DNA-binding sites of cohesin and condensin almost overlap with those of CTCF
A previous study found that the DNA binding sites of cohesin and condensin complexes almost completely overlap22,25. To further elucidate the relationship between cohesin and condensin, we analyzed the binding sites of mitotic and meiotic cohesin, condensin, and CCCTC-binding factor (CTCF) using previously published ChIP-seq datasets. ChIP-seq binding profiles revealed that mitotic cohesin components (SMC3, RAD21, and SMC1), the meiotic cohesin component (REC8), the condensin core factor (SMC4), and CTCF all bind to similar genomic locations (Fig. 2a). These findings suggest that, in addition to cohesin, condensin localizes to CTCF binding sites and play critical roles in maintaining the topological structure of chromatin and inducing DNA looping. Furthermore, both cohesin and condensin share binding sites with OCT4 and NANOG (Fig. 2a). These findings suggest that stem cell differentiation may be regulated by cohesin or condensin, supporting a previous report by Choi et al., which demonstrated that the depletion of SMC3, RAD21, or REC8 induces stem cell differentiation22. We confirmed the abundant expression of mitotic/meiotic cohesin and condensin components in ES cells using immunoblotting and RNA sequencing (Fig. 1b). We further characterized the expression patterns of cohesin and condensin factors in both asynchronous ES cells and those synchronized at the G1/S phase checkpoint using a double-thymidine block, after which the cells were released from the block and allowed to progress through the cell cycle (Fig. 2b and S2). The protein expression levels of SMC3, RAD21, REC8, and SMC4 were consistently maintained during the cell cycle (Fig. 2c, left). Next, co-immunoprecipitation (co-IP) assays were conducted to determine whether the mitotic/meiotic cohesin and condensin components were linked to CTCF, which creates boundaries between topologically associating domains in chromosomes and facilitates interactions between transcription regulatory sequences. Using antibodies against SMC3, RAD21, REC8, and SMC4, we analyzed whether cohesin and condensin components interacted with DNA looping sites. It was found that SMC3, RAD21, REC8, and SMC4 strongly interacted with CTCF independent of the cell cycle in ES cells (Fig. 2c, right). These results imply that the meiotic factor REC8 and the condensin core factor SMC4 may be essential for chromosome organization in ES cells.
Whole genome occupancy of cohesin and condensin. (A) ChIP-Seq binding profiles for cohesin components (SMC3, SMC1, RAD21, and REC8), condensin component (SMC4), CCCTC-binding factor (CTCF), and stemness factors (OCT4 and NANOG) at the Chr.17 39,401,345–39,505,768 loci, Chr.6 4,412,260-4,523,412, and Chr.12 90,010,212 − 90,148,525 in ES cells. ChIP-Seq data are reported as reads per million with the y-axis floor set to 0.2 reads per million. Previously published ChIP-seq datasets from ES cells were reanalyzed in the encyclopedia of DNA elements25,29. Scale bars are depicted above the profiles. (B) Scheme of the cell cycle progression. ES cells were synchronized by double-thymidine block (DTB) and released for cell cycle progression. (C) Immunoprecipitation (IP) analysis in ES cells. After a DTB, cells were released for cell cycle progression as shown in (B). Cohesin and condensin components were pulled down the using anti-CTCF antibody from whole-cell lysates in ES cells.
Protein expression and DNA loading of SMC4 after SMC3 knockdown
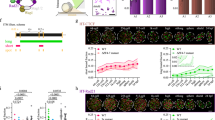
We inhibited the expression of cohesin components using siRNA and demonstrated through RNA sequencing, that this suppression affected SMC4 expression. Notably, SMC3 depletion resulted in the most significant reduction in SMC4 levels (Fig. 1g). Based on these findings, we further investigated the relationship between cohesin SMC3 and condensin SMC4 by analyzing the expression patterns and interactions of SMC3 and SMC4 in the cytosol and nucleus during SMC3 depletion. To optimize SMC3 knockdown efficiency using siRNA, we designed three siRNAs targeting different exons of SMC3. The siRNAs were administered to cells at concentrations of 50, 100, and 150 µM for 48 h, and their gene-silencing efficiencies were subsequently analyzed (Fig. 3a). siSmc3 #1, targeting exon 3, exhibited the highest efficiency, achieving suppression rates of 64.1% at 50 µM, 79.0% at 100 µM, and 68.6% at 150 µM, with 100 µM yielding the most effective transcriptional suppression of SMC3 (Fig. 3b). Therefore, we used siSmc3 #1 at a concentration of 100 µM for all subsequent experiments for SMC3 knockdown.
SMC4 expression changes in the cytosol and nucleus after SMC3 knockdown. (A) SMC3 protein expression in ES cells was suppressed by transfection with an siRNA pool against SMC3 (siSmc3). Cell transfected with 100 nM non-targeting RNA (siCtrl) were included. (B) The level of SMC3 relative to α-tubulin was normalize to the siRNA-transfected cell. (C) The cell-cycle profiles of ES cells treated with siSmc3 as characterized using FACS analysis. (D) Quantitative analysis of (C) in response to SMC3 knockdown. The percentages of siCtrl-transfected and SMC3-deficient cells in G1, S, and G2-phase were quantified using FlowJo software. Three independent experiments were performed. Error bars denote the mean ± SD (n = 3). (E) IP analysis for physical interaction between CTCF, cohesin, and condensin components. (F) Expression analysis of cohesin and condensin components in the presence or absence of SMC3. α-tubulin and Lamin B were used as cytoplasmic and nuclear protein loading markers, respectively. (G) Quantification of SMC3, SMC4, and RAD21 levels in cytoplasm and nucleus; Cy, cytosol; Nu, nucleus. The error bars are the mean ± SD from the biological replicates. (H) Analysis of SMC4 expression pattern during cell cycle progression. After a DTB, cells were released for cell cycle progression as shown in (Fig. 2B). α-tubulin and Lamin B were used as cytosol and nucleus markers, respectively. (I) Quantification of SMC4 expression levels of (H). The error bars denote the mean ± SD from three independent biological replicates.
We analyzed the cell cycle alteration induced by SMC3 knockdown. Compared with the normal cell state, SMC3 knockdown increased the proportion of cells in the G2/M phase by 37.3%, indicating that SMC3 depletion leads to G2/M arrest (Fig. 3c and d). Furthermore, we examined the protein interaction levels of CTCF, the cohesin component RAD21, and the key condensin factor SMC4. Consistent with RNA sequencing results, SMC3 depletion reduced the expression of SMC4 (Fig. 3e). Additionally, compared to treatment with siCtrl (Fig. 3e). SMC3 knockdown reduces CTCF binding efficiency to SMC4 by 50.74% and to RAD21 by 44.79% (Fig. 3S) However, we did not find direct evidence that SMC3 regulates SMC4; moreover, the G2/M-phase arrest caused by SMC3 depletion does not sufficiently explain the observed reduction in SMC4 expression. We analyzed SMC4 protein levels by separating the cytosol and nucleus of ES cells. Interestingly, under SMC3 knockdown, SMC4 expression was increased in the nucleus, RAD21 protein levels were unaffected (Fig. 3f and g). In contrast, SMC4 expression in the cytosol decreased (Fig. 3f and g). To further understand the differences in SMC4 protein expression between the cytosol and nucleus, we analyzed the expression pattern of SMC3 and SMC4 throughout the cell cycle using a double thymidine block (DTB). Cohesin is expressed from interphase and plays a crucial role in cohesion establishment and maintenance after the G1 phase. Following the G2 phase, it undergoes separation. The condensin complex is generally localized in both the cytoplasm and nucleus during interphase. From the S to G2 phase, it appears as nuclear speckles. After the G2 phase, condensin progressively accumulates in the nucleus, and following nuclear envelope breakdown (NEB), it becomes predominantly localized in the nucleus. This suggests that condensin plays a critical role in regulating chromosome condensation and segregation after the G2 phase.
In the cytosol, SMC4 expression continuously decreased for 10 h following DTB release, with a more rapid decline observed with siSmc3 treatment than with siCtrl treatment (Fig. 3h and i). Conversely, in the nucleus, SMC4 expression steadily increased from 0 h post-DTB release. Notably, under siSmc3, SMC4 levels increased sharply, reaching peak protein expression by 6 h post DTB-release (Fig. 3h and i). In a previous study, Kim and colleagues demonstrated that cohesin and condensin can share DNA binding sites and act as regulators in stabilizing and maintaining chromosome structure22. Thus, our findings suggest that cohesin and condensin play essential roles in chromosomal organization throughout the cell cycle in ES cells.
Expression of meiotic cohesin component REC8, and effect of RAD21 knockdown on REC8
We confirmed that the meiotic cohesin component REC8 is abundantly expressed in ES cells (Fig. 1b). Based on this finding, we hypothesized that in addition to the mitotic cohesin complex comprising of RAD21, a meiotic cohesin complex involving REC8 may also exist (Fig. 4a). To test this hypothesis, we analyzed the protein interactions of the cohesin core factor SMC3 with RAD21 and REC8. As expected, SMC3 was found to interact strongly with both RAD21 and REC8 in ES cells, and exhibited a strong interaction exclusively with REC8 in mouse testis (Fig. 4b). However, SMC3 did not interact with REC8 in MEFs. To gain further insight into the composition of the cohesin complex, we analyzed the interaction between REC8 and RAD21 using immunoprecipitation (IP) (Fig. 4c). The results did not reveal any interaction between RAD21 and REC8 in ES cells (Fig. 4c). These findings demonstrate that distinct cohesin complexes compodxsed of either RAD21 or REC8 exist independently in ES cells.
Function of meiotic cohesin consist of REC8 in ES cells (A) Mitotic cohesin and meiotic cohesin complexes. (B) IP analysis in ES cells, MEF, and mouse testis (mTestis) of C57BL/6J mice. Cohesin components were pulled down the using anti-cohesin (SMC3, RAD21, and REC8) antibodies from whole-cell lysates and tissues. (C) IP analysis in ES cells. Cohesin components were pulled down the using anti-cohesin (RAD21 and REC8) antibodies from whole-cell lysates. (D) IP analysis with cohesin components in cells treated with siCtrl and siRad21. PCNA was used as a house keeping gene. (E) Quantification of SMC3 and REC8 expression levels. The error bars are the mean ± SD from the biological replicates. (F) Immunofluorescence analysis in ES cells. Nuclei were stained with anti-REC8 antibody under siCtrl and knockdown condition using siRNA pool against RAD21 and REC8. Scale bar = 2.5 μm (G) Quantitative analysis of REC8 intensity in response to RAD21 or REC8 knockdown. Intensity levels in each condition were quantified. The scatter plots illustrate intensity of REC8 per individual cell (mean ± SD), and a minimum of 50 nuclei were counted for each experiment. Intensity level per nucleus was analyzed using NIS advanced software (Nikon). (H) Protein expression analysis of RAD21 and REC8 in the presence or absence of RAD21. α-tubulin and PCNA were used as cytoplasmic and nuclear protein loading markers, respectively. (I) Quantification of RAD21 and REC8 protein levels in WCL, cytoplasm, and nucleus; WCL, whole cell lysate. The error bars are the mean ± SD from the biological replicates. (J) (i) Distinct localization of mitotic (RAD21) and meiotic (REC8) cohesins at DNA looping sites in ES cells. (ii) Regulation of chromosome structure by cohesin and condensin. In the absence of SMC3, condensin is accumulated in nucleus.
To better understand the relationship between REC8 and RAD21, we analyzed the interaction between SMC3 and REC8, as well as their protein expression levels, following RAD21 depletion (Fig. 4d). Interestingly, RAD21 depletion resulted in a 1.67-fold increase in REC8 expression, while the expression of SMC3 remained unaffected (Fig. 4e). Additionally, the interaction between SMC3 and REC8 increased by 1.92-fold compared with treatment with siCtrl (Fig. 4e). Immunostaining was performed to validate the results of the IP analysis. Consistent with the IP results, RAD21 depletion led to a 1.94-fold increase in nuclear REC8 levels (Fig. 4f and g). Furthermore, fractionation of cytosolic and nuclear proteins from ES cells revealed changes in REC8 levels in both compartments (Fig. 4h). Upon RAD21 knockdown, REC8 levels in the nucleus and cytosol increased by 4.79-fold and 1.50-fold, respectively (Fig. 4i).
Discussion
In this study, we elucidated the interplay between cohesin and condensin complexes in ES cells, focusing on their shared DNA-binding sites and interactions with the insulator protein CTCF. In addition, we found that REC8 was specifically expressed in ES cells and formed a cohesin complex with SMC3, which was distinct from the RAD21-containing cohesin complex (Fig. 4j (i)).
In the cell cycle, cohesins bound to interphase chromatin are released during the prophase, coinciding with their association with condensin complexes31,32. They also play a critical role in chromosome compaction, as they mediate the alignment of sister chromatids prior to condensin action. Condensins, which are typically active during the mitotic phase, contribute to chromosome assembly in early prophase and facilitate chromosome compaction in preparation for mitotic segregation33,34. Despite their crucial roles, the mechanisms underlying the timely compaction of the entire genome by condensins remain poorly understood. Our findings revealed that cohesin (SMC3, RAD21, and REC8) and condensin (SMC4) complexes share binding sites enriched in CTCF-associated regions, supporting the results of previous studies indicating their roles in chromatin insulation and maintenance of topological domains23,25. Notably, SMC3 knockdown resulted in a significant nuclear accumulation of SMC4, with the formation of punctate structures observed on compacted chromosomes. This suggests that cohesin depletion facilitates the redistribution of condensin to cohesin-binding sites on the genome (Fig. 4j (ii)). These findings support previous work demonstrating the ability of condensin to bind cohesin-deficient chromatin regions and reorganize chromosome topology22,29. Thus, our findings prove that cohesin depletion provides condensin (SMC4) with an opportunity to occupy cohesin binding sites prior to the mitotic phase, offering an additional clue as to why SMC4 remains persistently accumulated in the nucleus.
Furthermore, we observed that REC8, traditionally considered a meiotic cohesin component, was specifically expressed in mitotic ES cells. IP analysis indicated the presence of cohesin complexes comprising SMC3–RAD21 and SMC3–REC8. Considering the essential role of RAD21- and REC8-containing cohesin in ES cells, we performed RNA-seq to identify the genes commonly regulated by RAD21 and REC8. Under RAD21 and REC8 knockdown conditions, 1,118 and 462 genes were commonly upregulated and downregulated, respectively (Fig. S1). The upregulated genes were associated with apoptotic processes, cell death, and inflammatory responses, whereas the top downregulated genes were enriched in translation, DNA repair, chromosome organization, and cell differentiation. These findings suggest that REC8, along with RAD21, play a critical role in the maintenance of chromosomal structure, chromatid cohesion, and cohesin-mediated DNA repair in ES cells. RAD21 knockdown increased the proportion of SMC3–REC8 complexes, underscoring the adaptability of cohesin composition in response to mitotic stress. This finding is consistent with recent studies demonstrating the flexibility of cohesin complexes in mediating chromosomal interactions in stem cells24,27.
The dual roles of mitotic and meiotic cohesin in pluripotency and chromosomal maintenance were further evidenced by the suppression of the stemness-associated genes, NANOG and OCT4 upon cohesin depletion. Young and colleagues reported that the genome-wide occupancy of two cohesin core complex proteins, SMC1a and SMC3, whose knockdown resulted in the loss of OCT4 via ChIP-sequencing29. As expected, cohesin occupies sites bound by CTCF but also occupies the enhancer and core promoter sites bound by cohesin factors in many active promoters of ES cells. This aligns with studies reporting that cohesin and condensin regulate transcriptional programs that are critical for maintaining stem cell identity22,29. Our findings suggest that in addition to mitotic cohesin, meiotic cohesin may also play a key role in regulating transcriptional programs critical for maintaining stem cell identity.
Despite these findings, several questions unanswered. First, the precise molecular mechanisms underlying the accumulation of SMC4 upon SMC3 depletion warrant further investigation.
Second, RNA-seq analysis revealed that REC8 may play a role similar to that of RAD21 in ES cells, as evidenced by the increased REC8–SMC3 binding ratio under RAD21 knockdown conditions (Fig. 4d; Fig. S1). However, it remains unclear whether this observation results from the release of RAD21–SMC3 cohesin complexes, leading to a relatively higher proportion of REC8–SMC3 complexes, or from REC8 binding to cohesin sites exposed under RAD21 depletion. Kim and colleagues showed that RAD21 and REC8 localize to distinct sites along the chromosomal axis and adjacent regions, with minimal overlap during WAPL depletion22. These findings suggest that RAD21 and REC8 may perform coordinated functions.
We observed a strong REC8 signal in certain cells in the absence of RAD21. ChIP-seq analysis revealed that RAD21 and REC8 occupy similar DNA binding regions, suggesting that the absence of RAD21 may provide an opportunity for REC8-containing cohesin complexes to bind22,25,29. Although the underlying mechanism remains unclear, this phenomenon may be influenced by uneven siRNA efficiency. In cells where RAD21 is strongly depleted, enhanced REC8 binding efficiency could lead to a stronger REC8 signal. However, analyzing REC8 signal intensity, protein expression levels, and transcriptional changes in RAD21 knockout ES cells using the CRISPR-Cas9 system could serve as a fundamental approach to elucidate the relationship between RAD21 and REC8.
Based on these results, we aim to further investigate the relationship between RAD21 and REC8 in chromosome topology and transcriptional regulation in ES cells.
These studies provide a novel research direction for investigating meiotic cohesin factors in mitotic ES cells and offer new insights into disease modeling and potential therapeutic approaches for cohesinopathies, such as Cornelia de Lange syndrome35,36.
Conclusions
This study provides novel insights into the dual roles of cohesin and condensin complexes in ES cells, emphasizing the significance of meiotic cohesin (REC8) in mitotic contexts. The adaptability of cohesin composition in response to stress and its role in transcriptional regulation highlight its importance in stem cell maintenance. Future investigations into the functional interplay between RAD21 and REC8 will enhance our understanding of chromosome topology and cohesinopathies, offering new therapeutic avenues for cohesin and condensin related-diseases.
Data availability
The data presented in this study are available on request from the corresponding author. The RNA sequencing datasets generated and/or analyzed during the current study are available in the BioProject repository (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA338725), (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1055310), accession number: PRJNA338725, PRJNA1055310.
References
Qi-Long, Y. et al. The ground state of embryonic stem cell self-renewal. Nature 453, 519–523 (2008).
Liu, L., Michowski, W., Kolodziejczyk, A. & Sicinski, P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell. Biol. 21, 1060–1067 (2019).
Barrington, C., Pezic, D. & Hadjur, S. Chromosome structure dynamics during the cell cycle: a structure to fit every phase. EMBO J. 36, 2661–2663 (2017).
Bell, J. C. & Straight, A. F. Condensin chromosome condensation. Nat. Cell. Biol. 17, 964–965 (2015).
Caemer, T. & Cremer, C. Chromosome territories, nuclear architecture and gene regulation in mammalian cells. Nat. Rev. Gene. 2, 292–301 (2001).
Neganova, I. & Lako, M. G1 to S phase cell cycle transition in somatic and embryonic stem cells. J. Anat. 213, 30–44 (2008).
Choi, E. H., Yoon, S. & Kim, K. P. Combined ectopic expression of homologous recombination factors promotes embryonic stem cell differentiation. Mol. Ther. 26, 1154–1165 (2018).
Nishiyama, T. Cohesion and cohesin-dependent chromatin organization. Curr. Opin. Cell. Biol. 58, 8–14 (2018).
Peters, J. M., Tedeschi, A. & Schmitz, J. The cohesin complex and its roles in chromosome biology. Genes Dev. 22, 3089–3114 (2008).
Klein, F. et al. Kim. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98, 91–103 (1999).
Uhlmann, F. Chromosome cohesion and separation: from men and molecules. Curr. Biol. 13, 104–114 (2003).
Sumara, I. et al. The dissociation of cohesin from chromosomes in prophase is regulated by polo-like kinase. Mol. Cell. 9, 515–525 (2002).
Rosen, L. E. et al. Morgan D. O. Cohesin clevage by separase is enhanced by a substrate motif distinct from the cleavage site. Nat. Comm. 15 https://doi.org/10.1038/s41467-019-13209-y (2019).
Katis, V., Galova, L., Rabitsch, M., Gregan, K. P., Nasmyth, K. & J, and Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a Kinetochore-Associated protein related to MEI-S332. Curr. Biol. 14, 560–572 (2004).
Kakui, Y. et al. Fission yeast condensin contributes to interphase chromatin organization and prevents transcription-coupled DNA damage. Genome Biol. 21 https://doi.org/10.1186/s13059-020-02183-0 (2020).
Freeman, L., Aragon-Alcide, L., Strunnikov, A. & & The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J. Cell. Biol. 149, 811–824 (2000).
Kim, E., Kerssemaker, J., Shaltiel, I. A., Haering, C. H. & Dekker, C. DNA-loop extruding condensin complexes can traverse one another. Nature 579, 438–442 (2020).
Kong, M. et al. Human condensin I and II drive extensive ATP-dependent compaction of nucleosome-bound DNA. Mol. Cell. 79, 99–114 (2020).
Wilhelm, L. et al. eLife 7, (2015). https://doi.org/10.7554/eLife.06659
Eeftens, J. M. et al. Condensin Smc2-Smc4 dimers are flexible and dynamic. Cell. Rep. 14, 1813–1818 (2016).
Cuylen, S., Metz, J. & Haering, C. H. Condensin structures chromosomal DNA through topological links. Nat. Struc Mol. Biol. 18, 894–901 (2011).
Choi, E. H. et al. Meiosis-specific cohesin complexes display essential and distinct roles in mitotic embryonic stem cell chromosomes. Genome Biol. 23 https://doi.org/10.1186/s13059-022-02632-y (2022).
Lazar-Stefanita, L. et al. Cohesins and condensins orchestrate the 4D dynamics of yeast chromosomes during the cell cycle. EMBO J. 36, 684–697 (2017).
Abramo, K. et al. A chromosome folding intermediate at the condensin-to-cohesin transition during telophase. Nat. Cell. Biol. 21, 1393–1402 (2019).
Dowen, J. M. et al. Multiple structural maintenance of chromosome complexes at transcriptional regulatory elements. Stem Cell. Rep. 1, 371–378 (2013).
Wendt, K. S. et al. Cohesin mediates transcriptionsal insulation by CCCTC-binding factor. Nature 451, 796–801 (2008).
Davidson, I. F. et al. CTCF is a DNA-tension-dependent barrier to cohesin-mediated loop extrusion. Nautre 616, 822–827 (2023).
Guo, Y., Monahan, K., Wu, H. & Wu, O. CTCF/cohesin-mediated DNA looping is required for Protocadherin a promoter choice. Proc. Nat. Aca Sci. 109, 21081–21086 (2012).
Kagey, M. H. et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 (2010).
Park, S. J., Park, S. J., Kwon, Y. W. & Choi, E. H. Synergistic combination of RAD51-SCR7 improves CRISPR-Cas9 genome editing efficiency by preventing R-loop accumulation. Mol. Ther. Nuc Aci. 35 https://doi.org/10.1016/j.omtn.2024.102274 (2024).
Kim, B. Human cohesin extrudes interphase DNA to make loops. Nat. Rev. Mol. Cell. Biol. 21 https://doi.org/10.1038/s41580-019-0198-z (2020).
Koji, N., Yoh, A. & Mitsuhiro, Y. Separase-mediated cleavage of cohesin at interphase is required for DNA repair. Nature 430, 1044–1048 (2004).
Hudson, D. F., Marshall, K. M. & Earnshaw, W. C. Condensin: Architect of mitotic chromosomes. Chromosome Res. 17, 131–144 (2009).
Thadani, R., Uhlmann, F. & Heeger, S. Condensin, chromatin crossbarring and chromosome condensation. Curr. Biol. 22, 1012–1021 (2012).
Liu, J. & Krantz, I. D. Cornelia de Lange syndrome, cohesin, and beyond. Clin. Genet. 76, 303–314 (2009).
Nolen, L. D., Boyle, S., Ansari, M., Pritchard, E. & Bickmore, W. A. Regional chromatin decompaction in Cornelia de Lange syndrome associated with NIPBL disruption can be uncoupled from cohesin and CTCF. Hum. Mol. Genet. 22, 4180–4193 (2013).
Acknowledgements
This work was supported by grants from the National Research Foundation of Korea (NRF) funded by the Korea government (MSIT) (RS-2023-00247618; RS-2023-00208191) and the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2022M3A9J4079468).
Author information
Authors and Affiliations
Contributions
E.H.C designed and structured the experiments. E.H.C. performed the experiments. E.H.C. and K. P. K analyzed data and wrote the manuscript. All authors had given final approval of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Choi, EH., Kim, K.P. Cohesin and condensin regulate chromosome topology and play an essential role in maintaining pluripotency in embryonic stem cells. Sci Rep 15, 9918 (2025). https://doi.org/10.1038/s41598-025-94533-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-94533-w